Abstract
AIM: To investigate the presence of Helicobacter species by nested PCR of 16S rRNA genes followed by the presence of Helicobacter pylori(H pylori)16S rRNA, ureA, cagA genes in bile obtained at endoscopic retrograde cholangio-pancreatography (ERCP) from 60 Indian subjects.
METHODS: Sixty bile samples were obtained from patients diagnosed with various hepato-biliary diseases and control subjects at ERCP. PCR analysis was carried out using primers for Helicobacter genus 16S rRNA gene and H pylori (16S rRNA, ureA and cagA) genes. Gastric H pylori status was also assessed from biopsies obtained at endoscopy from patients with various hepato-biliary diseases and controls. The control group mainly consisted of subjects with gastric disorders. Sequencing analysis was performed to confirm that PCR products with 16S rRNA and cagA primers were derived from H pylori.
RESULTS No Helicobacters were grown in culture from the bile samples. Helicobacter DNA was detected in bile of 96.7% and 6.6% of groups I and II respectively. Ten from group I were positive for 16S rRNA and ureA and 9 were positive for cagA gene. In contrast of the 2 from the control, 1 amplified with 16S rRNA, ureA and cagA primers used. The sequences of the 16S rRNA genes and cagA were 99% similar to Helicobacter pylori.
CONCLUSION: Helicobacters are associated with the pathogenesis of various hepato-biliary disorders.
Keywords: Helicobacter pylori; Bile; Hepato-biliary diseases; PCR, Sequence analysis
INTRODUCTION
The re-discovery of Helicobacter pylori (H pylori) as a curved bacterium in the stomach by histological examination of gastric biopsies[1,2] and its subsequent first isolation by Warren & Marshall in 1983 have probably etched new avenues in the management of various gastro-duodenal disorders. Since its discovery, this microaerophilic Gram negative pathogen has been linked to various gastric pathologies including gastric carcinoma and mucosa associated lymphoid tissue (MALT) lymphoma[3,4]. Numerous other Helicobacter species along with Helicobacter pylori have subsequently been isolated from sites other than the stomach, including oral cavity, liver and biliary tree of animals and humans[5-7]. Recent studies have implicated the association between Helicobacter infections with certain diseases of the liver of some animal species such as H canis in dogs[8] and H hepaticus & H bilis in mice[9-11].
In humans, other Helicobacter species including H pylori DNA has been detected in the liver of patients suffering from cholestatic diseases[12,13]. In one study, a high frequency of H pylori and H pullorum sequences were detected by PCR, in the liver of patients with cirrhosis and superimposed hepatocellular carcinoma[14]. More recently, a study by Pellicano et al suggests that presence of Helicobacter spp in liver samples could possibly serve as a co-factor in the development of end-stage of liver disease in humans[15]. These concerns have spurred considerable interest in determining the mechanisms by which these extra cellular bacteria and the associated inflammatory response endorse hepatic and biliary disease.
Therefore, we investigated the presence of Helicobacter spp genomes in the bile specimens obtained from patients with different hepato-biliary diseases and among control group (without any hepatic and biliary disease but with different gastric disorders) at endoscopic retrograde cholangio-pancreatography (ERCP). We also evaluated the association between the presence of Helicobacter species with various hepato-biliary disorders.
MATERIALS AND METHODS
Subjects
The study population consisted of 60 subjects categorized into two groups (thirty in each) viz., Group I (those with hepato-biliary disorders) and Group II (those with no pathologically proven liver or biliary disease but with different gastric disorders, served as control). Patients of both sexes and age range: 23-68 years, average age: 48.1 years were included in the study. Subjects from either group underwent ERCP followed by upper gastrointestinal endoscopy at the Department of Gastroenterology, Deccan College of Medical Sciences, Hyderabad. The study protocol was approved by the Institutional Review Board (IRB) and Institutional Ethical Committee (IEC). Approval was obtained from IEC for the initiation of the study. Informed consent was obtained from all the patients before their enrolling in the study according to the Helsinki Declaration. None of the patients included for the study were on antibiotics prior to ERCP or endoscopy.
