Abstract
AIM: To study the amino acid substitutions in the carboxy (C)-terminal part of E2 protein and in the interferon (IFN) sensitivity determining region (ISDR) and their correlation with response to IFN and viral load in 85 hepatitis C virus (HCV)-1b-infected patients treated with IFN.
METHODS: The C-terminal part of E2 (codons 617-711) including PKR/eIF2α phosphorylation homology domain (PePHD) and ISDR was sequenced in 85 HCV-1b-infected patients treated by IFN monotherapy.
RESULTS: The amino acid substitutions in PePHD detected only in 4 of 85 patients were not correlated either with response to IFN or with viral load. The presence of substitutions in a N-terminal variable region (codons 617-641) in the C-terminal part of E2 was significantly correlated with both small viral load (33.9% vs 13.8%, P = 0.0394) and sustained response to IFN (25.0% vs 6.9%, P = 0.0429). Four or more substitutions in ISDR were significantly correlated with both small viral load (78.6% vs 16.2%, P < 0.0001) and sustained response to IFN (85.7% vs 2.9%, P < 0.0001). In multivariate analysis, ISDR in nonstructural (NS) 5A (OR = 0.39, P < 0.0001) and N-terminal variable region (OR = 0.51, P = 0.039) was selected as the independent predictors for small viral load, and ISDR (OR = 39.0, P < 0.0001) was selected as the only independent predictor for sustained response.
CONCLUSION: The N-terminal variable region in the C-terminal part of E2 correlates with both response to IFN monotherapy and viral load and is one of the factors independently associated with a small viral load.
Keywords: E2, Genotype, HCV, Interferon, ISDR, NS5A, PePHD, PKR, SVR
INTRODUCTION
Hepatitis C virus (HCV) is one of the most frequent and important causes of chronic viral hepatitis and approximately 170 million people are infected with the virus worldwide[1]. HCV usually causes chronic infection, which can result in chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma[2].
Interferon (IFN) monotherapy and combination therapy of IFN and ribavirin have been shown to be effective for eradicating chronic HCV infection[3,4]. Genotype 1b is resistant against antiviral therapy[5], and viral load is known as a predictor for response to IFN mono therapy[6]. While the combination therapy has significantly improved the effectiveness for patients with large viral load of genotype 1b, viral load still determines the outcome of treatment[7].
RNA-activated protein kinase (PKR) is a mediator of IFN-induced antiviral resistance. PKR-binding domain (PKR-bD) comprising the interferon sensitivity determining region (ISDR) within the nonstructural (NS) 5A protein is known to be potentially important for antiviral therapy outcome. However, reported results concerning the correlation of the substitutions within PKR-bD/ISDR to treatment outcome are conflicting[8-12].
A PKR/eIF2α phosphorylation homology domain (PePHD, codons 659-670) within the carboxy (C)-terminal part of E2 protein has been found to interact with PKR and inhibit PKR in vitro, suggesting a possible mechanism of HCV to evade the antiviral effects of IFN[13]. Several studies have concluded that a correlation of substitutions in PePHD does not exist[14-17], while only 3 others have confirmed it[18-20]. PePHD-deleted HCV-1 mutants remain capable of binding to PKR to some extent, while C-terminal-truncated E2 protein loses activity of inhibiting PKR. Thus regions other than PePHD within C-terminal part of E2 protein may interact with PKR. It is possible that the mutations in regions other than PePHD within C-terminal part of E2 protein may correlate with response to IFN. Sarrazin et al[21] reported that substitutions in C-terminal part of E2 are linked with response to antiviral therapy.
In the present study, we investigated the amino acid substitutions in the C-terminal part of E2 protein and in ISDR and their correlation with response to IFN and viral load in 85 HCV-1b infected patients treated with IFN.
MATERIALS AND METHODS
Patients
Eighty-five patients infected with HCV genotype 1b, were retrospectively analyzed (Table 1). The patients received various IFN preparations in various schedules of administration. Thirty-six patients received lymphoblastoid IFNα, 14 IFNα 2a, 18 natural IFNβ, 3 IFNα 2b and 14 both lymphoblastoid IFNα and natural IFNβ. Total dose of IFN was 200-400MU in 32, 400-600MU in 40, and 600MU or more in 13 patients. All patients were positive for both serum anti-HCV antibody (second generation enzyme immunoassay, Abbott laboratories, North Chicago, IL) and serum HCV RNA (Amplicor HCV test, Nippon Roche, Tokyo). HCV genotype was determined as previously described[22] and classified according to Simmonds’ the classification of simmonds et al[23]. Viral load was measured by branched DNA assay (Quantiplex HCV RNA assay, version 2, Chiron Corp., Emeryville, CA). Histological evaluations were done according to the grading and staging suggested by Desmet et al[24].
