Abstract
AIM: To characterize the immunogenicity of a hepatitis C virus (HCV) E2 DNA vaccine alone or with a protein vaccine boost in murine and porcine animal models.
METHODS: A DNA vaccine expressing a secreted form of HCV E2 protein was constructed and used to vaccinate mice and piglets with or without boosting with a recombinant E2 protein vaccine formulated with CpG ODN and 10% Emulsigen. The immunogenicity of HCV E2 vaccines was analyzed by ELISA for antibody responses, MTT assay for lymphocyte proliferation, ELISPOT for the number of interferon-γ secreting cells, and cytotoxic T lymphocyte assays.
RESULTS: Intradermal injection of E2 DNA vaccine induced strong Th1-like immune responses in mice. In piglets, E2 DNA vaccine elicited moderate and more balanced immune responses. A DNA vaccine prime and protein boost vaccination strategy induced significantly higher E2-specific antibody levels and shifted the immune response towards Th2-like ones in piglets.
CONCLUSION: A DNA vaccine expressing a secreted form of HCV E2 protein elicited E2-specific immune responses in mice and piglets. Recombinant E2 protein vaccination following DNA immunization significantly increased the antibody response in piglets. These HCV E2 vaccines may represent promising hepatitis C vaccine candidates for further investigations.
Keywords: Hepatitis C virus E2, DNA vaccine, DNA vaccine prime-protein boost
INTRODUCTION
Hepatitis C continues to be a severe health threat to a large population with about 123 million people being affected globally[1]. The etiologic agent, hepatitis C virus (HCV)[2] is able to establish persistent infections in up to 85% of infected individuals with severe clinical consequences[3]. Current therapy with pegylated interferon and ribavirin is only effective in about 50% of the patients[4]. Although vaccines represent one of the most effective means to combat infectious diseases, there is no vaccine available for hepatitis C[5]. Hence, evaluating different vaccination strategies that can induce HCV-specific immunity is critical for the development of effective vaccines to reduce HCV-related mortality and morbidity.
HCV is the only member of the Hepacivirus genus in the Flaviviridae family[6]. The positive-sense, single-stranded RNA genome encodes a polyprotein of about 3100 amino acids in length[7]. Processing of the polyprotein by cellular or viral proteases generates up to 11 viral proteins, including three structural proteins (core, envelope proteins E1 and E2) and six non-structural proteins (NS-2, -3, -4A, -4B, -5A, and -5B). As the major envelope protein in HCV particles[8,9], the E2 protein is likely to be critical for inducing antibody responses against HCV infections. In line with this notion, anti-E2 antibodies have been consistently detected in hepatitis C patients[10-13]. Furthermore, there is evidence to suggest that anti-E2 antibodies can inhibit HCV infections in both in vivo and in vitro settings[14-17]. However, generation of E2-specific antibodies in hepatitis C patients is usually delayed and of low magnitude, which may be one of the reasons for such a high rate of persistent HCV infections[12,13]. These findings indicate that E2-specific antibodies are beneficial and induction of these antibodies should be taken into consideration when designing a vaccine against hepatitis C.
Although the correlations for a successful immune response that can resolve HCV infections have not been well characterized, previous studies suggest a rapid, vigorous, and broadly targeted cell-mediated immune response tends to be associated with HCV clearance[18-22]. In line with this notion, E2-specific cellular immune responses have been detected in hepatitis C patients as demonstrated by E2-specific lymphocyte proliferation, cytotoxic T lymphocyte, and ELISPOT assays[23-25]. More importantly, it has been demonstrated that a stronger E2-specific cell-mediated response is associated with better response to interferon therapy and viral clearance[25]. These findings indicate that it is desirable for a candidate HCV E2 vaccine to induce cell-mediated immune responses.
Delivery of transgenes by plasmid DNA is a novel platform technology for vaccine development. DNA vaccines tend to induce a Th1-biased response in the host[26-30]. Manipulation of protein subcellular localization may enhance antibody responses to the antigen. For instance, directing antigenic expression to secretion pathways by adding a signal peptide sequence may increase B-cell mediated responses[31]. In addition, boosting with a protein subunit vaccine following DNA vaccination is another feasible means for inducing strong antigen-specific humoral responses[32-36].