Patients’ details and diagnoses were recorded and 5-10 mL bile samples were collected in glycerol by aspiration during the ERCP procedure and 3 gastric biopsies were collected during endoscopy from the same patient: one in urea solution for rapid urease test (RUT), one in brucella broth supplemented with fetal calf serum (FCS) for culture and 1 in phosphate buffered saline (PBS) for DNA isolation. Aspirated bile samples and the biopsy collected in PBS were stored at -80 oC until DNA was isolated.
Helicobacter culture
The gastric biopsy collected in supplemented brucella broth was immediately transported to the laboratory and streaked on the chocolate brucella agar supplemented with 70 mL/L sheep blood and 6 mg/L-vancomycin, 2 mg/L-amphoteracin-B and polymixin-B 2500/L (Sigma Chemicals, USA) and incubated at 37 oC in microaerobic conditions. A small aliquot (about 0.5 mL) of the collected bile sample was also instantaneously homogenized within half an hour of collection in 0.5 to 1 mL brucella broth (Becton and Dickinson, USA) containing 50 mL/L fetal calf serum (Gibco BRL, Germany) and streaked over the same medium for primary isolation of H pylori and incubated as described above. Approximately 50 μL of the sample was plated onto each plate and the remaining sample was used for DNA isolation.
DNA extraction
The genomic DNA from the gastric tissue, isolated culture and bile samples was isolated as per the standard protocol previously described[16]. In case of bile sample, briefly 450 μL of the sample was diluted with equal volume of PBS and centrifuged at 15 000 g for 20 min. The supernatant was discarded and the pellet was again subsequently mixed with 250 μL of the PBS and DNA isolated by modified cetyl trimethyl ammonium bromide (CTAB) method. The DNA was extracted and preserved at -20oC until amplification was performed. Appropriate care was taken during extraction to remove the PCR inhibiting substances present in the bile[17]. Briefly as Helicobacter DNA was isolated from an unusual source, there is possibility of existence of specific inhibitors and competing substrates. For such situations, dilution of inhibited samples provides a rapid and straightforward way of permitting amplification. This dilution exploits the sensitivity of PCR by reducing the concentration of inhibitors relative to target DNA.
PCR amplification
Amplification was performed as per standard protocol described previously[13] with minor modifications. All primers were synthesized at Bioserve Biotechnologies Pvt Ltd, Hyderabad, India. Amplification was performed in a PTC 100 thermocycler (M J Research Inc. Water town, USA).
PCR amplification with Helicobacter genus-specific primers
Nested PCR was carried out using two oligonucleotide pairs previously reported by Pellicano et al[15] and designated as Heli in Table 1. The primers (Helinest-S & R, Heli-S & R) used in our study were reported to amplify 26 species of Helicobacter genus[15]. At each amplification, H pylori DNA was used as a positive control, while water instead of DNA served as a negative control.
Table 1.
List of primers used for the study
| Primer | Sequence(5’→3’) | Productsize |
| (bp) | ||
| Heli-nestS | 5’ATTAGTGGCGCACGGGTGAGTAA3’ | 1 300 |
| Heli-nestR | 5’TTTAGCATCCCGACTTAAGGC3’ | |
| Heli-S | 5’GAACCTTACCTAGGCTTGACATTG3’ | 480 |
| Heli-R | 5’GGTGAGTACAAGACCCGGGAA3’ | |
| 16S-rRNA.F | 5’TAAGAGATCAGCCTATATGTCC3’ | 534 |
| 16S-rRNA.R | 5’TCCCACGCTTTAAGCGCAAT3’ | |
| UreA-S | 5’GCCAATGGTAAATTAGTT3’ | 411 |
| UreA-R | 5’CTCCTTAATTGTTTTTAC3’ | |
| CagA-S | 5’CCATGAATTTTTGATCCGTTCGG3’ | 349 |
| CagA-R | 5’GATAACAGGCAAGCTTTTGAGAGGGA3’ | |
First amplification
Amplification was carried out in a total volume of 20 μL containing 0.5 μL DNA, PCR buffer (1×), 200 μmol/L dNTPs, 1.5 mmol/L Mg 2+, 0.2 μmol/L primers (Heli-nest-S and Heli-nest-R), 1U Taq DNA polymerase (Invitrogen Life Technologies, Germany). Amplification conditions were optimized and are enlisted in Table 2. A sample was scored positive if an amplification product of 1 300 bp could be resolved after electrophoresis on 15 g/L agarose gel.
