Abstract
AIM: Different strains of bifidobacteria were analysed for their effects on HT-29 intestinal epithelial cells (IECs) in in vitro models both of the non-inflamed and inflamed intestinal epithelium.
METHODS: A reporter gene system in HT-29 cells was used to measure levels of NF-κB activation after challenge with bifidobacteria or after bacterial pre-treatment following LPS challenge. IL-8 protein and pro-inflammatory gene expression was investigated using normal HT-29 cells.
RESULTS: None of the bifidobacteria tested induced activation of nuclear factor κB (NF-κB) indicating that bifidobacteria themselves do not induce inflammatory events in IECs. However, six out of eight bifidobacteria tested inhibited lipopolysaccharide- (LPS-) induced NF-κB activation in a dose- and strain-dependent manner. In contrast, NF-κB activation in response to challenge with tumor necrosis factor-α (TNF-α) was affected by none of the tested bifidobacteria, indicating that the inhibitory effect of bifidobacteria is specific for LPS-induced inflammation in IECs. As shown with two of the six inhibition-positive bifidobacteria, LPS-induced inhibition of NF-κB activation was accompanied by a dose-dependent decrease of interleukin 8 (IL-8) secretion and by lower mRNA levels for IL-8, TNF-α, cyclooxygenase 2 (Cox-2), and intercellular adhesion molecule 1 (ICAM-1).
CONCLUSION: Some strains of bifidobacteria are effective in inhibiting LPS-induced inflammation and thus might be appropriate candidates for probiotic intervention in chronic intestinal inflammation.
Keywords: NF-κB, Bifidobacteria, Anti-inflammatory, LPS
INTRODUCTION
Inflammatory bowel diseases such as Crohn’s disease (CD) and ulcerative colitis (UC) are multifactorial disorders characterized by chronic inflammation of the intestinal epithelium. In addition to the genetic predisposition[1], the intestinal microflora seems to be involved in triggering inflammation, as observed in several animal models, where colitis was not induced in germ-free animals[2-4]. It has been suggested, that breakdown of tolerance towards the intestinal bacterial flora and dysfunction of regulatory T-cells are implicated in the development of chronic intestinal inflammation[5-8].
One of the central transcription factors mediating inflammatory responses is nuclear factor κB. Upon activation, through a wide range of stimuli such as TNF-α, IL-1β, or LPS, NF-κB translocates to the nucleus[9,10], where it regulates the transcription of a series of genes involved in acute responses to injury and in chronic intestinal inflammation including the genes for IL-1β, TNF-α, IL-6, IL-8, IL-12, Cox-2, ICAM-1, vascular endothelial growth factor-1, T-cell receptor-α, and major histocompatibility complex class II molecules[11,12]. LPS activates NF-κB through toll-like receptor 4 (TLR4)[13]. The events preceding the actual binding of LPS to TLR4 involve LPS-binding protein (LBP) and the intermediate receptor CD14[14,15]. LBP conveys LPS to CD14, which then promotes binding of LPS to the LPS receptor complex composed of TLR4 and MD-2, a co-receptor essential for LPS signalling via TLR4[16]. On monocytes and macrophages CD14 was shown to be membrane-bound by a glycosylphophoinositol anchor[17]. Stimulation of endothelial and epithelial cells with LPS requires the second form of CD14, i.e. soluble CD14 (sCD14)[18].
There is increasing evidence that in chronic intestinal inflammation expression of TLR4 and CD14 on IECs is abnormal. Under normal conditions, TLR4 and CD14 are expressed in IECs at very low levels in vivo[19,20] and are confined to undifferentiated cells of the crypts, which are not exposed to the overwhelming quantity of luminal antigens[21]. By contrast, expression of TLR4 and CD14 is significantly increased in IECs of both, animal models of colitis[22,23] and IBD patients[19]. A possible explanation for this increased expression could come from genetic studies. Higher frequencies of the Asp299Gly allele in the extracellular domain of TLR4 and the T allele and TT genotype at position -159 in the gene for CD14 have been positively associated with CD and UC[24-27]. Abnormal LPS signalling through increased levels of TLR4 and CD14 in IECs could contribute to the sustained mucosal inflammation in IBD.
Bifidobacteria are Gram-positive, anaerobic micro-organisms that inhabit mainly the colon of healthy infants and adults. In breast-fed infants, soon after birth, up to 90% of all bacteria in faecal samples detected by fluorescence in situ hybridisation are bifidobacteria[28] and they still make up 3%-5% of the adult microflora[29]. Thus, bifidobacteria undoubtedly constitute one of the predominant species of the human colonic microflora[30]. The analysis and annotation of the genome sequence of B. longum NCC2705 illustrated the close relationship of this bacterium with its human host[31]. Several beneficial health effects have been claimed to be related to the presence of bifidobacteria in the colon[32-41]. Based on these properties, bifidobacteria have become increasingly interesting for probiotic use, both in pharmaceutical application and dairy products[42]. One of these applications is their use in probiotic intervention in chronic intestinal inflammation. Probiotics containing bifidobacteria have been shown to be effective in reducing the severity of inflammation in several rodent models and patients with IBD[38,43-47].