Table 1.
Characteristics of patients with different response to IFN therapy
| Sustained responders | Non-responders | P | |
| Patients (n) | 16 | 69 | |
| Age (yr) | 51.2 ± 10.7 | 52.4 ± 11.1 | NS |
| Male/Female | 14/2 | 47/22 | NS |
| History of blood | 5/11 | 21/48 | NS |
| transfusion (+/-) | |||
| ALT (IU/I) | 110.4 ± 88.0 | 95.6 ± 61.0 | NS |
| Viral load (Meq/mL) | 0.9 ± 0.9 | 6.9 ± 7.8 | 0.0027 |
| Histology | |||
| Grade | 1.9 ± 0.7 | 2.3 ± 0.8 | NS |
| Stage | 1.9 ± 0.7 | 2.5 ± 0.9 | 0.0154 |
| Number of amino acid | |||
| Substitutions in | |||
| ISDR | 5.0 ± 1.2 | 0.7 ± 1.1 | < 0.0001 |
| C-terminal part of E2 | 3.4 ± 1.7 | 2.5 ± 1.2 | 0.0149 |
| N-terminal variable region | 1.6 ± 1.1 | 0.9 ± 0.9 | 0.0148 |
| Substitution of aa 625 (+/-) | 6/10 | 8/61 | 0.0213 |
Patients who obtained normalization of alanine aminotransferase (ALT) levels and HCV RNA clearance for more than 6 mo after IFN therapy were considered as sustained virological responders (SVR). The other patients were grouped as nonresponders.
This study was approved by the local committee on medical ethics and adhered to the guidelines of the 1975 Declarations of Helsinki.
Sequencing of the C-terminal part of E2 protein comprising PePHD
HCV RNA was extracted from 100 μL of patients’ sera before IFN therapy by Sepa Gene RV-RTM (Sanko Junyaku Co. Inc., Tokyo). cDNA was prepared from extracted RNA by reverse transcription using the random primer (Takara, Ohtsu, Japan). Nested polymerase chain reaction (PCR) was done for amplifying the C-terminal part of the E2 protein comprising PePHD (nt 2178-2462). Four primers designed from the nucleotide sequences of HCV-J were used for the nested PCR. The 1st round of PCR was performed using the external primers (sense primer; nt 2127-2143; 5’-GGGCCCTGGTTGACACC-3’, and anti-sense primer; nt 2508-2524; 5’-GCGGACGCGCGCGTCTG-3’) and the 2nd round of PCR was performed using the internal primers (sense primer; nt 2157-2174; 5’-GACTACCCATACAGGCTC-3’, and anti-sense primer; nt 2496-2515; 5’-TTCCTTCTTCTGGCGGA-3’). All rounds of PCR were done with Taq polymerase (Takara, Ohtsu, Japan) by the following protocol: 35 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C, and a final extension for 7 min at 72°C.
Sequencing was performed by the PCR direct sequencing method. Amplified DNA was isolated by agarose gel electrophoresis and purified by EASYTRAPTM Ver.2 (Takara, Ohtsu, Japan). The nucleotide sequence was determined by the dideoxy method using a dye terminator cycle sequencing kit (Perkin-Elmer Japan, Tokyo, Japan).