Table 2.
Conditions of polymerase chain reactions used in the study
| Target gene | Initial denaturation step | Temperature o denaturation, annealing and | Cycle | Final extension Step extension |
| Helicobacter | 94 °C for 5 min | 94°C for 30 s | ||
| Spp,16S rRNA | 55°C for 30 s | 35 | 72°C for 7 min | |
| 72°C for 1.5 min | ||||
| Second | 94°C for 5 min | 94°C for 30 s | ||
| amplification | 60°C for 30 s | 35 | 72°C for 7 min | |
| step | 72°C for 30 s | |||
| Helicobacter | 95°C f | 94°C for 30 s | ||
| pylori, 16S rRNA | 56°C for 30 s | 40 | 72°C for 5 min | |
| 2°C for 1 min | ||||
| ureA | 95°C for 5 min | 94°C for 30 s | 35 | 72°C for 6 min |
| 52°C for 30 s | 72°C for 1 min | |||
| cagA | 95°C for 5 min | 94°C for 1 min | ||
| 52°C for 1 min | 35 | 72°C for 10 min | ||
| 72°C for 2 min |
Second amplification
One microliter of amplicon from the first amplification step was used with primers Heli-S and Heli-R (Table 1) and amplification was repeated with minor alterations (Table 2). The expected product size of the amplicon was 480 bp.
Specificity test of genus specific primers
The specificity of genus specific primers viz., Helinest-S, Helinest-R, Heli-S and Heli-R as well as primers specific to H pylori was assessed by using 6 bacterial strains. This included two different Helicobacter pylori strains along with 4 other enteric bacteria commonly residing the stomach.
H pylori specific 16S rRNA PCR
The samples positive with Helicobacter genus PCR were further analyzed for the presence of H pylori DNA in the culture, biopsy and bile samples by targeting the 16S rRNA gene using primers enlisted in Table 1. Amplification was carried out as per the mentioned protocol (Table 2). The amplification product size was 534 bp typical of H pylori.
Amplification of ureA gene
Samples positive for H pylori were subsequently analyzed with a different set of primers designated ureA, in Table 1. The sense and anti-sense primers of this gene were used for PCR as per the mentioned protocol in Table 2. The amplification product size was 411 bp typical of H pylori.
Amplification of cagA gene
Samples positive for H pylori were subsequently screened for the presence of cagA gene using specific primers (Table 1). Amplification was carried out as per the program given in Table 2 with minor alterations increasing the annealing time to 1 min and extension time to 2 min. The expected product size of the primers used was 349 bp.
Sequencing of the 16S rRNA products and cagA amplification product
The H pylori positive DNA fragments from the bile samples were sequenced. Comparison of DNA sequences of the genomic 16S rRNA amplicons and cagA amplicons with those of H pylori was performed using sequence alignment with the BLAST programme. The presence of H pylori sequence was thus confirmed in these bile samples.
Statistical analysis
Helicobacter genus positivity and negativity was compared using the Fisher’s exact test. P < 0.05 was considered as significant.
RESULTS
Culture
Of the 60 gastric biopsies streaked, colonies could be isolated from 54(90%) subjects (26 from group I and 28 from group II). No Helicobacter pylori colonies could be grown of the 60 bile samples streaked, even after prolonged incubation for up to 2 wk under microaerophilic conditions.