In this study, the effects of a selection of bifidobacteria on IECs were investigated using an in vitro approach mimicking the situation of increased responsiveness of IECs to LPS. In this model, the antagonistic potential of several bifidobacteria on LPS-induced inflammatory responses in IECs was shown to be mediated by blocking NF-κB activation. This suggests that several Bifidobacterium strains, especially of B. bifidum, might be good candidates for probiotic intervention in IBD.
MATERIALS AND METHODS
Bacterial strains, cell lines and culture conditions
All bacterial strains used are shown in Table 1. For experiments, bacteria were cultured for 16 h at 37°C (bifidobacteria: de Man-Rogosa Sharpe medium with 0.5 g/L cysteine, anaerobic; E. coli: brain heart infusion medium, aerobic; all media BD Difco, Basel, Switzerland). HT-29 cells were maintained at 50% CO2 in DMEM (4.5 g/L glucose; Amimed, Basel, Switzerland) supplemented with 100 mL/L fetal calf serum (FCS; Amimed), 10 mL/L non-essential amino acids (NEAA, Sigma, Basel, Switzerland), 10 mL/L penicillin-streptomycin solution (pen-strep, Sigma). HT-29 clone 34 cells were obtained by stable transfection of the HT-29 cell line with the pNF-κB-SEAP-NPT plasmid[48]. This plasmid harbours the reporter gene for secreted alkaline phosphatase (SEAP). It contains the κB4 enhancer element fused to a TATA-like promoter region from the Herpes simplex virus thymidine kinase promoter, which permits the expression of the reporter gene following activation of the NF-κB signalling pathway. The SEAP coding sequence is followed by the SV40 late polyadenylation signal to ensure proper and efficient processing of the SEAP transcript in eukaryotic cells. The vector backbone also contains a pUC origin of replication and an ampicillin resistance gene for propagation and selection in E. coli. In addition, it contains the neomycin phosphotransferase gene for the dominant selection marker of geneticin resistance in eukaryotic cells. After amplification in E. coli TG1, the plasmid was transfected into HT-29 cells using lipofectamine (Invitrogen, Basel, Switzerland) as described by Moon et al[48], and the stable clone 34, showing good kinetics of reporter gene activation using recombinant human TNF-α (10 μg/L, R&D Systems, Oxon, England) as a stimulus, was used for reporter gene assays. HT-29 clone 34 cells were cultured in DMEM with 100 mL/L FCS, 10 mL/L NEAA, and geneticin (0.5 g/L; Invitrogen). For experiments, both HT-29 and HT-29 clone 34 cells were seeded at 2 × 105 cells/mL in 12-well format cell culture plates (Falcon®, Milian, Geneva, Switzerland) and cultured in the respective medium until they reached 90%-100% confluence. At this stage, approximately 1 × 106 cells/well were counted.
Table 1.
Bacterial strains used in this study
| Strain or oligonucleotides | Relevant characteristics | Source or PCR product |
| E. coli | ||
| TG1 | Cloning host | DSMZ1 |
| D2241 | Non-pathogenic intestinal isolate, control | NCC2 |
| Bifidobacterium | ||
| NCC362 | B. lactis, type strain | NCC2 |
| NCC2705 | B. longum, type strain | NCC2 |
| NCC251 | B. adolescentis, type strain | NCC2 |
| NCC189 | B. bifidum | NCC2 |
| S16 | B. bifidum, intestinal isolate from a breast-fed infant | [57] |
| S17 | B. bifidum, intestinal isolate from a breast-fed infant | [57] |
| E18 | B. infantis/longum, intestinal isolate from an adult | [57] |
| MB226 | B. breve, type strain | D. Matteuzzi |
DSMZ: Deutsche Sammlung von Mikroorganismen und Zellkulturen; 2 NCC: Nestlé Culture Collection.
LPS and TNF-α challenge of IEC lines
HT-29 or HT-29 clone 34 cells were grown as described above. At 90%-100% confluence, cells were washed and the medium was changed to 0.9 mL of DMEM with 100 mL/L FCS, 10 mL/L NEAA, 50 mmol/L Hepes (Invirogen), and penicillin G (100 kU/L; Fluka). Bacteria were grown as described above and added at the indicated multiplicity of infection (moi) in 0.1 mL of cell culture medium. In some experiments, cells were pre-incubated for 1 h with bacteria and subsequently stimulated with TNF-α (10 μg/L) or LPS (10 μg/L, serotype B55:O5; Sigma). Where indicated, a challenge was performed in the presence of 50 mL/L of human milk (HM). Milk was sampled from different mothers (d 20 of lactation) and pooled. The pool of HM used for this study contained 26 mg/L of sCD14 (determined by ELISA; P. Serrant, personal communication). After 16 h of incubation, supernatants were collected by centrifugation and used for quantification of SEAP or IL-8. Viability of the eukaryotic cells was continuously checked by microscopic examination of trypan blue (Sigma) exclusion and was > 95% in all experiments.