In this study, a DNA vaccine was designed to induce expression of a secreted form of HCV E2 protein. Immunogenicity studies using inbred mice showed that this HCV E2 DNA vaccine elicited strong Th1-like immune responses. In piglets, the E2 DNA vaccine elicited a moderate and more Th1-Th2 balanced response. E2 protein vaccination after DNA immunization had a more pronounced boosting effect in piglets than in mice by significantly increasing E2-specific antibody response, causing a shift of the immune response towards a Th2-type.
MATERIALS AND METHODS
HCV E2 DNA vaccine construction
To generate a DNA vaccine encoding a secreted form of HCV E2 protein, a portion of the E2 coding sequence (amino acid residues 412-661 of HCV polyprotein) without the hypervariable region 1 (HVR1) and the hydrophobic region was amplified by polymerase chain reaction (PCR). The template was plasmid pDM22, a cDNA clone of the BK isolate (genotype 1b; kindly provided by Dr. A. Takamizawa)[37]. The primers were E2-412-Nhe-Sense (5’-AATTGCTAGCCAGCTTATAAACACCAATGGG-3’, NheI site is underlined) and E2-661-Bgl-AS (5’-AATTAGATCTTCACTCCGGCCTATCCCTGTC-3’, a stop codon TGA was added followed by a BglII site which is underlined). The PCR product was cloned into a plasmid vector pSLIA-tPAs[38] with NheI and BglII (New England Biolabs), allowing the addition of the signal peptide sequence of the tissue plasminogen activator (tPA) to the amino-terminus of the truncated E2 protein. The identity of PCR amplified E2 gene was confirmed by DNA sequencing experiments. Subsequently, the HindIII-BglII fragment containing the tPAs-tE2 fusion gene was cloned into a CpG enriched DNA vaccine vector pBISIA24[39], which contains 24 copies of a Th1-promoting CpG motif (GTCGTT). An unexpected extension of the open reading frame resulted from the NdeI recognition sequence in the multiple cloning site of plasmid pBISIA24 was removed by deleting the small fragment after PstI digestion and subsequent ligation of the large fragment. The final DNA vaccine construct was designated pBISIA24-tPAs-tE2 (Figure 1). Plasmid prepared by endotoxin-free plasmid purification reagents (Qiagen) was used for immunization.
Figure 1.
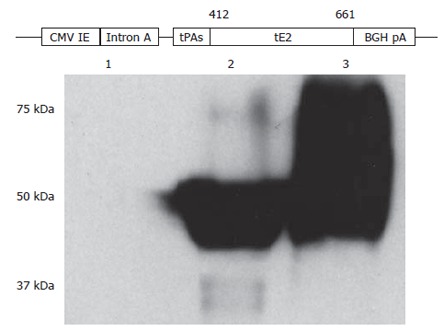
Construction and characterization of HCV E2 DNA vaccine. A secreted form of HCV E2 protein generated by removing the carboxyl-terminal hydrophobic region and replacing the hypervariable region 1 with the signal peptide sequence of the tissue plasminogen activator (tPAs) was cloned into a DNA vaccine vector pBISIA24. The construction of E2 DNA vaccine, pBISIA24-tPAs-tE2, is schematically presented. TE 671 cells were transfected with pBISIA24 (lane 1) or pBISIA24-tPAs-tE2 (lanes 2 and 3). Cell lysates (lanes 1 and 2) and culture medium (lane 3) were analyzed in immunoblotting using an E2-specific antibody.
Transfection and immunoblotting analysis
To determine whether the constructed DNA vaccine plasmid expressed the E2 protein, TE671 cells, a human rhabdomyosarcoma cell line, were transfected with plasmids pBISIA24-tPAs-tE2 and pBISIA24 using the calcium phosphate method[40]. At 24 h after transfection, culture medium was collected and cells were lyzed in a lysis buffer (1% SDS, 10 mmol/L Tris-HCl, pH8.0). The protein concentration was determined by the Bradford assay (Bio-Rad) using bovine serum albumin (BSA) (Sigma) as a standard. Twenty-five micrograms of protein were loaded on a 10% SDS protein gel and transferred to a PVDF membrane (GE Healthcare). The membrane was incubated with a monoclonal antibody against HCV E2 protein (H52, kindly provided by Dr. J. Dubuisson). After incubation with an HRP-conjugated secondary antibody, immuno-reactive protein bands were visualized by the enhanced chemiluminescence system (ECLPlus, GE Healthcare).