Specificity test
Helicobacter genus specificity was tested using a panel of H pylori strains by PCR with specific primer sets used for this study. All the H pylori DNA gave a positive amplification with the expected product size. The non-Helicobacter species DNA extracts did not yield any result with the oligonucleotide primers used.
PCR amplification with Helicobacter genus-specific primers
Helicobacter DNA was detected by nested-PCR in 29(96.7%) of the 30 bile samples collected from group I patients and 2(6.6%) of the subjects from group II sub-group respectively. All the 29 samples from group I and 2 from group II amplified at both first and second amplification reactions. The amplification product sizes of both the PCR are shown in Figures 1A and 1B respectively. DNA isolated from 60 biopsy and 54 cultures, gave a positive amplification with the expected product size.
Figure 1.
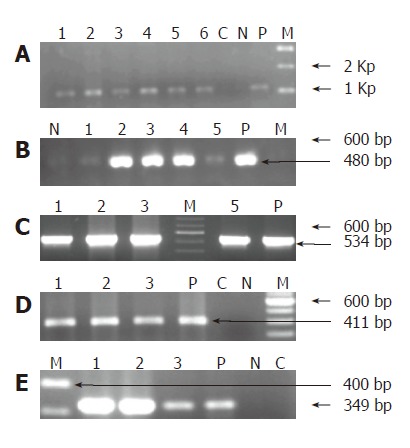
A schematic representation of the PCR products of the Helicobacter spp DNA. A: Gel image showing first amplification products of 16S rRNA PCR with Helicobacter genus specific primers at 1 300 bp. Lanes 1 to 6 represent Helicobacter DNA isolated from bile samples, ‘P’ represents positive control andLanes C, N and M represents negative control, reaction negative control and 1 Kb molecular weight marker. B: Second amplification products of 16S rRNA PCR with Helicobacter genus specific primers at 480 bp. Lanes 1 to 5 represent bile DNA, ‘P’ represents positive control. Lanes N and M represent reaction negative control and 100 bp molecular weight marker ladder respectively. C: Gel picture showing 16S rRNA amplification products specific to H pylori of bile samples at 534 bp. Lanes 1, 2, 3 and 5 represent bile DNA samples while Lanes M and P represent 100 bp ladder and positive control respectively. D: ureA amplification products at 411 bp. Lanes 1 to 3 represent bile DNA, Lane ‘P’ represents positive control and Lanes C, N and M represent negative control, reaction negative control and 100bp molecular weight ladder respectively. E: Gel picture illustrating the cagA amplification products at 349 bp. Lanes 1, 2 and 3 represent bile DNA while Lanes M, P, N and C represent 100bp molecular weight ladder, positive and reaction negative control, negative control respectively.
Screening for H pylori DNA in bile
Of the 29 Helicobacter genus positive samples from group I subjects, 10(33.3%) bile samples were amplified with the H pylori specific 16S rRNA primers, yielding a product size of 534 bp on electrophoresis (Figure 1C). While of the 2 Helicobacter genus positive from group II, only 1 was amplified with the H pylori 16S rRNA primers used (Table 3).
Table 3.
Details of Helicobacter genus, H pylori 16S rRNA, ureA, cagA positivity in the study subjects
| No | Category (n=60) | Helicobacter Genus positivity n(%) | Hpylori16S rRNA positive n(%) | Ure A positive n(%) | Cag A positive n(%) |
| 1 | Group I | 29(96.7) | 10(33.3) | 10(33.3) | 09(30) |
| 2 | Group II | 02(6.6) | 01(3.3) | 01(3.3) | 01(3.3) |
Confirmation of gastric H pylori colonization
Screening of the 60 biopsy DNA and 54 culture DNA from both the study groups gave positive amplification with the specified primers used, yielding a product size of 534 bp.
Analysis of ureA and cagA gene
Of the 29 subjects analyzed for the presence of ureA and cagA sequences from group I sub-group, we found that 10 (33.5%) were amplified with ureA gene while 9 (30%) amplified with the cagA primer. On the other hand, 1 sample which was amplified for H pylori 16S rRNA gene in group II, also gave positive amplification for ureA and cagA respectively (Table 3). The amplified products of these genes are represented in Figure 1D, E.