Detection of SEAP and IL-8 protein
Phosphatase activity of SEAP was quantified by bioluminescence using the PhosphaLight™ kit (Tropix, MA, USA). Bioluminescence was measured as relative light units (RLU) in a TECAN SPECTRAFluor Plus spectrometer with an integration time of 100 milliseconds and gain set to 100. Data were analysed using the XFlour software version 4.40. RLU varied between experiments due to the nature of the assay. Where more than one experiment was used to present figures and for statistical analysis, correction for these day to day variations was performed by normalizing data to set 100 RLU units for the positive control (LPS + HM). In one experiment, levels of NF-κB activation were similar for 10 μg/L of TNF-α and 10 μg/L of LPS in combination with HM.
Secreted IL-8 protein was quantified by ELISA using the IL-8 Eli-pair kit (Diaclone, CT, USA). Absorbance was measured in a Dynex MRX microplate reader (Dynex Technologies, Worthing, England) at 450 nm.
Real-time PCR
For real-time PCR, RNA of HT-29 cells was isolated after bacterial pre-treatment and 4 h of LPS challenge in the presence of 5% (v/v) HM using the NucleoSpin® RNAII kit (Macherey-Nagel, Düren, Germany). RNA was quantified using Ribogreen® RNA quantification kit (Molecular Probes, Basel, Switzerland). Quality of RNA was verified using Agilent RNA 6000 Nano Assays and the 2100 Bioanalyzer (Agilent Technologies, CA, USA) with corresponding software (version A.01.16). 1 μg of total RNA was transcribed to cDNA using TaqMan Reverse Transcription Reagents. Real time reactions were set up using 5 μL TaqMan® 2 x PCR Master Mix, 0.5 μL of TaqMan® assays-on-demand primer/probe mixture (glyceraldehyde-3 phosphate dehydrogenase (GAPDH): Hs99999905_mL; IL-8: Hs00174103_mL; TNF-α: Hs00174128_mL; COX-2: Hs00153133_mL; ICAM-1: Hs00164932_mL; all reagents were purchased from Applied Biosystems) and 4.5 μL H2O containing cDNA corresponding to 10-20 ng RNA, depending on the target gene. Real-time PCR was performed in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) and analysed with the SDS software package version 2.1. Results of the different RNA samples were normalised for RNA quantity using the gene for GAPDH.
RESULTS
Bifidobacteria do not induce NF-κB-dependent reporter gene activity in HT-29 clone 34 cells
To study the effect of bifidobacteria on inflammatory events in IECs, i.e. on NF-κB activation, a reporter gene assay in HT-29 clone 34 cells was used. Cells were incubated with bacteria (moi = 100) for 16 h. Subsequently, activity of the NF-κB-driven reporter protein SEAP was measured in culture supernatants (Figure 1). While non-pathogenic E. coli D2241 induced significant reporter gene activity, all bifidobacteria tested failed to induce any activation of NF-κB above background levels. The same cell-based assay was used to investigate the effect of bifidobacteria on LPS-induced inflammation in IECs. HT-29 clone 34 cells were challenged with LPS for 16 h and subsequently SEAP activity was measured in the culture supernatants (Table 2). Only a 2.5-fold induction of NF-κB activation by LPS alone compared to un-stimulated cells was observed. To mimic the situation in the intestinal epithelium of IBD patients where expression of CD14 is increased, 50 mL/L of HM, containing sCD14, were added to the assay. Addition of HM led to a 50-fold increase in LPS-induced NF-κB activation, whereas HM alone did not result in activation of NF-κB. Also, in the presence of HM none of the tested bifidobacteria induced NF-κB-dependent reporter gene activity above background levels while stimulation with E. coli D2241 was enhanced about 200-fold (data not shown), further supporting the results indicating that bifidobacteria do not induce inflammatory events in IECs.
Figure 1.
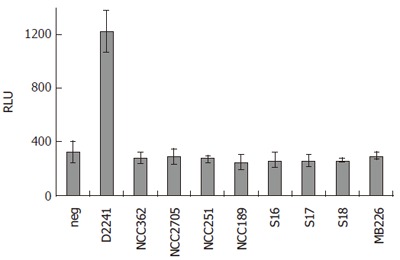
Quantification of NF-κB-driven SEAP activity in the supernatants of HT-29 clone 34 cells challenged with different bacterial strains at moi = 100 for 16 h. Results are shown as RLU and are mean ± standard deviation (SD) of triplicate measurements of one representative of three independent experiments. neg: negative control, no bacteria.