Recombinant E2 protein expression and purification
To generate a recombinant E2 protein vaccine, HCV E2 protein was expressed and purified as a fusion protein with a poly-histidine tag. For this purpose, the coding sequence of HCV E2 was amplified by PCR using plasmid pDM22 as the template. The primers for the PCR reaction were E2-Bam-Nhe-Sense (5’-GGGGGGGGATCCGCTAGCGATACCCACGTGACAGGGG-3’, BamHI and NheI sites are underlined) and E2-746-Bgl-AS (5’-GGGGGGAGATCTCAGGCCTCGGCCTGGGCTA-3’, a stop codon TGA was added followed by a BglII site which is underlined). The PCR fragment was cloned into an expression vector pRSETA (Invitrogen) using restriction enzymes BamHI and BglII, allowing the addition of a poly-histidine tag at the amino-terminus of the E2 protein. The resultant plasmid, designated pRSET-E2, was confirmed by restriction analysis and DNA sequencing. The NotI-BglII fragment of plasmid pRSET-E2 was substituted by the NotI-BglII fragment of pBISIA24-tPAs-tE2, generating plasmid pRSET-tE2 encoding a poly-histidine-tagged E2 protein without the carboxyl hydrophobic region. Plasmid pRSET-tE2 was then transformed into E. coli BL21 (pLysS) and expression of E2 protein was induced by isopropylthio-β-galactoside (IPTG, Invitrogen). The induced poly-histidine-tagged tE2 protein was purified under denaturing conditions in the presence of 8 mol/L urea using Ni-NTA agarose (Qiagen) as per manufacturer’s instructions. The urea was removed from the protein preparation after dialysis against phosphate buffered saline (PBS). The level of endotoxin was determined by the Limulus amoebocyte test (QCL-1000 Chromogenic Limulus amebocyte lysate kit, Cambrex).
Mouse and piglet immunization
The experimental protocols were approved by the Committee of Animal Care and Supply, University of Saskatchewan. In the mouse trial, 24 six-week old B6C3F1 (H-2d) female mice were randomly divided into three groups. Groups of mice were immunized either three times with 40 μL saline subcutaneously (s.c.), three times with 50 μg of DNA vaccine pBISIA24-tPAs-tE2 intradermally (i.d.), or twice with 50 μg of DNA vaccine pBISIA24-tPAs-tE2 i.d. followed by one subcutaneous vaccination with 5 μg of recombinant tE2 protein. The recombinant truncated E2 protein vaccine was formulated with 10 μg of CpG oligonucleotide (ODN) 1826 (5’-TCCATGACGTTCCTGACGTT-3’, CpG motifs are underlined; kindly provided by Merial Limited) and 10% Emulsigen (MVP Laboratories) per dose. This formulation has been shown to elicit strong immune responses with a superior safety profile[41]. The vaccines were given three weeks apart. Blood was sampled two weeks after each immunization and spleens were collected two weeks after final immunization for analyzing immune responses.
In the piglet trial, 24 out-bred piglets (Landrace cross, 4-5 wk old, Prairie Swine Center) were randomly allocated into three groups with eight piglets in each group. The vaccination schedule was the same as that in the mouse trial but the doses were 10-fold of those used for mice. In addition, CpG ODN 2007 (5’-TCGTCGTTGTCGTTTTGGTCGTT-3’, kindly provided by Merial Limited), which can stimulate porcine peripheral blood mononuclear cells (PBMCs)[42], was used for protein vaccine formulation.