Sequence analysis of PCR product from bile DNA with 16S rRNA and cagA primers
To confirm that the PCR product obtained with the 16S rRNA and cagA primers belonged to H pylori, we examined the sequence of the PCR product in 4 bile samples (3 from Group I and 1 from Group II). The nucleotide sequence of the amplified products shared 99% identity with the 16S rRNA and cagA gene of H pylori respectively.
DISCUSSION
The presence of Helicobacter species DNA in the bile samples of patients with different hepato-biliary diseases is interesting since some reports in the past have suggested a positive association of Helicobacter and the evolution of liver diseases[9,15,18,19]. Recent studies on Helicobacter spp in different diseases of liver and bile ducts have shown that Helicobacter can be detected not only in the extremely hostile milieu of the stomach but also in human bile[20,19]. Although none of the previous studies including the present study have not been able to isolate H pylori in vitro, it has been proven that some of Helicobacter species live in the gall bladder. In a recent study[15], by PCR and subsequent sequencing of the 16S rDNA and amplification of cagA gene, Helicobacter spp was detected in 17 of 20 patients operated for hepatocellular carcinoma (HCC). The same study reported the presence of a 290 bp product of 128 KDa CagA protein specific only for type I H pylori. Further, it has been shown that several Helicobacter spp. secrete a liver specific toxin that causes hepatocyte necrosis in cell culture and might also be involved in damaging liver parenchyma in vivo[15]. In contrast, other authors did not detect any Helicobacter or H pylori DNA in the patients with similar diseases[21,28]. These observations impelled us to explore a possible association of Helicobacter and hepato-biliary disease among Indian patients.
In the present study, which comprised of two groups (I & II), of the 30 subjects from group I with various hepato-biliary ailments, we could detect Helicobacter genus specific 16S rRNA sequence in 29 bile samples by nested PCR and only 1 sample did not give any amplification with the specific primers used. By contrast, in Group II subjects, i.e. those with no significant hepato-biliary disease but with various gastric disorders (Control subjects) only 2 subjects gave positive amplification with the Helicobacter genus specific primer used yielding the desired fragment. We carried out 16S rRNA amplification specific to H pylori on 29 subjects positive from group I and 2 from group II for Helicobacter genus, followed by subsequent sequencing of the amplified products to confirm the presence of H pylori DNA in the bile samples. We found that 33.3% samples from group I and 3.33% from group II were amplified giving a product of 534 bp (Figure 1C). Further, we also investigated the presence of ureA and cagA gene in all the bile specimens followed by sequencing of the 16S rRNA and cagA amplified products of 4 bile samples (3 from group I and 1 from group II). Sequence comparison of the sequenced samples confirmed the presence of H pylori DNA sequence in the bile samples.
The usage of less invasive ERCP procedure to obtain bile is an adequate method for this purpose compared with other invasive approaches currently in practice. This procedure also avoids contamination with H pylori colonizing the stomach, as the sampling devices are inserted inside the endoscope and hence never traverse the stomach[28].