Table 2.
Quantification of NF-κB-driven SEAP activity in the supernatants of HT-29 clone 34 cells
| Treatment | RLU ± SD 1 |
| None (untreated control) | 2 ± 1 |
| HM | 2 ± 1 |
| LPS | 5 ± 1 |
| LPS + HM | 100 ± 6 |
Values are mean ± SD of at least 3 independent experiments for each condition performed in triplicate. For each individual experiment luminescence measurements were normalized to give 100 RLU for stimulation with LPS + HM.
Strain- and dose-dependent inhibition of LPS-induced NF-κB activation by bifidobacteria
Several reports suggest anti-inflammatory effects of probiotics containing bifidobacteria[38,44,49]. Therefore, all bifidobacteria used in this study were assessed for their potential inhibitory effect on LPS-induced NF-κB activation at different bacterial doses. At a moi of 1, none of the bifidobacteria tested had an effect on NFκB-dependent reporter gene activity (data not shown). As shown in Figure 2, at a moi = 10, however, pre-treatment with B. lactis NCC362 and B. bifidum NCC189, S16, and S17 significantly decreased NF-κB-dependent reporter gene activity to 47%-71% of the positive control. When the bacterial dose for pre-treatment was further increased to 100 bacteria per cell (moi = 100), the inhibitory effect of B. bifidum NCC189 and that of B. bifidum S17 was almost complete (5% of positive control), while that of B. lactis NCC362, B. longum NCC2705, B. adolescentis NCC251 and B. bifidum S16 was intermediate (20%-52% of positive control). In contrast, B. longum/infantis E18 and B. breve MB226 had no or only very slight inhibitory effect on SEAP activation at either moi. To assess whether physical presence of bifidobacteria was required for their inhibitory effect, further experiments were carried out using cell culture medium pre-treated for 16 h with bifidobacteria followed by the removal of bifidobacterial cells using filter sterilization. Use of this conditioned medium did not result in any inhibition of LPS-induced NF-κB activation in HT-29 clone 34 cells (data not shown). These results show that six out of eight bifidobacteria tested significantly inhibited LPS-induced NF-κB activation in IECs and that this inhibitory capacity of bifidobacteria is strain- and dose-dependent. Furthermore, presence of bifidobacterial cells during LPS challenge is required for their anti-inflammatory effects indicating that this inhibition is not due to secreted compounds. To prove that decreased activity of the reporter protein was not a consequence of apoptosis due to high bacterial doses in combination with LPS, viability of the cells was confirmed by trypan blue exclusion. Microscopic examination of cells pre-treated with two representative strains (B. longum NCC2705 and B. bifidum S17), challenged with LPS + HM showed that viability was > 95% in all experiments demonstrating that inhibition of LPS-induced reporter gene activity by bifidobacteria is not due to cell death (Figure 3).
Figure 2.
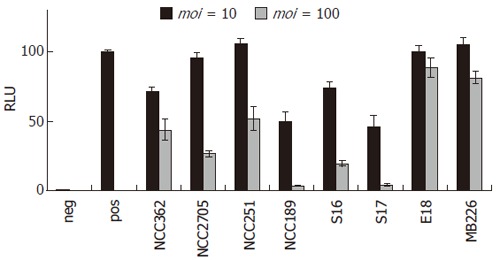
Dose- and strain-dependent inhibition of LPS-induced NF-κB activation by different bifidobacteria. SEAP activity was quantified in the supernatants of HT-29 clone 34 cells pre-incubated with different bifidobacteria at moi = 10 (dark grey bars) or moi = 100 (light grey bars) and subsequently stimulated with LPS + HM for 16 h. Cells treated with medium only served as negative control (neg; white bar) and positive controls (pos; black bar) were cells challenged with LPS + HM without bacterial pre-treatment. Results are means ± standard error of the mean (SEM) of three independent experiments performed in triplicate. RLU of indicidual experiments were normalised to give 100 RLU for the positive control.
Figure 3.
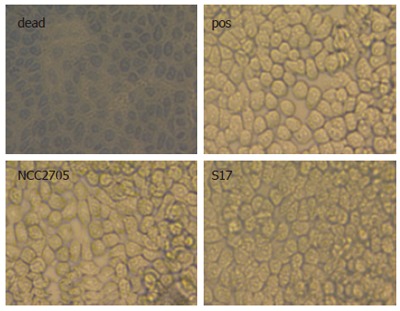
Microscopic examination of cellular viability. Trypan blue exclusion was monitored in HT-29 clone 34 cells challenged with LPS + HM for 16 after pre-incubation with bifidobacteria (B. longum NCC2705 or B. bifidum S17; moi = 100). Positive control (pos) was cells incubated with LPS + HM without bacterial pre-treatment. As a control for trypan blue staining cells were incubated for 16 h in cell culture medium at pH 4 (dead).