Sequencing of ISDR in NS5A
cDNA of ISDR in NS5A was prepared by reverse transcription using 5ASRAW primer (nt 7177-7197; 5’-GTGCCCATATGGGCAACGCTG-3’) with RNA extracted from 50 μL of serum and then amplified by nested PCR with a DNA thermal cycler (Lobocycler 40; Stratagene, La Jolla, CA). The 1st round of PCR was performed using the external primers (sense primer 5SPDV; nt 6825-6844; 5'-CCGGATGTGGCAGTGCTCAC-3', and anti-sense primer 5ASRAW. The second round of PCR was performed using the internal primers (sense primer 5STDV; nt 6853-6874; 5'-TCACCGACCCCTCTCATATTAC-3', and biotin-labeled anti-sense primer 5IASPKR; nt 7141-7161; 5'-GTTTTCGCAGGATCTCCGCCG-3'). Twenty-nine cycles of PCR were done as follows: denaturation for 1 min at 94°C, annealing for 1.5 min at 56°C, and extension for 1.5 min at 72°C. The PCR products labeled with biotin were recovered with streptavidin-coated magnetic beads (Dynal, Oslo, Norway). The sequencing reaction was performed using a thermo sequenase fluorescent-labeled primer cycle sequencing kit (Amersham, Buckinghamshire, England) with Cy5-labeled 5ISKRR fluorescent primer (nt6887-6906; 5’-CAAGCGTAGGCTGGCCAGGG-3’) under the conditions recommended by the kit supplier. The reaction mixture was applied to the ALFII DNA sequencer (Amersham Pharmacia Biotech, Tokyo, Japan). Identification of the substituted amino acid was performed using a KINOP alignment software system (Otsuka-Fujitsu Collaborative Development, Tokyo, Japan).
Phylogenetic tree construction
A phylogenetic tree was constructed to determine the genetic relation using the Neighbour-joining method[25].
Statistical analysis
Data were analyzed by Student’s t test, chi-square test, and linear regression test for each applicable analysis. Multivariate analysis was done by discriminant analysis by stepwise forward selection method. P < 0.05 was considered statistically significant.
RESULTS
Response to IFN therapy
Sixteen of 85 patients had sustained virological response (Table 1). Viral load was significantly smaller in sustained virological responders than in non-responders (P = 0.0027). Stage of liver histology was significantly lower in sustained virological responders than in non-responders (P = 0.0154).
Sequences of C-terminal part of E2 protein
All of the obtained sequences were aligned. The most popular amino acid at each position was determined as the consensus sequence. Since there were two amino acids (isoleucine and valine) equally popular at codon 705, we considered both of them as the consensus sequences. The number of amino acid substitutions in the C-terminal part of E2 protein (codons 617-711) was 0 in 2, 1 in 13, 2 in 30, 3 in 22, 4 in 10, 5 in 3 and 6 in 5 patients respectively (Figures 1A and B).
Figure 1.
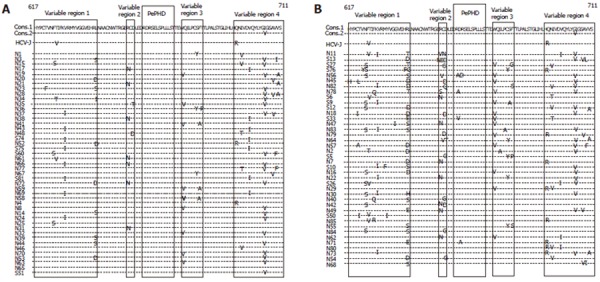
Amino acid sequences of C-terminal part of E2 protein with 0-2 substitutions (A) and 3-6 substitutions (B) in 45 and 40 HCV 1b-infected patients treated by IFN therapy respectively.
Amino acid substitutions in PePHD were seen only in 4 out of 85 patients and did not correlate either with viral load or with response to IFN (Figures 1A and B).
The number of substitutions in the C-terminal part of E2 protein was significantly larger in sustained virological responders than in non-responders (P = 0.0149, Table 1). The patients with 4 or more substitutions in the C-terminal part of E2 protein had sustained virological response to IFN therapy [38.9% (7/18)] more frequently than those with less than 4 substitutions [13.4% (9/67)]( P = 0.0358).
In the molecular-evolutional analysis with phylogenetic tree, we could not find any specific clustering either in sustained virological responders or in non-responders (Data not shown).
N-terminal region in the C-terminal part of E2 protein
In the C-terminal part of E2 protein, there were four variable regions where amino acid sequences were variable (Figures 1A and B). The regions correlating with response to IFN and viral load were searched for. The number of substitutions in N-terminal variable region designated as variable region 1 was significantly larger in sustained virological responders than in non-responders (P = 0.0148, Table 1). The patients with substitutions in N-terminal variable region had sustained virological response to IFN therapy [25.0% (14/56)] more frequently than those without substitutions [6.9% (2/29), P = 0.0429]. The number of substitutions in N-terminal variable region was significantly larger in the patients with small viral load (HCV-RNA < 1Meq/mL) than in those with large viral load (P = 0.0107, Table 2). The patients with substitutions in N-terminal variable region had small viral load [33.9% (19/56)] more frequently than those without substitutions [13.8% (4/29), P = 0.0394]. Amino acid substitutions in other parts of C-terminal of E2 protein were not correlated with either viral load or sustained response to IFN therapy (Data not shown).
