Abstract
Crohn’s disease and ulcerative colitis are inflammatory disorders of the gastrointestinal tract with a substantial heritable component. The IBD5 region on chromosome 5q31 is one of only two loci widely confirmed to be associated with Crohn’s disease in multiple independent cohorts. Although many populations have demonstrated association with IBD5, there remains uncertainty as to the causal variant within the region. A recent report identified polymorphisms in SLC22A4 (OCTN1) and SLC22A5 (OCTN2) as being responsible for the IBD5 association, however, these findings have not been replicated. This review discusses the data evaluating the IBD5 locus and the OCTN genes and their relationship to inflammatory bowel disease. Several other genes, including IRF1 and P4HA2 may be equally as likely to contain the IBD5 causal variant as the OCTN genes.
Keywords: IBD5, Inflammatory bowel disease
INTRODUCTION
The pathogenesis of inflammatory bowel disease (IBD) is believed to involve exposure to an as yet unidentified environmental trigger in a genetically susceptible individual[1]. The importance of genetic factors in the pathophysiology of IBD has been repeatedly established by the substantially increased relative risk to siblings of affected individuals and higher monozygotic versus dizygotic twin concordance rates[2-6]. While the heritability of Crohn’s disease (CD) seems higher than that for ulcerative colitis (UC), the available epidemiological and genetic data suggest that there are common susceptibility factors for both diseases and it is likely that gene-gene and gene-environmental interactions determine the specific phenotype[6].
Significant progress has been made in identifying genetic factors responsible for IBD with numerous replicated susceptibility loci identified. However, among the confirmed susceptibility loci identified through linkage for IBD (IBD1-IBD6)[7-12] only the susceptibility gene within the IBD1 locus has been identified[13,14]. The caspase recruitment domain-containing protein 15 (CARD15) gene was described simultaneously by a positional and candidate gene approach and data from numerous populations have confirmed an association between three variants within the CARD15 (NOD2) gene and CD[13-15]. Sub-phenotypic analysis shows that mutations in the CARD15 (NOD2) gene are particularly associated with the development of small bowel CD[15]. Despite widespread replication of the CARD15 (NOD2) findings, there is still important ethnic and geographic heterogeneity. For example, despite general genetic similarity between Ashkenazi Jews (AJ) and Non-Jewish (NJ) Caucasians, the allele frequencies and odds ratios of the CARD15 (NOD2) variants for AJ are slightly different when compared to those found in NJ populations and in Asian individuals affected by CD, CARD15 (NOD2) variants are extremely rare and not associated with CD[15-18].
IDENTIFICATION OF THE IBD5 LOCUS
The IBD5 locus on chromosome 5q31 is the only other locus clearly demonstrated to confer increased risk for CD through association. Following an initial finding of significant linkage in a Canadian population[12], subsequent studies identified a 250 kb risk haplotype within the IBD5 locus that is significantly associated with CD[19]. As part of this effort, the region was sequenced in eight individuals and was intensively evaluated with dense genotyping in more than 250 trios which led to the discovery of long segments of complete linkage disequilibrium (LD), described as “haplotype blocks”, which were separated by apparent “hotspots” of recombination[20]. This original study described 11 haplotype tagging (single nucleotide polymorphisms) SNPs in this region which were equally associated with CD. The 250 kb region has since been the subject of intensive study as a number of highly relevant potential candidate genes are located within and nearby to it. These include interferon regulatory factor 1 (IRF1), solute carrier family 22, member 4 (SLC22A4) also known as organic cation transporter 1 (OCTN1); solute carrier family 22, member 5 (SLC22A5), also known as organic cation transporter 2 (OCTN2); and prolyl 4-hyroxylase (P4HA2) among others and there are also a number of important cytokine genes (IL-4, IL-13, IL-5, IL-3) located adjacent to the immediate 250 kb risk haplotype. Since the initial identification of this risk haplotype, numerous studies have replicated the association with CD and a few have demonstrated an association between IBD5 and UC as well[21-25], establishing this as one of a still limited number of bona fide associations in human complex disease genetics and the second in IBD specifically.