Inhibition of LPS-induced inflammatory events by B. bifidum S17 and B. longum NCC2705
To further investigate the effect of bifidobacteria on inflammatory events following NF-κB activation, IL-8 secretion and mRNA level of the genes encoding IL-8, TNF-α, COX-2, and ICAM-1 were monitored in normal HT-29 cells after bacterial pre-treatment with B. bifidum S17 and B. longum NCC2705 and a subsequent LPS challenge. Pre-incubation with B. bifidum S17 reduced IL-8 secretion of the IECs to about 20% of the positive control already at a moi = 10 and was able to completely inhibit IL-8 secretion at 100 bacteria per cell (Table 3). Effective inhibition with B. longum NCC2705 was only observed at a moi = 100. Thus, the dose-response of the IECs for IL-8 secretion was similar to that observed for NF-κB-driven SEAP reporter gene activity.
Table 3.
IL-8 protein in the supernatants of HT-29 cells (mean ± SD, n = 3)
| Treatment | IL-8 [μg/L] 1 | |
| moi = 10 | moi = 100 | |
| None (untreated control) | 2 ± 1 | |
| LPS+HM | 36 ± 3 | |
| NCC2705/LPS+HM 1 | 30 ± 2 | 7 ± 3 |
| S17/LPS+HM 1 | 7 ± 2 | 2 ± 0 |
Pre-incubation with B. longum NCC2705 or B. bifidum S17 at moi = 10 or 100 was followed by stimulation with LPS + HM for 16 h.
The effect of pre-treatment of HT-29 cells with bifidobacteria on mRNA levels for genes known to be regulated by NF-κB was investigated by real-time PCR. Pre-treatment with B. longum NCC2705 and B. bifidum S17 at a moi = 100 severely inhibited transcriptional activation of the genes for IL-8, TNF-α, COX-2, and ICAM-1 in HT-29 cells challenged with LPS in the presence of HM (Table 4). Again, B. bifidum S17 was more effective than B. longum NCC2705. Taken together, these results indicate that selected strains of bifidobacteria are able to inhibit LPS-induced inflammatory events in IECs.
Table 4.
Real-time quantification of mRNA levels of several genes in HT-29 cells
| Treatment |
Relative mRNA level 1 ± SD of gene for |
|||
| IL-8 | TNF-α | COX-2 | ICAM-1 | |
| LPS+HM (positive control) | 40.6 ± 17.4 | 37.4 ± 12.6 | 5.9 ± 1.7 | 28.0 ± 10.7 |
| B. longum NCC2705/ LPS+HM 2 | 11.4 ± 1.1 a | 12.7 ± 1.8 a | 2.5 ± 1.6 | 12.2 ± 9.2 |
| B. bifidum S17/ LPS+HM 2 | 3.5 ± 2.6 a | 2.9 ± 2.4 a | 1.4 ± 0.6 a | 2.0 ± 0.8 a |
Results are mRNA levels relative to the negative control (no bacteria, no LPS) and are mean ± standard deviation (SD) of three independent pools of RNA for each condition.
Pre-incubation with B. longum NCC2705 or B. bifidum S17 at moi = 100 was followed by stimulation with LPS + HM for 4 h.
P < 0.05 vs positive control; Student’s t-test.
TNF-α-induced NF-κB activation is not inhibited by bifidobacteria
NF-κB is activated through a wide range of stimuli other than LPS (see introduction). Therefore, it was interesting to test whether a similar inhibition of bifidobacteria could be observed for a pro-inflammatory stimulus other than LPS, such as TNF-α. At moi = 100, none of the tested bifidobacteria had any inhibitory effect on NF-κB-dependent reporter gene activity when 10 μg/L TNF-α was used to challenge HT-29 clone 34 cells (Figure 4). Further experiments were performed using a lower dose of TNF-α (1 μg/L) as stimulus and different doses of bacteria (moi = 10 and 100). In none of the tested conditions, an inhibitory effect of bifidobacteria on TNF-α induced NF-κB activation was observed (data not shown). These results suggest that the inhibitory capacity of bifidobacteria is specific for LPS-induced NF-κB activation in IECs.
Figure 4.
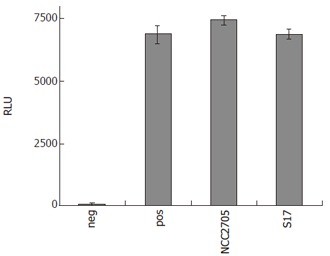
NF-κB-driven SEAP activity in the supernatants of HT-29 clone 34 cells pre-incubated with different bifidobacteria and subsequent stimulation with TNF-α (10 μg/L) for 16 h in the presence of 50 mL/L HM. Cells treated with medium only served as negative control (neg) and positive controls (pos) were cells challenged with TNF-α + HM without bacterial pre-treatment. Results are given as RLU and are means ± SD of triplicate measurements of a representative of two independent experiments.