Enzyme-linked immunosorbent assay (ELISA)
To analyze antibody levels in murine or porcine sera after immunization, 96-well polystyrene plates (Immulon 2, Dynatech Laboratories) were coated with purified E2 protein at 100 ng/well in a carbonate buffer (pH9.6) overnight at 4°C. Serially diluted murine or porcine sera were added to each well and incubated for 2 h at room temperature (RT). To determine the total IgG levels, biotinylated goat anti-mouse IgG antibody (Caltag Laboratories) at a dilution of 1:10000 or alkaline phosphatase labeled goat anti-porcine IgG (KPL) at a dilution of 1:2500 was applied to detect bound IgG. To determine E2-specific IgG1 and IgG2a antibody levels in murine sera, bound antibodies were incubated with biotinylated goat anti-mouse IgG1 or IgG2a antibodies (Caltag Laboratories) at a dilution of 1:10 000, respectively, followed by streptavidin-AP (Jackson ImmunoResearch Laboratories) at a dilution of 1:5000. To determine E2-specific IgG1 and IgG2 antibody levels in porcine sera, bound antibodies were incubated with mouse anti-porcine IgG1 or IgG2 antibodies (Serotec) at a dilution of 1:100, respectively, followed by biotinylated goat anti-mouse IgG antibody (Caltag Laboratories) at a dilution of 1:10 000. The reactions were developed by adding p-nitrophenyl phosphate (PNPP) (Sigma) at 100 ng/well and the optical density was recorded at 405 nm.
Enzyme-linked immunospot (ELISPOT) assay
ELISPOT assay was performed to determine the frequency of interferon-γ (IFN-γ) secreting cells in mouse splenocytes or porcine PBMCs after vaccination. Microplate Devices Unifilter 96-well plates (Waterman) were coated with anti-mouse IFN-γ antibody (125 ng/well) (BioSource International) or anti-porcine IFN-γ monoclonal antibody (500 ng/well) (Endogen) overnight at 4°C. After washing and blocking, 1 × 106 murine splenocytes or porcine PBMCs were added along with 200 ng of purified E2 protein into each well. The plates were incubated at 37°C and 5% CO2 for 40 h. For mouse sera, biotinylated anti-mouse IFN-γ antibody (125 ng/well) was added and incubated at 37°C and 5% CO2 for 3 h. For porcine sera, rabbit anti-porcine IFN-γ antibody (200 ng/well) (Endogen) was added and incubated at RT for 4 h followed by the addition of biotinylated goat rabbit IgG (Zymed) at a dilution of 1:5000 for 2 h at RT. All the plates were then incubated for 1.5 h at RT with streptavidin-AP (Jackson ImmunoResearch Laboratories) at a dilution of 1:500 and developed with 5-bromo-4-chloro-3-indolylphospohate and nitroblue tetrazolium substrate tablets (Sigma). The plates were dried and the spots were recorded.
Lymphocyte proliferation-MTT assay
Splenocytes (3 × 105 cells) isolated from each group of mice were seeded into each well of round bottom tissue culture 96-well plates (Nunc). Purified E2 protein (100 ng) was added to each well and incubated at 37°C and 5% CO2 for 72 h. Eighty ng of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT, Sigma) was added to each well and incubated at 37°C and 5% CO2 for 3 h. The plates were centrifuged at 1000 × g for 10 min. One hundred μL of the supernatant was mixed with 100 μL of acidified isopropanol (0.375% HCl in isopropanol) and the optical density was recorded at 590 nm.
Cytotoxic T lymphocyte (CTL) assay
Effector cell preparation and stimulation: To prepare effector cells, splenocytes were isolated from each group of mice and pooled 14 d after vaccination. To generate stimulating cells, splenocytes isolated from naïve, syngenic mice were infected with a recombinant vaccinia virus expressing HCV BK E2 protein (VP1478, kindly provided by Sanofi Pasteur MSD) at an m.o.i. of 10 for 1 h at 37°C followed by an irradiation at 3000 rads. The splenocytes from experimental groups were cultured with irradiated stimulating cells for four days at 37°C and 5% CO2 before they were used as effector cells in CTL assays.
Target cell preparation: To generate target cells, a syngeneic mastocytoma cell line P1-HTR-TK+[43] (H-2d, a highly transfectable variant of P815 cells, kindly provided by Dr. T. Boon) was transfected with a plasmid expressing HCV BK E2. Stable transfectants, designated P1-E2 cells, were selected with 800 μg/mL of Geneticin (Invitrogen). The expression of HCV E2 protein was confirmed by immunohistochemical staining using an E2-specific polyclonal antibody as previously described[44]. In brief, after fixation and blocking, P1-E2 cells or plasmid vector-transfected P1 cells were incubated with an anti-E2 antibody at a dilution of 1:1000 in PBS for 30 min. Cells were then washed and incubated with a biotinylated secondary antibody for 30 min. The reaction was developed with 3, 3’-diaminobenzidine tetrahydrochloride (DAB) (Vector Laboratories).