THE IBD5 SUSCEPTIBILITY GENES
In 2004 Peltekova and colleagues postulated that functional polymorphisms, L503F (rs1050152) and G-207C (rs2631367) in the SLC22A4 (OCTN1) and SLC22A5 (OCTN2) genes, respectively, comprised a two locus risk haplotype that accounted for the association findings at IBD5[26]. The “TC haplotype” described was found in 54% of CD patients versus 42% of affected controls (P = 0.0003). Moreover, the odds ration for those homozygous for the TC haplotype was 3.43-5.14 for CD with an even higher odds ratio for those carrying the TC haplotype and a CARD15 risk allele (7.28-10.5). No association was found with ulcerative colitis. In this study, the TC haplotype was independently associated with CD as compared to the background risk haplotype where the marker IGR2078 was utilized as a tagging SNP to represent the IBD5 risk haplotype.
That functional polymorphisms in significant linkage disequilibrium with each other in adjacent, highly homologous genes may act as a functional cassette to increase disease risk would represent a novel, important paradigm in complex disorders. In particular, SLC22A5 (OCTN2) transports carnitine, which plays a key role in mitochondrial transport of long chain fatty acids; furthermore, SLC22A5 (OCTN2)-deficient mice develop intestinal lymphocytic infiltration. However, establishing a functional role for SNPs located within conserved elements which subtly alter gene expression can be difficult compared to establishing altered activity for amino acid polymorphisms.
REPLICATION STUDIES AND GENOTYPE PHENOTYPE STUDIES OF THE IBD5 LOCUS
As described above, there have been numerous studies to confirm the association of CD with the IBD5 risk haplotype[21-25]. These studies initially tested varying markers selected as tagging markers for the risk haplotype on chromosome 5 due to the high degree of linkage disequilibrium in the region. Statistical support for a relatively unique contribution of the two-locus OCTN haplotype was initially described, but not examined in large enough cohorts to confidently refine localization in the IBD5 region[27]. Further studies performed subsequent to the identification of the OCTN variants have primarily tested these markers alone although more recent reports have also tested additional SNPs within the IBD5 risk haplotype.
A well designed study including 374 patients with CD from Noble and colleagues in Edinburgh demonstrated that while the OCTN1 and OCTN2 variants are associated with CD, this association is not independent of the background risk haplotype in the region[28]. This study assessed not only the OCTN variants but also a number of surrounding SNPs from the list of SNPs comprising the IBD5 risk haplotype (IGR2096, IGR 2198 and IGR2230). Utilizing these SNPs they could not demonstrate independent association of the OCTN variants and suggest that the IGR2078 SNP used by Peltekova and colleagues is one haplotype block removed from the OCTN variants and therefore was not truly indicative of an independent association (Figure 1). A number of additional such reports have been presented in abstract form with similar findings (DDW 2005).
Figure 1.
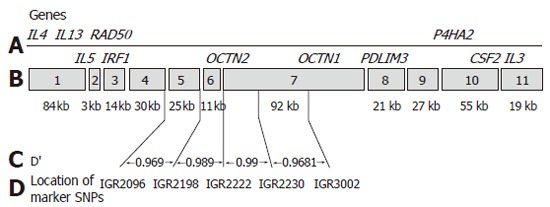
IBD5 map demonstrating the location of SNPs and regional candidate genes (Taken from Noble et al ref 28 - permission for reproduction being sought).