DISCUSSION
Previous studies have shown that the intestinal microflora is implicated in the pathogenesis of IBD[2-4]. Increased expression of CD14 and TLR4 in IECs[19] might contribute to the sustained inflammation in the epithelium of patients of IBD. On the other hand, for several probiotic formulations, effects on inflammation in cultured IEC lines and in mouse models of chronic intestinal inflammation have been reported, yet there is a significant lack in our understanding of the molecular mechanisms by which probiotics downmodulate intestinal inflammation. Most studies on anti-inflammatory effects of probiotics were performed using mixtures of different probiotic strains, mainly lactobacilli and bifidobacteria[50]. For HT-29 and T84 cells, it was shown that incubation of these cells with VSL#3, a formulation containing eight different strains including 3 strains of bifidobacteria, resulted in decreased secretion of IL-8 in response to challenge with non-pathogenic E. coli or pathogenic Salmonella dublin[51]. In the IL-10 knock-out model of colitis, introduction of VSL#3 in the diet had an inhibitory effect on basal and LPS-stimulated TNF-α secretion in biopsies obtained from the colonic mucosa[44]. In the same mouse model, feeding a diet containing B. infantis 35624 was superior to L. salivarius UCC118 in effectively reducing the total gastrointestinal inflammatory score[38]. Finally, VSL#3 has been successfully used in the treatment of some IBDs[43,45].
Here, the effects of bifidobacteria on IECs were investigated with particular emphasis on LPS-induced inflammation. To our knowledge this represents the first attempt to characterize anti-inflammatory effects of whole cells of a single bifidobacterial strain. None of the tested bifidobacteria induced activation of NF-κB in HT-29 clone 34 cells, whereas a challenge with E. coli D2241 resulted in significant induction of NF-κB driven reporter gene activity in this cell line. Challenge of HT-29 clone 34 cells with LPS alone resulted in only a 2.5-fold induction of NF-κB-dependent reporter gene activity above background levels. This is consistent with results from other groups, which showed that colonic epithelial cells are poorly responsive to LPS[20]. However, using HM as a source of sCD14[52], the NF-κB response of HT-29 IECs to LPS was dramatically increased. This result supports previous studies showing increased responsiveness of HT-29 cells to LPS in the presence of HM[52-54]. This increased responsiveness of HT-29 cells to LPS in the presence of milk could be blocked by an anti-CD14 antibody[52].
NF-κB-dependent reporter gene activation by LPS in the presence of milk could be inhibited by pre-incubation with bifidobacteria in a strain- and dose-dependent manner, with the three B. bifidum strains (NCC189, S16, and S17) being most effective. To achieve this inhibitory effect, the presence of bifidobacterial cells was required, as filter-sterilized cell culture medium pre-treated with bifidobacteria did not show any inhibition of LPS-induced NF-κB activation. This indicates that the anti-inflammatory compound is not actively secreted by bifidobacteria or released from lysed cells. Furthermore, incubation of HT-29 cells with B. longum NCC2705 and B. bifidum S17 prior to LPS challenge reduced IL-8 secretion and had a significant inhibitory effect on mRNA levels of the TNF-α, IL-8, ICAM-1, and COX-2 genes, all of which have been shown to be regulated by NF-κB[9]. In a similar study, VSL#3 inhibited NF-κB reporter gene activity and MCP-1 secretion after a challenge with TNF-α, in conditionally immortalised mouse colon cells[49]. In the presented experiments, none of the tested strains of bifidobacteria inhibited NF-κB activation in response to TNF-α. These results suggest that bifidobacteria interfere with pro-inflammatory signals upstream of the pathway of NF-κB activation common for LPS and TNF-α.
Lipoteichoic acids (LTA) of two lactobacilli strains were able to inhibit IL-8 secretion in response to LPS in HT-29 cells[54]. In a recent study, it was shown that the composition of LTA have an impact on the immunogenicity and anti-inflammatory capacity of L. plantarum[55]. Of note, a mutant lacking D-alanine substitutions to the back bone of LTA showed dramatically improved anti-inflammatory properties compared to the wild type. The mechanism by which bifidobacteria exert their inhibitory effect remains to be elucidated. One study investigating DNA from the VSL#3 mixture showed that the bifidobacterial strains’ DNA was effective in limiting epithelial pro-inflammatory responses in IL-10-deficient mice and in HT-29 cells challenged with TNF-α. However, the pro-inflammatory response to LPS in HT-29 cells was not inhibited by the probiotic DNA[56]. This rules out, to a certain degree, that the effects observed in the presented experiments stem from DNA leaking from lysed bifidobacteria. Furthermore, it suggests that the mechanisms by which whole cells of bifidobacteria inhibit inflammatory responses are distinct from those described for their DNA.