Table 2.
Characteristics of patients with small or large viral load
| Patients with small viral load | Patients with large viral load | P | |
| Patients (n) | 24 | 61 | |
| Age (yr) | 52.5 ± 11.0 | 52.0 ± 11.1 | NS |
| Male/female | 19/5 | 42/19 | NS |
| History of blood | |||
| transfusion (+/-) | 8/16 | 18/43 | NS |
| ALT (IU/I) | 89.3 ± 64.1 | 102.0 ± 67.6 | NS |
| Viral load (Meq/mL) | 0.5 ± 0.1 | 7.8 ± 7.9 | 0.0001 |
| Histology | |||
| Grade | 2.2 ± 0.8 | 2.3 ± 0.8 | NS |
| Stage | 2.3 ± 0.8 | 2.4 ± 0.9 | NS |
| Number of amino acid | |||
| Substitutions in | |||
| ISDR | 3.3 ± 2.5 | 0.8 ± 1.2 | < 0.0001 |
| C-terminal part of E2 | 3.1 ± 1.7 | 2.5 ± 1.2 | NS |
| N-terminal variable region | 1.5 ± 1.0 | 0.9 ± 0.9 | 0.0090 |
| Substitution of aa 625 (+/-) | 7/17 | 7/54 | NS |
Specific amino acid substitutions correlated with viral load or with response to IFN therapy were searched for. Amino acid substitutions of threonine to serine at codon 625 were found to be significantly correlated with sustained virological response to IFN therapy (P = 0.0213) (Table 1).
Changes in sequences of C-termianl part of E2 after IFN therapy
The sequences of C-terminal part of E2 after IFN therapy were determined in 16 non-responders (Figure 2). In 8 patients, the sequences did not change before and after IFN therapy, while in the other 8 patients, the sequences changed. The number of substitutions in C-terminal part of E2 decreased in 3 patients (from 5 to 2 in N45, from 3 to 2 in N7, and from 2 to 1 in N3), increased in 4 patients (from 2 to 3 in N28 and N36, and from 1 to 3 in N31 and N46), and did not changed in N38.
Figure 2.
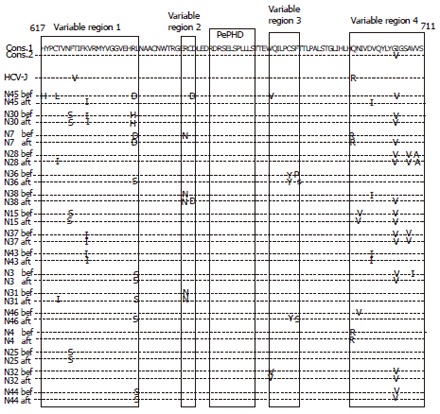
Amino acid sequences of C-terminal part of E2 before and after IFN therapy in 16 non-responders.
Sequences of ISDR
The sequences of ISDR were determined in 82 patients (Figure 3). The number of substitutions in ISDR was significantly larger in sustained virological responders than in non-responders (P < 0.0001, Table 1). The patients with 4 or more substitutions in ISDR had sustained virological response to IFN therapy [85.7% (12/14)] more frequently than those with less than 4 substitutions [2.9% (2/68), P < 0.0001]. The number of substitutions in ISDR was significantly larger in the patients with small viral load than in those with large viral load (P < 0.0001, Table 2). The patients with 4 or more substitutions in ISDR had small viral load [78.6% (11/14)] more frequently than those with less than 4 [16.2% (11/68), P < 0.0001].
Figure 3.

Amino acid sequences of ISDR in 82 HCV-infected patients.
Correlation between substitutions in C-terminal part of E2 and ISDR
The substitutions in C-terminal part of E2 and ISDR were significantly correlated with each other (P = 0.026, r = 0.246) (Figure 4). The number of substitutions in N-terminal variable region did not correlate with that in ISDR.
Figure 4.