This is the first Indian study to simultaneously investigate the presence of Helicobacter DNA in bile specimens and gastric tissues and underscore the association of Helicobacter in bile obtained from patients with various hepato-biliary disorders. We detected Helicobacter DNA by nested PCR using two sets of primer. Further, our study also successfully demonstrated the presence of ureA and virulence genes such as cagA specific to H pylori in the DNA isolated from bile samples. As evident from the results, 33.3% carried ureA gene whereas 30% were amplified with the primers used for cagA detection (Table 3). Of the 9 positive for H pylori cagA gene, 5 belonged to cholangio-carcinoma, 2 belonged to common bile duct stones and the remaining subjects had pancreatico-biliary malignancies (Data not shown). In addition, we also found that among 10 subjects positive for H pylori in group I, ureA and cagA were simultaneously detected in 9 subjects, 1 subject gave amplification only for ureA gene and the only sample which was amplified with the 16S rRNA specific primer of H pylori from group II was found to possess both genes respectively (data not shown). The only subject whose bile sample gave positive results with 16S rRNA, ureA and cagA primers in group II was found to suffer from antral gastritis endoscopically and was co-incidentally positive for H pylori by culture and PCR of both the biopsy and culture DNA (data not shown). Every possible precautionary measure was taken to assure that laboratory contamination did not account for the positive amplification results such as diluting the bile with sterile distilled water, centrifuging the samples at 15 000 × g for 20 min, and the supernatant being discarded, thus by concentrating the bacterial cells in the pellet, enabling removal of some of the inhibitors predominantly present in the bile. Collection of bile in glycerol and immediate plating nevertheless did not prove to be successful for in vitro isolation of Helicobacters from any of the bile samples as we could not get any growth even after extended incubations for up to 15 d.
The present study also investigated the gastric H pylori status of the patients enrolled in both the sub-groups to see if at all the gastric H pylori status had any impact on the etiology of the hepato-biliary diseases. Unfortunately from the results we could neither associate the severity of the hepato-biliary disease with that of the gastric diseases nor could we link the gastric H pylori status and the detection of H pylori DNA in bile of the hepato-biliary diseases. Even though 29 subjects from group I showed the presence of Helicobacter species DNA, only 10 showed the presence of H pylori DNA in the bile thus signifying the possibility of the presence of the other bile-resistant Helicobacters that normally reside in the liver and biliary tract. Besides the above results obtained in this study, we noticed that patients with various hepato-biliary disorders had much greater probability of positive Helicobacter species DNA compared to those with no pathologically proven liver and biliary diseases. Why these intestinal Helicobacters have been identified in Chilean patients[26] and H pylori has been identified in patients in other geographical regions, as we observed in this study warrant further investigations. In fact, the typical finding of the present study, correlates well with those of Kuroki et al[22], who recently demonstrated that the level of epithelium proliferation was higher in Helicobacter-positive biliary epithelium than in bacterium-negative epithelium.
The results of our study also suggest that besides Helicobacter spp sequences, even sequences pertaining to the virulence genes of H pylori are consistently found at a high frequency in bile samples and that they may be a significant cause of biliary diseases. Several hypotheses have been proposed[23-25] , by which these Helicobacters find their way into the liver and bile duct. But these hypotheses entail strong evidences to underline the precise mechanism by which the Helicobacter species anchor the liver and aggravate the clinical outcome.
The findings of this study are in concert to those obtained by Fox et al[26] and Linn et al[18] who reported Helicobacter spp in gall bladder tissue from Chileans with chronic cholecystitis but different from those of Fallone et al[28] and Mendez-Sanchez et al[21] who were not able to detect Helicobacter sequences in the bile samples of North Americans and Mexicans respectively. The raison d’être for these discordant results could be the regional variations in the distribution of bile-resistant Helicobacter species[20]. The other alternative reason for the inconsistent results in their studies could be the methodology used, the selection of primers used, as both the studies had used a single primer set for the amplification of the conserved 16S rRNA gene. In contrast, our study used nested PCR for 16S rRNA amplification of Helicobacter genus, which according to Stark et al[27] is 104 times more sensitive than a 1-step PCR. Though we used only one set of oligonucleotide primers each for amplifying ureA and cagA, the specificity and sensitivity of the selected primers were previously determined during our routine screening. The primers were found to possess the sensitivity and specificity of 90% and 95% for ureA and 89% and 85% for cagA, particularly in this geographical area.