Despite the controversy that still exists over the true causal gene within the IBD5 locus, studies performed both before and after the identification of the OCTN variants have attempted to further dissect the specific phenotype associated with this region. Just as is the case for the causal genetic variant there is also inconsistent data regarding the phenotype associated. Early on, it was reported that the IBD5 locus was associated with perianal CD in a UK study[21]. This report analyzed several of the SNPs on the IBD5 risk haplotype in a case-control fashion. More recently, association of the IBD5 region has been found to be associated with markers of more severe disease such as progression to stricturing or penetrating CD and need for surgery in an Edinburgh population[28]. No association was found for any specific disease location or with UC in this report. A study from the same investigators in a pediatric population also confirms the association of the OCTN variants with CD and UC but the variants were not independently associated from the surrounding SNPs of the IBD5 risk haplotype[29]. This study also demonstrated that the OCTN TC haplotype may be a marker for more severe CD with an association with lower height, weight and body mass index (BMI) at the time of diagnosis. A report evaluating IBD patients from Italy also demonstrated IBD5 to be related to more severe CD behaviour although by multiple regression it was felt that this effect was essentially conferred by the presence of CARD15 risk alleles rather than by IBD5 loci independently[30].
A study of more than 981 IBD patients from Leuven could not demonstrate an association of the OCTN1 and OCTN2 variants with IBD, CD or UC globally but did show an association with perianal and penetrating CD[31]. These association findings have yet to be replicated and their significance has yet to be determined. One may speculate that the IBD5 locus represents a “phenotype-determining” genetic effect, however, the lack of an overall positive association with CD in the Belgian population makes this interpretation somewhat precarious. There have been inconsistent reports describing a role of IBD5 and UC with only a few describing positive findings[22,23,32]. A study of 187 German families with UC was the first to describe an association of IBD5 with UC by family-based association testing and, in particular, the association was strongest in those individuals carrying one of the CARD15 risk alleles[22]. This potential epistatic interaction was also reported in a cohort from the UK[23]. Although these results require further investigation in larger sample sizes, it is not altogether unexpected that there may be some effect of IBD5 on the development of UC. There is an increased risk of UC developing in the relatives of those with CD and the reverse is also true suggesting there are genes which confer overlapping susceptibility[6]. In addition, the clinical heterogeneity among the two disorders could result in misspecification of disease type in those with colonic involvement. Moreover, it has been previously noted that there are subsets of CD which have a more “UC-like” appearance raising further suspicion for overlapping genetic susceptibility factors[33]. One might also speculate on the possibility of a unique “IBD5 positive”, genetically and clinically unique subset of IBD patients, that would require further testing to be better characterized. As is the case with CARD15, the IBD5 region does show some variation with respect to the population studied. Studies from Asian populations show no evidence for IBD5 as a contributing factor to IBD susceptibility[34,35]. In addition, a study with a significant Jewish population report no evidence for a contribution of IBD5 to susceptibility in this high risk group[36].
In conclusions, while common alleles of modest effect play a very important role in the architecture of complex diseases and can be readily detected in some cases with modest sample sizes, it is now widely recognized that extremely large sample sizes are required in order to conclusively confirm these factors, resolve their location through fine-mapping and identify specific subphenotypes that are either preferentially associated or not-associated with disease. While the two allele risk haplotype of the SLC22A4 (OCTN1) and SLC22A5 (OCTN2) genes may thus far be the best candidates for the IBD5 association findings, there is as yet insufficient evidence to make concrete conclusions regarding their significance. Current data suggests that other, nearby SNPs, can be considered statistically equivalent in terms of their association to CD and the extensive linkage disequilibrium in this region suggests that caution must be exerted in asserting which of the SNPs in this region is potentially causal. Further functional evaluation of these variants is certainly warranted. In addition, the inconsistent findings in UC illustrate the importance of better powered UC genetics studies to further delineate the role of IBD5 and UC susceptibility and phenotype. In complex disorders, the IBD5 association represents one of the best replicated genomic regions, and therefore warrants comprehensive studies in large, well-phenotyped cohorts to define those population subsets and functional polymorphisms which most likely contribute to disease risk. To further advance the understanding of the IBD5 locus and its role in IBD susceptibility will require even larger population studies than those already performed, that will have adequate power to dissect the high degree of linkage disequilibrium found in this region in conjunction with functional studies that will demonstrate mechanisms. Due to the modest but important genetic relative risk of this locus, large consortia with prospectively collected, well-phenotyped cohorts will be required in order to refine the information regarding this region.