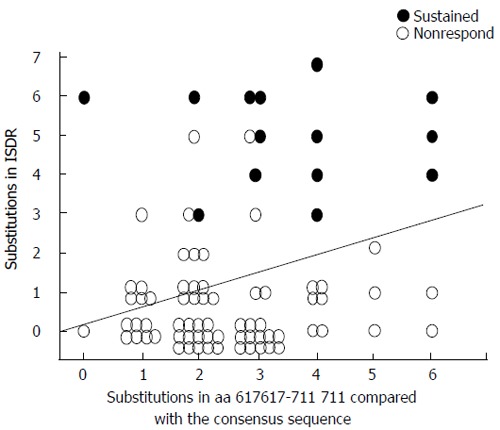
Correlation between the number of amino acid substitutions in ISDR and C-terminal part of E2 protein.
Multivariate analysis for factors independently associated with response to IFN therapy and viral load
In multivariate analysis, the number of substitutions in ISDR was selected as the only independent factor associated with sustained virological response to IFN therapy (OR = 39.0, P < 0.0001). The substitutions in ISDR (OR = 0.39, P < 0.0001) and N-terminal variable region (OR = 0.51, P = 0.039) were selected as the independent factors correlated with small viral load.
DISCUSSION
In the present study, the substitutions in C-terminal part of E2 were found to be significantly correlated with response to IFN therapy. Especially, the substitutions in N-terminal variable region were significantly correlated with response to IFN therapy and one of the factors independently associated with small viral load. So far, only Sarrazin et al[21] have suggested the correlation of the substitutions in C-terminal part of E2 with response to antiviral therapy. They reported that normalized Shannon entropy as well as mean Hamming distances in C-terminal part of E2 are significantly lower in sustained responders than in non-responders by calculation of genetic complexity and diversity of amino acid sequences, although no significant correlation of specific mutations or number of mutations within C-terminal part of E2 with response to IFN therapy has been found[21]. Further studies are needed to assess the correlation of sequences of C-terminal part of E2 with response to the combination therapy of IFN and ribavirin as well as viral load.
Tayler et al[13] described that PePHD-deleted HCV-1 mutants remain capable of binding to PKR to some extent, while C-terminal-truncated E2 protein loses activity of inhibiting PKR. This finding indicates that regions other than PePHD within C-terminal part of E2 interact with PKR and play an important role in suppressing PKR, a mediator of IFN-induced antiviral resistance. The correlation of C-terminal part of E2 with response to IFN therapy and small viral load may suggest that the substitutions in the region abrogate the activity of suppressing PKR, resulting in susceptibility to IFN therapy and small viral load.
When the sequences of C-terminal part of E2 were compared before and after IFN therapy, the decrease in substitutions was observed in only 3 of 16 patients, indicating that resistant HCV becomes predominant after IFN therapy. The factors other than the number of substitutions of C-terminal part of E2 also play a role in resistance to IFN therapy.
The present study showed that PePHD was highly conserved and that there were no significant correlations of the substitutions of PePHD with the response to IFN therapy or with viral load. It was reported that PePHD has similar sequences with PKR and eIF2α in E2 region of HCV genotype 1a[13]. Other studies also showed that PePHD is highly conserved and that there are no significant correlations between the substitutions in PePHD and the response to IFN therapy[14-17]. Only three reports on genotypes 1, 2, and 3 have reported a correlation between substitutions of PePHD and the effect of IFN therapy[18-20]. Recently Bagaglio et al[26] reported that substitutions in PePHD correlate with hepatocellular carcinoma. Further studies on the correlation of substitutions of PePHD with clinical aspects of chronic HCV infection are needed.
In the present study, the substitution of threonine to serine at codon 625 significantly correlated with sustained response. No reports are available on the correlation of a specific amino acid substitution with response to IFN therapy. Both threonine and serine have side chains containing hydroxylic groups which are similar to each other. Thus this substitution makes only a slight difference in the structure of E2 protein. Further studies are necessary to elucidate the correlation of the substitution with response to IFN therapy.
The correlation of ISDR with viral load and response to IFN therapy was confirmed in the present study. Gale et al[27] reported that NS5A represses PKR through direct interaction and inhibits IFN-induced antiviral response. Multiple ISDR mutations probably abrogate this action of NS5A on inhibiting PKR[28]. This is probably the reason why patients with mutant ISDR are likely to obtain sustained virological response and have small viral load. Inhibition of antiviral activity of IFN by NS5A has been shown in mammalian cells using encephalomyocarditis virus and vesicular stomatitis virus[29]. However inconsistent association between type of ISDR and IFN sensitivity in vitro has been also reported[30].