In conclusion, this study demonstrated that Helicobacter spp DNA can be detected in bile by PCR and that gastric presence of H pylori in patients with proven hepato-biliary disease had no clinical correlation with the hepatic and biliary disease. However, like other previous studies, we were unable to isolate the bacterium in culture. There are different reasons to justify this finding. Firstly, this could be due to the bacterial conversion from viable helicals to non-viable coccoids in an adverse bile rich environment. It is also possible that the number of bacterium is very few and that they may have been partially inhibited by unfavorable environment that exists in the biliary milieu. In addition, our study also confirmed by DNA sequencing that sequences specific to H pylori (16S rRNA, cagA) can be found at a high frequency in the bile samples, thus instilling strong evidence that presence of Helicobacter spp may in someway aggravate the etio-pathogenesis of hepato-biliary diseases. However, mere detection of Helicobacter DNA from patients with different hepatic and biliary disease does not confirm the precise role played by these organisms. Further, future studies unraveling the molecular mechanisms by which these Helicobacter members contribute to the clinical outcome of hepato-biliary disorders would be helpful to assess the true impact of enterohepatic Helicobacters and its metabolites in the genesis of biliary diseases.
Footnotes
Supported by the Department of Biotechnology, Government of India
Co-correspondence: Aleem A Khan
S- Editor Guo SY L- Editor Zhu LH E- Editor Bai SH
References
- 1.Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–1275. [PubMed] [Google Scholar]
- 2.Buckley MJ, O'Morain CA. Helicobacter biology--discovery. Br Med Bull. 1998;54:7–16. doi: 10.1093/oxfordjournals.bmb.a011681. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 4.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338:1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- 5.Melito PL, Munro C, Chipman PR, Woodward DL, Booth TF, Rodgers FG. Helicobacter winghamensis sp. nov., a novel Helicobacter sp. isolated from patients with gastroenteritis. J Clin Microbiol. 2001;39:2412–2417. doi: 10.1128/JCM.39.7.2412-2417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiwari SK, Khan AA, Ahmed KS, Ali SM, Ahmed I, Habeeb A, Kauser F, Hussain MA, Ahmed N, Habibullah CM. Polymerase chain reaction based analysis of the cytotoxin associated gene pathogenicity island of Helicobacter pylori from saliva: an approach for rapid molecular genotyping in relation to disease status. J Gastroenterol Hepatol. 2005;20:1560–1566. doi: 10.1111/j.1440-1746.2005.03955.x. [DOI] [PubMed] [Google Scholar]
- 7.Tiwari SK, Khan AA, Ahmed KS, Ahmed I, Kauser F, Hussain MA, Ali SM, Alvi A, Habeeb A, Abid Z, et al. Rapid diagnosis of Helicobacter pylori infection in dyspeptic patients using salivary secretion: a non-invasive approach. Singapore Med J. 2005;46:224–228. [PubMed] [Google Scholar]
- 8.Fox JG, Drolet R, Higgins R, Messier S, Yan L, Coleman BE, Paster BJ, Dewhirst FE. Helicobacter canis isolated from a dog liver with multifocal necrotizing hepatitis. J Clin Microbiol. 1996;34:2479–2482. doi: 10.1128/jcm.34.10.2479-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ, Gorelick PL, Ward JM. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox JG, Li X, Yan L, Cahill RJ, Hurley R, Lewis R, Murphy JC. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of helicobacter-induced carcinogenesis. Infect Immun. 1996;64:1548–1558. doi: 10.1128/iai.64.5.1548-1558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox JG, Yan LL, Dewhirst FE, Paster BJ, Shames B, Murphy JC, Hayward A, Belcher JC, Mendes EN. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avenaud P, Marais A, Monteiro L, Le Bail B, Bioulac Sage P, Balabaud C, Megraud F. Detection of Helicobacter species in the liver of patients with and without primary liver carcinoma. Cancer. 2000;89:1431–1439. [PubMed] [Google Scholar]