CTL assay: CTL assays were performed based on the evaluation of the cytoplasmic lactate dehydrogenase (LDH) after cell lysis as described previously[45]. In brief, effector cells were harvested by centrifugation at 800 × g for 10 min and washed twice with medium. The cells were then adjusted to 2.0 × 107 cells/mL in AIM-V medium and incubated with target cells at standard effector:target (E:T) ratios in triplicates in a 96-well round bottom plate. After 4 h incubation, the plate was centrifuged at 250 × g for 4 min. Fifty μL of the supernatant was transferred to a 96-well flat bottom plate and mixed with the Substrate mix provided in the Cytotox 96 Non-radioactive Cytotoxicity Assay kit (Promega). After 30 min, the reaction was terminated by adding 50 μL of the Stop Solution into each well and the optical density was determined at 490 nm. Spontaneous and maximal LDH release by target cells was determined by incubating the target cells with medium alone or with medium plus the lysis buffer containing 0.9% Triton X-100 (Promega). HCV E2-specific lysis was calculated as [(experimental release-spontaneous release)/(maximal release-spontaneous release)] × 100.
Statistical analysis
The experimental data were analyzed by software programs Prism 4 (GraphPad) or Excel (Microsoft) and were expressed as mean ± SE. A P value of ≤ 0.05 determined by Student's t test was considered statistically significant.
RESULTS
Construction and characterization of a DNA vaccine expressing a secreted form of HCV E2 protein
We designed a DNA vaccine encoding a secreted form of HCV E2 protein by removing the transmembrane domain and replacing the hypervariable region 1 (HVR1) of the E2 protein with the tissue plasminogen activator signal peptide sequence (tPAs). The corresponding coding sequence was amplified by PCR and cloned into a DNA vaccine vector pBISIA24[39], resulting in the plasmid pBISIA24-tPAs-tE2 (Figure 1). To determine the expression of E2 protein, TE671 cells were transfected with pBISIA24 (vector control) and pBISIS24-tPAs-tE2. The presence of E2 protein in cell lysates and culture medium was analyzed by immunoblotting. As shown in Figure 1, an E2-specific antibody recognized proteins of about 50 kDa in cell lysates and of 50-75 kDa in culture medium after pBISIA24-tPAs-tE2 transfection, whereas these proteins were not detected after pBISIA24 vector transfection, indicating the expression and secretion of the E2 protein by pBISIA24-tPAs-tE2. The apparent molecular masses (50-75 kDa) of tE2 protein are larger than the calculated one (31 kDa) and the extracellular form contained larger protein species than the intracellular form, suggesting that E2 protein may have been modified post-translationally by glycosylation.
Recombinant tE2 protein expression and purification
The E2 protein used in immunization was produced in E. coli. The expression of poly-histidine tagged tE2 was induced by IPTG and purified by affinity chromatography (not shown). The purified poly-histidine tagged tE2 protein was dialyzed into phosphate-buffered saline before it was used in mouse and piglet immunization. The protein vaccine contained endotoxin at a concentration of 80 ng/mg of protein as determined by the Limulus amoebocyte test.
Antibody responses to HCV E2 DNA vaccine in mice
To determine whether the DNA vaccine expressing a secreted E2 protein elicited antibody responses in mice, mouse sera collected after vaccination were analyzed for E2-specific IgG titers by ELISA assays. As shown in Figure 2A, vaccinated-mice all developed E2-specific IgG antibody after intradermal DNA vaccination with the average IgG titer being 1.6 × 105. To test whether a protein vaccine could enhance the IgG response, mice were injected twice with the DNA vaccine followed by recombinant E2 protein boosting. Mice that received the protein boosting vaccination had about two-fold higher titers of E2-specific IgG (3.0 × 105) (Figure 2A). These results indicate that the DNA vaccine expressing a secreted form of HCV E2 protein induced strong antibody responses in mice, which were further increased by a protein vaccine.