Footnotes
S- Editor Pan BR E- Editor Ma WH
References
- 1.Rogler G. Update in inflammatory bowel disease pathogenesis. Curr Opin Gastroenterol. 2004;20:311–317. doi: 10.1097/00001574-200407000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Tysk C, Lindberg E, Järnerot G, Flodérus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996;312:95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subhani JMS, Ounder RE, Wakefield AJ. Concordance rates of twins and siblings in inflammatory bowel disease. Gut. 1998;Suppl 42:A40. [Google Scholar]
- 5.Halfvarson J, Bodin L, Tysk C, Lindberg E, Järnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–1773. doi: 10.1016/s0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 6.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sørensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 7.Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, et al. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 8.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger JD, Lathrop GM, Bell JI, Jewell DP. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 9.Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJ, Cardon LR, Sakul H, Harris TJ, Buckler A, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999;64:808–816. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, Targan SR, Fischel-Ghodsian N, Rotter JI, Yang H. A genome-wide search identifies potential new susceptibility loci for Crohn's disease. Inflamm Bowel Dis. 1999;5:271–278. doi: 10.1097/00054725-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Duerr RH, Barmada MM, Zhang L, Pfützer R, Weeks DE. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11-12. Am J Hum Genet. 2000;66:1857–1862. doi: 10.1086/302947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000;66:1863–1870. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 14.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 15.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 16.Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 17.Shifman S, Kuypers J, Kokoris M, Yakir B, Darvasi A. Linkage disequilibrium patterns of the human genome across populations. Hum Mol Genet. 2003;12:771–776. doi: 10.1093/hmg/ddg088. [DOI] [PubMed] [Google Scholar]
- 18.Sugimura K, Taylor KD, Lin YC, Hang T, Wang D, Tang YM, Fischel-Ghodsian N, Targan SR, Rotter JI, Yang H. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72:509–518. doi: 10.1086/367848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 20.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 21.Armuzzi A, Ahmad T, Ling KL, de Silva A, Cullen S, van Heel D, Orchard TR, Welsh KI, Marshall SE, Jewell DP. Genotype-phenotype analysis of the Crohn's disease susceptibility haplotype on chromosome 5q31. Gut. 2003;52:1133–1139. doi: 10.1136/gut.52.8.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giallourakis C, Stoll M, Miller K, Hampe J, Lander ES, Daly MJ, Schreiber S, Rioux JD. IBD5 is a general risk factor for inflammatory bowel disease: replication of association with Crohn disease and identification of a novel association with ulcerative colitis. Am J Hum Genet. 2003;73:205–211. doi: 10.1086/376417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGovern DP, Van Heel DA, Negoro K, Ahmad T, Jewell DP. Further evidence of IBD5/CARD15 (NOD2) epistasis in the susceptibility to ulcerative colitis. Am J Hum Genet. 2003;73:1465–1466. doi: 10.1086/379745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirza MM, Fisher SA, King K, Cuthbert AP, Hampe J, Sanderson J, Mansfield J, Donaldson P, Macpherson AJ, Forbes A, et al. Genetic evidence for interaction of the 5q31 cytokine locus and the CARD15 gene in Crohn disease. Am J Hum Genet. 