The present study demonstrated the correlation between the number of substitutions in the C-terminal part of E2 and ISDR. Both regions bind to PKR and inhibit its activity. It may be the reason why the mutations of the two regions occur simultaneously. They are probably adoptive mutations to the circumstances of individual hosts. The mutations may bring out some advantages for HCV to survive, while they may also make HCV susceptible to IFN or viral load smaller. Maekawa et al[31] reported that introduction of NS5A mutations enables HCV replicon derived from HC-J4 to replicate efficiently in Huh-7 cells[31]. The possible advantages of the mutations in the two regions require further elucidation. The mutations in ISDR also have been reported to correlate with development of hepatocellular carcinoma[32]. The role of the C-terminal part of E2 in the development of hepatocellular carcinoma is needed to be future studied.
In conclusion, N-terminal variable region in the C-terminal part of E2 correlates with both response to IFN therapy and viral load, and it is one of the factors independently associated with small viral load.
Footnotes
S- Editor Wang J L- Editor Wang XL E- Editor Bai SH
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell RH. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 3.Davis GL, Balart LA, Schiff ER, Lindsay K, Bodenheimer HC Jr, Perrillo RP, Carey W, Jacobson IM, Payne J, Dienstag JL. Treatment of chronic hepatitis C with recombinant alpha-interferon. A multicentre randomized, controlled trial. The Hepatitis Interventional Therapy Group. J Hepatol. 1990;11 Suppl 1:S31–35. doi: 10.1016/0168-8278(90)90160-s. [DOI] [PubMed] [Google Scholar]
- 4.Brillanti S, Garson J, Foli M, Whitby K, Deaville R, Masci C, Miglioli M, Barbara L. A pilot study of combination therapy with ribavirin plus interferon alfa for interferon alfa-resistant chronic hepatitis C. Gastroenterology. 1994;107:812–817. doi: 10.1016/0016-5085(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 5.Yoshioka K, Kakumu S, Wakita T, Ishikawa T, Itoh Y, Takayanagi M, Higashi Y, Shibata M, Morishima T. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-alpha therapy: relationship to genotypes of hepatitis C virus. Hepatology. 1992;16:293–299. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]
- 6.Lau JY, Davis GL, Kniffen J, Qian KP, Urdea MS, Chan CS, Mizokami M, Neuwald PD, Wilber JC. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 7.Di Bisceglie AM, Thompson J, Smith-Wilkaitis N, Brunt EM, Bacon BR. Combination of interferon and ribavirin in chronic hepatitis C: re-treatment of nonresponders to interferon. Hepatology. 2001;33:704–707. doi: 10.1053/jhep.2001.22346. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 9.Sarrazin C, Berg T, Lee JH, Teuber G, Dietrich CF, Roth WK, Zeuzem S. Improved correlation between multiple mutations within the NS5A region and virological response in European patients chronically infected with hepatitis C virus type 1b undergoing combination therapy. J Hepatol. 1999;30:1004–1013. doi: 10.1016/s0168-8278(99)80253-0. [DOI] [PubMed] [Google Scholar]
- 10.Yoshioka K, Kobayashi M, Orito E, Watanabe K, Yano M, Sameshima Y, Kusakabe A, Hirofuji H, Fuji A, Kuriki J, et al. Biochemical response to interferon therapy correlates with interferon sensitivity-determining region in hepatitis C virus genotype 1b infection. J Viral Hepat. 2001;8:421–429. doi: 10.1046/j.1365-2893.2001.00323.x. [DOI] [PubMed] [Google Scholar]
- 11.Squadrito G, Orlando ME, Cacciola I, Rumi MG, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to "interferon sensitivity determining region" variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30:1023–1027. doi: 10.1016/s0168-8278(99)80255-4. [DOI] [PubMed] [Google Scholar]
- 12.Nousbaum J, Polyak SJ, Ray SC, Sullivan DG, Larson AM, Carithers RL Jr, Gretch DR. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74:9028–9038. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 14.Gerotto M, Dal Pero F, Pontisso P, Noventa F, Gatta A, Alberti A. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology. 2000;119:1649–1655. doi: 10.1053/gast.2000.20230. [DOI] [PubMed] [Google Scholar]