Figure 2.
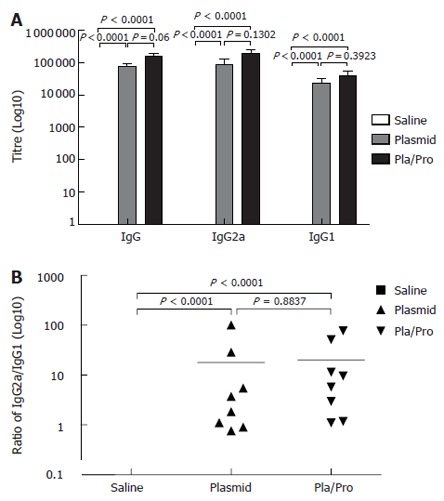
Antibody responses to HCV E2 vaccines in mice. B6C3F1 mice were intradermally injected with pBISIA24-tPAs-tE2 three times or pBISIA24-tPAs-tE2 twice followed by subcutaneous vaccination with E2 protein formulated with CpG ODN 1826 and 10% Emulsigen at 3-wk intervals. E2-specific IgG, IgG1, and IgG2a levels were determined by ELISA assays (A). The ratios of IgG2a to IgG1 were determined (B) (P < 0.0001, vs saline).
Since the relative levels of IgG subclasses are an indicator for evaluating the quality of the immune response[46,47], the titers of IgG1 and IgG2a were determined. E2 DNA vaccine induced higher IgG2a titers than IgG1 (Figure 2), indicating a Th1-biased response. Boosting with E2 protein formulated with CpG ODN and 10% Emulsigen did not change this pattern, although the IgG1 and IgG2a levels were elevated (Figure 2).
Cell-mediated responses to HCV E2 DNA vaccine in mice
To determine whether the HCV E2 vaccine induced cell-mediated immune responses in mice, we analyzed the responses of mouse splenocytes after vaccination by measuring cell proliferation and interferon-γ secretion upon antigen re-stimulation in vitro.
In lymphocyte proliferation assay, splenocytes from saline vaccine group showed detectable proliferation after antigen stimulation (Figure 3A). Splenocytes isolated from mice after DNA vaccination expanded to a significantly higher degree after E2 antigen re-stimulation than medium control (P < 0.001, Figure 3A). Protein boost immunization did not change E2-specific lymphocyte proliferation (DNA vs DNA/Protein, P = 0.47, Figure 3A). The results of IFN-γ ELISPOT assay demonstrated that DNA vaccination elicited a strong IFN-γ response (DNA vs saline, P < 0.001, Figure 3B), whereas a protein boost immunization induced fewer IFN-γ secreting spots (DNA vs DNA/Protein, P = 0.03, Figure 3B). These results indicate that stronger E2-specific Th1-type immune responses were induced in mice that received the DNA vaccine three times than in mice that received twice DNA vaccination followed by a protein boosting injection.
Figure 3.
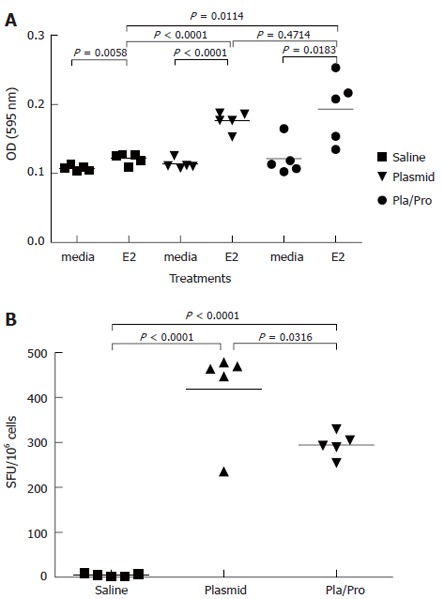
Cell-mediated immune responses to HCV E2 vaccines in mice. Murine splenocytes were isolated 2 wk after last immunization and were re-stimulated with E2 protein. Cell proliferation was analyzed by MTT assay (A). (P < 0.0001 vs saline; P < 0.05, P < 0.01 vs media). The number of interferon-γ secreting cells was determined by ELISPOT assay (B). (P < 0.05 vs plasmid; P < 0.0001 vs saline).