Although anti-inflammatory activity of probiotics has been reported previously, to our knowledge, this is the first report on a specific inhibitory effect of bifidobacteria on LPS-induced inflammatory events in IECs, suggesting a role for bifidobacteria in down-modulation of pro-inflammatory cytokines[57]. Blocking of LPS-induced NF-κB activation by bifidobacteria could, at least in part, explain their positive effects on chronic intestinal inflammation in vivo through prevention of further amplification of the pro-inflammatory signal after exposure to LPS. Our results suggest that the capacity to inhibit LPS-induced NF-κB activation is strain-dependent. Especially, the strains of B. bifidum are promising candidates for probiotic intervention in inflammatory disorders of the gastrointestinal tract.
ACKNOWLEDGMENTS
C.U.R was funded by a Nestlé PhD fellowship. B. breve MB226 was generously provided by D. Matteuzzi. P. Serrant kindly provided the pool human milk.
Footnotes
S- Editor Pan BR L- Editor Lakatos PL E- Editor Ma WH
References
- 1.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 2.Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- 3.Contractor NV, Bassiri H, Reya T, Park AY, Baumgart DC, Wasik MA, Emerson SG, Carding SR. Lymphoid hyperplasia, autoimmunity, and compromised intestinal intraepithelial lymphocyte development in colitis-free gnotobiotic IL-2-deficient mice. J Immunol. 1998;160:385–394. [PubMed] [Google Scholar]
- 4.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 6.Duchmann R, Kaiser I, Hermann E, Mayet W, Ewe K, Meyer zum Büschenfelde KH. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD) Clin Exp Immunol. 1995;102:448–455. doi: 10.1111/j.1365-2249.1995.tb03836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duchmann R, Schmitt E, Knolle P, Meyer zum Büschenfelde KH, Neurath M. Tolerance towards resident intestinal flora in mice is abrogated in experimental colitis and restored by treatment with interleukin-10 or antibodies to interleukin-12. Eur J Immunol. 1996;26:934–938. doi: 10.1002/eji.1830260432. [DOI] [PubMed] [Google Scholar]
- 8.Neurath MF, Fuss I, Kelsall BL, Presky DH, Waegell W, Strober W. Experimental granulomatous colitis in mice is abrogated by induction of TGF-beta-mediated oral tolerance. J Exp Med. 1996;183:2605–2616. doi: 10.1084/jem.183.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 10.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 11.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 12.Neurath MF, Becker C, Barbulescu K. Role of NF-kappaB in immune and inflammatory responses in the gut. Gut. 1998;43:856–860. doi: 10.1136/gut.43.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 14.Wright SD, Tobias PS, Ulevitch RJ, Ramos RA. Lipopolysaccharide (LPS) binding protein opsonizes LPS-bearing particles for recognition by a novel receptor on macrophages. J Exp Med. 1989;170:1231–1241. doi: 10.1084/jem.170.4.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, Tobias PS, Ulevitch RJ. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 16.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- 18.Pugin J, Schürer-Maly CC, Leturcq D, Moriarty A, Ulevitch RJ, Tobias PS. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci U S A. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 21.Furrie E, Macfarlane S, Thomson G, Macfarlane GT. Toll-like receptors-2, -3 and -4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology. 2005;115:565–574. doi: 10.1111/j.1365-2567.2005.02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meijssen MA, Brandwein SL, Reinecker HC, Bhan AK, Podolsky DK. Alteration of gene expression by intestinal epithelial cells precedes colitis in interleukin-2-deficient mice. Am J Physiol. 1998;274:G472–G479. doi: 10.1152/ajpgi.1998.274.3.G472. [DOI] [PubMed] [Google Scholar]
- 23.Ortega-Cava CF, Ishihara S, Rumi MA, Kawashima K, Ishimura N, Kazumori H, Udagawa J, Kadowaki Y, Kinoshita Y. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J Immunol. 2003;170:3977–3985. doi: 10.4049/jimmunol.170.8.3977. [DOI] [PubMed] [Google Scholar]
- 24.Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Duerig N, Epplen JT. A polymorphism in the CD14 gene is associated with Crohn disease. Scand J Gastroenterol. 2002;37:189–191. doi: 10.1080/003655202753416867. [DOI] [PubMed] [Google Scholar]
- 25.Obana N, Takahashi S, Kinouchi Y, Negoro K, Takagi S, Hiwatashi N, Shimosegawa T. Ulcerative colitis is associated with a promoter polymorphism of lipopolysaccharide receptor gene, CD14. Scand J Gastroenterol. 2002;37:699–704. doi: 10.1080/00365520212504. [DOI] [PubMed] [Google Scholar]