- 15.Berg T, Mas Marques A, Höhne M, Wiedenmann B, Hopf U, Schreier E. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology. 2000;32:1386–1395. doi: 10.1053/jhep.2000.20527. [DOI] [PubMed] [Google Scholar]
- 16.Chayama K, Suzuki F, Tsubota A, Kobayashi M, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Takahashi N, et al. Association of amino acid sequence in the PKR-eIF2 phosphorylation homology domain and response to interferon therapy. Hepatology. 2000;32:1138–1144. doi: 10.1053/jhep.2000.19364. [DOI] [PubMed] [Google Scholar]
- 17.Hung CH, Lee CM, Lu SN, Lee JF, Wang JH, Tung HD, Chen TM, Hu TH, Chen WJ, Changchien CS. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J Viral Hepat. 2003;10:87–94. doi: 10.1046/j.1365-2893.2003.00414.x. [DOI] [PubMed] [Google Scholar]
- 18.Sarrazin C, Kornetzky I, Rüster B, Lee JH, Kronenberger B, Bruch K, Roth WK, Zeuzem S. Mutations within the E2 and NS5A protein in patients infected with hepatitis C virus type 3a and correlation with treatment response. Hepatology. 2000;31:1360–1370. doi: 10.1053/jhep.2000.7987. [DOI] [PubMed] [Google Scholar]
- 19.Lo S, Lin HH. Variations within hepatitis C virus E2 protein and response to interferon treatment. Virus Res. 2001;75:107–112. doi: 10.1016/s0168-1702(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, Ito T, Ishiko H, Yonaha M, Morikawa K, Miyokawa A, Mitamura K. Sequence analysis of PePHD within HCV E2 region and correlation with resistance of interferon therapy in Japanese patients infected with HCV genotypes 2a and 2b. Am J Gastroenterol. 2003;98:1377–1383. doi: 10.1111/j.1572-0241.2003.07469.x. [DOI] [PubMed] [Google Scholar]
- 21.Sarrazin C, Bruckner M, Herrmann E, Rüster B, Bruch K, Roth WK, Zeuzem S. Quasispecies heterogeneity of the carboxy-terminal part of the E2 gene including the PePHD and sensitivity of hepatitis C virus 1b isolates to antiviral therapy. Virology. 2001;289:150–163. doi: 10.1006/viro.2001.1092. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73(Pt 3):673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 23.Simmonds P, McOmish F, Yap PL, Chan SW, Lin CK, Dusheiko G, Saeed AA, Holmes EC. Sequence variability in the 5' non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74(Pt 4):661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 24.Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513–1520. [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Bagaglio S, De Mitri MS, Lodrini S, Paties C, Cassini R, Bianchi G, Bernardi M, Lazzarin A, Morsica G. Mutations in the E2-PePHD region of hepatitis C virus type 1b in patients with hepatocellular carcinoma. J Viral Hepat. 2005;12:243–250. doi: 10.1111/j.1365-2893.2005.00589.x. [DOI] [PubMed] [Google Scholar]
- 27.Gale MJ Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 28.Gale M Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gale M Jr, Kwieciszewski B, Dossett M, Nakao H, Katze MG. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson M, Laxton CD, Thomas HC, Ackrill AM, Foster GR. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology. 1999;117:1187–1197. doi: 10.1016/s0016-5085(99)70405-1. [DOI] [PubMed] [Google Scholar]
- 31.Maekawa S, Enomoto N, Sakamoto N, Kurosaki M, Ueda E, Kohashi T, Watanabe H, Chen CH, Yamashiro T, Tanabe Y, et al. Introduction of NS5A mutations enables subgenomic HCV replicon derived from chimpanzee-infectious HC-J4 isolate to replicate efficiently in Huh-7 cells. J Viral Hepat. 2004;11:394–403. doi: 10.1111/j.1365-2893.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- 32.Giménez-Barcons M, Franco S, Suárez Y, Forns X, Ampurdanès S, Puig-Basagoiti F, Sánchez-Fueyo A, Barrera JM, Llovet JM, Bruix J, et al. High amino acid variability within the NS5A of hepatitis C virus (HCV) is associated with hepatocellular carcinoma in patients with HCV-1b-related cirrhosis. Hepatology. 2001;34:158–167. doi: 10.1053/jhep.2001.25512. [DOI] [PubMed] [Google Scholar]