Since cytotoxic T lymphocytes are one of the major effectors in cell-mediated immune responses, we also determined the presence of E2-specific cytotoxic T lymphocytes after vaccination. For this purpose, a syngeneic cell line expressing HCV E2 protein was generated. Immunohistochemistry staining using an anti-E2 antibody detected specific signal in P1-HTR cells stably transfected by the E2-expressing plasmid, but not in vector-transfected cells, demonstrating the expression of the E2 protein by the stable transfectant (not shown). When the E2-expressing cell line was used as target cells in the CTL assay, effector cells from DNA vaccine- or DNA and protein vaccine-immunized mice showed 36% or 31% specific lysis at an effector to target ratio of 100:1, respectively (Figure 4). No CTL activity was detected after saline immunization (Figure 4). These results demonstrate the elicitation of E2-specific cytotoxic T lymphocytes in mice immunized with E2 DNA vaccine or DNA vaccine followed by a protein boost.
Figure 4.
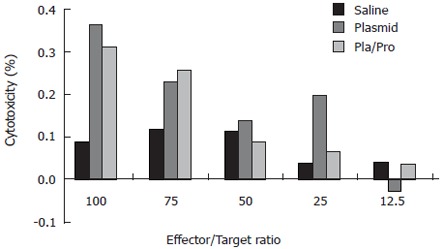
Cytotoxic T lymphocyte (CTL) response to HCV E2 vaccines in mice. Murine splenocytes isolated from groups of mice two weeks after final immunization were pooled and stimulated with feeder cells infected with recombinant vaccinia virus expressing HCV E2 protein. The cytotoxic activity was determined by measuring the released lactate dehydrogenase (LDH) by the target cells after incubation with the effectors at different ratios.
Immune responses to HCV E2 vaccines in piglets
Since vaccine efficacy can be different in various test species[31] and pigs represent a promising model for human biology because of its body size and physiology[48], we evaluated our E2 vaccines in piglets. As shown in Figure 5A, all the piglets receiving the E2 DNA vaccine three times intradermally developed E2-specific IgG titers with the average titer being 1 × 103. Vaccination with the E2 protein after two DNA vaccine priming injections significantly boosted the E2-specific IgG titers (DNA vaccine vs DNA/Protein, P = 0.0156, Figure 5A). These results indicate that E2 DNA vaccine was also effective in inducing antigen-specific antibody response in piglets, which was boosted significantly by a protein vaccine.
Figure 5.
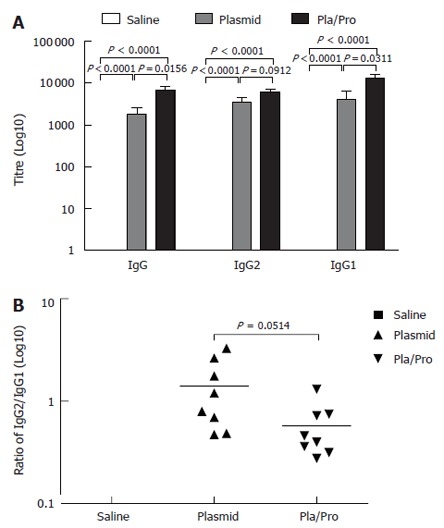
Antibody responses of piglets to HCV E2 vaccines. Piglets were immunized by saline, E2 DNA vaccine three times, or E2 DNA vaccine twice followed by subcutaneous vaccination with E2 protein formulated with CpG ODN 2007 and 10% Emulsigen at three-week intervals. ELISA assays were performed to determine E2-specific IgG, IgG1, and IgG2 levels (A), as well as the ratios of IgG2 to IgG1 (B) (P < 0.05 vs plasmid; P < 0.0001 vs saline).
To analyze the type of the immune responses, we determined the ratios of E2-specific IgG1 to IgG2 levels in the porcine serum samples after vaccination. As illustrated in Figure 5B, DNA vaccination induced a slightly Th1-biased immune response with the IgG2 to IgG1 ratio being 1.4, whereas protein boosting after DNA vaccination shifted the immune response towards Th2 direction as demonstrated by a reduced IgG2/IgG1 ratio (0.58).