2003;72:1018–1022. doi: 10.1086/373880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negoro K, McGovern DP, Kinouchi Y, Takahashi S, Lench NJ, Shimosegawa T, Carey A, Cardon LR, Jewell DP, van Heel DA. Analysis of the IBD5 locus and potential gene-gene interactions in Crohn's disease. Gut. 2003;52:541–546. doi: 10.1136/gut.52.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 27.Török HP, Glas J, Tonenchi L, Lohse P, Müller-Myhsok B, Limbersky O, Neugebauer C, Schnitzler F, Seiderer J, Tillack C, et al. Polymorphisms in the DLG5 and OCTN cation transporter genes in Crohn's disease. Gut. 2005;54:1421–1427. doi: 10.1136/gut.2005.066340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble CL, Nimmo ER, Drummond H, Ho GT, Tenesa A, Smith L, Anderson N, Arnott ID, Satsangi J. The contribution of OCTN1/2 variants within the IBD5 locus to disease susceptibility and severity in Crohn's disease. Gastroenterology. 2005;129:1854–1864. doi: 10.1053/j.gastro.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 29.Russell RK, Drummond HE, Nimmo ER, Anderson NH, Noble CL, Wilson DC, Gillett PM, McGrogan P, Hassan K, Weaver LT, et al. Analysis of the influence of OCTN1/2 variants within the IBD5 locus on disease susceptibility and growth indices in early onset inflammatory bowel disease. Gut. 2006;55:1114–1123. doi: 10.1136/gut.2005.082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latiano A, Palmieri O, Valvano RM, D'Incà R, Vecchi M, Ferraris A, Sturniolo GC, Spina L, Lombardi G, Dallapiccola B, et al. Contribution of IBD5 locus to clinical features of IBD patients. Am J Gastroenterol. 2006;101:318–325. doi: 10.1111/j.1572-0241.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 31.Vermeire S, Pierik M, Hlavaty T, Claessens G, van Schuerbeeck N, Joossens S, Ferrante M, Henckaerts L, Bueno de Mesquita M, Vlietinck R, et al. Association of organic cation transporter risk haplotype with perianal penetrating Crohn's disease but not with susceptibility to IBD. Gastroenterology. 2005;129:1845–1853. doi: 10.1053/j.gastro.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri O, Latiano A, Valvano R, D'Incà R, Vecchi M, Sturniolo GC, Saibeni S, Peyvandi F, Bossa F, Zagaria C, et al. Variants of OCTN1-2 cation transporter genes are associated with both Crohn's disease and ulcerative colitis. Aliment Pharmacol Ther. 2006;23:497–506. doi: 10.1111/j.1365-2036.2006.02780.x. [DOI] [PubMed] [Google Scholar]
- 33.Vasiliauskas EA, Plevy SE, Landers CJ, Binder SW, Ferguson DM, Yang H, Rotter JI, Vidrich A, Targan SR. Perinuclear antineutrophil cytoplasmic antibodies in patients with Crohn's disease define a clinical subgroup. Gastroenterology. 1996;110:1810–1819. doi: 10.1053/gast.1996.v110.pm8964407. [DOI] [PubMed] [Google Scholar]
- 34.Tosa M, Negoro K, Kinouchi Y, Abe H, Nomura E, Takagi S, Aihara H, Oomori S, Sugimura M, Takahashi K, et al. Lack of association between IBD5 and Crohn's disease in Japanese patients demonstrates population-specific differences in inflammatory bowel disease. Scand J Gastroenterol. 2006;41:48–53. doi: 10.1080/00365520510023864. [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki K, Takazoe M, Tanaka T, Ichimori T, Saito S, Iida A, Onouchi Y, Hata A, Nakamura Y. Association analysis of SLC22A4, SLC22A5 and DLG5 in Japanese patients with Crohn disease. J Hum Genet. 2004;49:664–668. doi: 10.1007/s10038-004-0204-x. [DOI] [PubMed] [Google Scholar]
- 36.Newman B, Gu X, Wintle R, Cescon D, Yazdanpanah M, Liu X, Peltekova V, Van Oene M, Amos CI, Siminovitch KA. A risk haplotype in the Solute Carrier Family 22A4/22A5 gene cluster influences phenotypic expression of Crohn's disease. Gastroenterology. 2005;128:260–269. doi: 10.1053/j.gastro.2004.11.056. [DOI] [PubMed] [Google Scholar]