- 26.Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Devière J, et al. Deficient host-bacteria interactions in inflammatory bowel disease The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut. 2004;53:987–992. doi: 10.1136/gut.2003.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, Archimandritis A, Ikonomopoulos J, Gorgoulis VG. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol. 2005;11:681–685. doi: 10.3748/wjg.v11.i5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Harmsen HJ, Raangs GC, He T, Degener JE, Welling GW. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol. 2002;68:2982–2990. doi: 10.1128/AEM.68.6.2982-2990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 31.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernet MF, Brassart D, Neeser JR, Servin AL. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl Environ Microbiol. 1993;59:4121–4128. doi: 10.1128/aem.59.12.4121-4128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–1049. doi: 10.1016/s0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- 34.Reddy BS. Possible mechanisms by which pro- and prebiotics influence colon carcinogenesis and tumor growth. J Nutr. 1999;129:1478S–1482S. doi: 10.1093/jn/129.7.1478S. [DOI] [PubMed] [Google Scholar]
- 35.Yildirim Z, Winters DK, Johnson MG. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J Appl Microbiol. 1999;86:45–54. doi: 10.1046/j.1365-2672.1999.00629.x. [DOI] [PubMed] [Google Scholar]
- 36.Brady LJ, Gallaher DD, Busta FF. The role of probiotic cultures in the prevention of colon cancer. J Nutr. 2000;130:410S–414S. doi: 10.1093/jn/130.2.410S. [DOI] [PubMed] [Google Scholar]
- 37.Asahara T, Nomoto K, Shimizu K, Watanuki M, Tanaka R. Increased resistance of mice to Salmonella enterica serovar Typhimurium infection by synbiotic administration of Bifidobacteria and transgalactosylated oligosaccharides. J Appl Microbiol. 2001;91:985–996. doi: 10.1046/j.1365-2672.2001.01461.x. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy J, O'Mahony L, O'Callaghan L, Sheil B, Vaughan EE, Fitzsimons N, Fitzgibbon J, O'Sullivan GC, Kiely B, Collins JK, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–980. doi: 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asahara T, Shimizu K, Nomoto K, Hamabata T, Ozawa A, Takeda Y. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157: H7. Infect Immun. 2004;72:2240–2247. doi: 10.1128/IAI.72.4.2240-2247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O'Sullivan GC, Kiely B, Collins JK, Shanahan F, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 42.Simmering R, Blaut M. Pro- and prebiotics--the tasty guardian angels. Appl Microbiol Biotechnol. 2001;55:19–28. doi: 10.1007/s002530000512. [DOI] [PubMed] [Google Scholar]
- 43.Gionchetti P, Rizzello F, Venturi A, Brigidi P, Matteuzzi D, Bazzocchi G, Poggioli G, Miglioli M, Campieri M. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305–309. doi: 10.1053/gast.2000.9370. [DOI] [PubMed] [Google Scholar]
- 44.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 45.Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 46.Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, De Simone C, Sartor RB. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100:1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 47.Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'neil DA, Macfarlane GT. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon KY, Hahn BS, Lee J, Kim YS. A cell-based assay system for monitoring NF-kappaB activity in human HaCat transfectant cells. Anal Biochem. 2001;292:17–21. doi: 10.1006/abio.2001.5059. [DOI] [PubMed] [Google Scholar]
- 49.Petrof EO, Kojima K, Ropeleski MJ, Musch MW, Tao Y, De Simone C, Chang EB. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127:1474–1487. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C. Review article: bifidobacteria as probiotic agents -- physiological effects and clinical benefits. Aliment Pharmacol Ther. 2005;22:495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 51.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 52.Labéta MO, Vidal K, Nores JE, Arias M, Vita N, Morgan BP, Guillemot JC, Loyaux D, Ferrara P, Schmid D, et al. Innate recognition of bacteria in human milk is mediated by a milk-derived highly expressed pattern recognition receptor, soluble CD14. J Exp Med. 2000;191:1807–1812. doi: 10.1084/jem.191.10.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal K, Labéta MO, Schiffrin EJ, Donnet-Hughes A. Soluble CD14 in human breast milk and its role in innate immune responses. Acta Odontol Scand. 2001;59:330–334. doi: 10.1080/000163501750541219. [DOI] [PubMed] [Google Scholar]
- 54.Vidal K, Donnet-Hughes A, Granato D. Lipoteichoic acids from Lactobacillus johnsonii strain La1 and Lactobacillus acidophilus strain La10 antagonize the responsiveness of human intestinal epithelial HT29 cells to lipopolysaccharide and gram-negative bacteria. Infect Immun. 2002;70:2057–2064. doi: 10.1128/IAI.70.4.2057-2064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci U S A. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jijon H, Backer J, Diaz H, Yeung H, Thiel D, McKaigney C, De Simone C, Madsen K. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology. 2004;126:1358–1373. doi: 10.1053/j.gastro.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Staudt C. Wechselwirkungen von bifidobakterien mit darmepithelzellen und mit extrazellulären matrix- und plasmaproteinen[Thesis] German: University of Ulm; 2002. [Google Scholar]