- 13.Nilsson HO, Taneera J, Castedal M, Glatz E, Olsson R, Wadström T. Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38:1072–1076. doi: 10.1128/jcm.38.3.1072-1076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponzetto A, Pellicano R, Leone N, Cutufia MA, Turrini F, Grigioni WF, D'Errico A, Mortimer P, Rizzetto M, Silengo L. Helicobacter infection and cirrhosis in hepatitis C virus carriage: is it an innocent bystander or a troublemaker. Med Hypotheses. 2000;54:275–277. doi: 10.1054/mehy.1999.0987. [DOI] [PubMed] [Google Scholar]
- 15.Pellicano R, Mazzaferro V, Grigioni WF, Cutufia MA, Fagoonee S, Silengo L, Rizzetto M, Ponzetto A. Helicobacter species sequences in liver samples from patients with and without hepatocellular carcinoma. World J Gastroenterol. 2004;10:598–601. doi: 10.3748/wjg.v10.i4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clayton CL, Mobley HLT. Methods in molecular medicine, Helicobacter pylori protocols, (3rdeds. ) Totowa: Humana Press Inc; 2002. p. 33. [Google Scholar]
- 17.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin TT, Yeh CT, Wu CS, Liaw YF. Detection and partial sequence analysis of Helicobacter pylori DNA in the bile samples. Dig Dis Sci. 1995;40:2214–2219. doi: 10.1007/BF02209009. [DOI] [PubMed] [Google Scholar]
- 19.Leong RW, Sung JJ. Review article: Helicobacter species and hepatobiliary diseases. Aliment Pharmacol Ther. 2002;16:1037–1045. doi: 10.1046/j.1365-2036.2002.01282.x. [DOI] [PubMed] [Google Scholar]
- 20.Bulajic M, Maisonneuve P, Schneider-Brachert W, Müller P, Reischl U, Stimec B, Lehn N, Lowenfels AB, Löhr M. Helicobacter pylori and the risk of benign and malignant biliary tract disease. Cancer. 2002;95:1946–1953. doi: 10.1002/cncr.10893. [DOI] [PubMed] [Google Scholar]
- 21.Méndez-Sánchez N, Pichardo R, González J, Sánchez H, Moreno M, Barquera F, Estevez HO, Uribe M. Lack of association between Helicobacter sp colonization and gallstone disease. J Clin Gastroenterol. 2001;32:138–141. doi: 10.1097/00004836-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Kuroki T, Fukuda K, Yamanouchi K, Kitajima T, Matsuzaki S, Tajima Y, Furui J, Kanematsu T. Helicobacter pylori accelerates the biliary epithelial cell proliferation activity in hepatolithiasis. Hepatogastroenterology. 2002;49:648–651. [PubMed] [Google Scholar]
- 23.Cassell GH. Infectious causes of chronic inflammatory diseases and cancer. Emerg Infect Dis. 1998;4:475–487. doi: 10.3201/eid0403.980339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crabtree J. Cytokine responses to Helicobacter pylori-induced infection. In: Riecken EO, Zeitz M, Stallmach A, Heise W, editors. Malignancy and chronic inflammation in the gastro-intestinal tract: new concepts. Kluwer Academic publishers, Lancaster; 2002. pp. 25–36. [Google Scholar]
- 25.Hornick RB. Enteric fever. In Blaser MJ, Smith PD, Ravdin JI, Greenberg HB and Guerrant RL (eds), Infections of the gastrointestinal tract. New York: Raven Press, Ltd; 1997. pp. 325–332. [Google Scholar]
- 26.Fox JG, Dewhirst FE, Shen Z, Feng Y, Taylor NS, Paster BJ, Ericson RL, Lau CN, Correa P, Araya JC, et al. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 27.Stärk KD, Nicolet J, Frey J. Detection of Mycoplasma hyopneumoniae by air sampling with a nested PCR assay. Appl Environ Microbiol. 1998;64:543–548. doi: 10.1128/aem.64.2.543-548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fallone CA, Tran S, Semret M, Discepola F, Behr M, Barkun AN. Helicobacter DNA in bile: correlation with hepato-biliary diseases. Aliment Pharmacol Ther. 2003;17:453–458. doi: 10.1046/j.1365-2036.2003.01424.x. [DOI] [PubMed] [Google Scholar]