To further characterize the immune responses of piglets to E2 vaccines, the number of IFN-γ secreting cells in porcine PBMCs was determined after vaccination. As shown in Figure 6, PBMCs from E2 vaccinated piglets contained significantly higher number of IFN-γ secreting cells than saline treated piglets after in vitro antigen re-stimulation in ELISPOT assay (DNA vs saline, P < 0.0001, DNA/Protein vs saline, P < 0.0001). No difference was detected between the DNA vaccine alone group and the DNA prime and protein boost group (Figure 6, DNA vs DNA/Protein, P = 0.4531).
Figure 6.
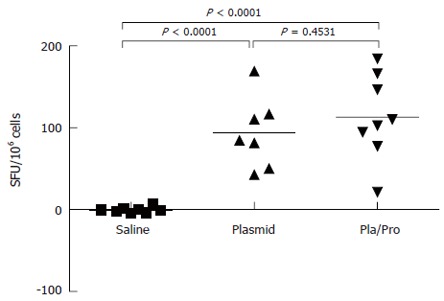
Cell-mediated immune response of piglets to HCV E2 vaccines. Porcine peripheral blood mononuclear cells (PBMCs) were isolated three weeks after final immunization. After re-stimulation with E2 antigen, the number of interferon-γ secreting cells was determined by ELISPOT assay (P < 0.0001 vs saline).
DISCUSSION
Numerous research groups have evaluated the potential of using E2 protein as a hepatitis C vaccine candidate through different strategies[5,36,49-74]. These studies have demonstrated E2-specific immune responses, mostly in mouse trials, when E2 alone or as part of the entire HCV structural region was delivered as a DNA vaccine, viral vectored vaccine, or subunit protein vaccine. The most important finding of these studies is that the vaccine-induced E2-specific immune responses were able to delay HCV infections in chimpanzees or humans[55,61,62,64]. While these studies have demonstrated the feasibility of inducing E2-specific immune responses through vaccination, the efficacy of E2 vaccines is less satisfactory in inducing sterilizing immunity. It is thus obvious that further improvement of E2 vaccines is required by evaluating additional vaccination strategies.
Given the importance of E2 protein in inducing host immune responses, we have chosen E2 as a vaccine candidate against hepatitis C. It has been established that HCV E2 expressed by a number of systems is an intracellular protein modified by high-mannose type oligosaccharides[75-78]. However, the envelope proteins on the hepatitis C virions have been shown to be modified by complex type oligosaccharides[79], suggesting secretion of E2 protein has occurred in HCV life cycle. In agreement with this notion, the secreted, complex form E2 protein possesses greater immunoreactivity against hepatitis C patient sera than the intracellular E2 protein when it is expressed in cell culture[78]. The retention of the expressed E2 protein is due to the presence of a membrane anchor domain (amino acids 718-746) at its carboxyl-terminus[80-82] and deletion of the transmembrane domain leads to E2 protein secretion[83]. However, a comparative study of a series of carboxyl-terminally truncated E2 proteins has demonstrated that truncation to amino acid residue 661 is necessary to achieve proper folding of the secreted E2 protein[83]. Hepatitis C virus is known to be able to quickly develop multiple sequence variants after infection within the host due to the lack of proof-reading activity of RNA-dependent RNA polymerase[84]. A short stretch of coding sequence (amino acids 384-411, the hypervariable region 1) at the very amino-terminus of E2 has shown extremely high variability[85,86] and may contribute to the generation of immune escape HCV variants. Although E2 HVR-1 may contain antigenic epitopes[15,16], rapid appearance of variable mutants and their interference with the development of cellular immunity in the host[87] make it a less favorable component in an E2 DNA vaccine. Based on the above findings and analysis, we designed an E2 DNA vaccine by deleting the carboxyl-terminal hydrophobic region after amino acid residue 661 and replacing HVR1 by a strong signal peptide sequence derived from tissue plasminogen activator[88]. Indeed, we showed that the engineered E2 protein was efficiently secreted into the culture medium (Figure 1). In addition, in agreement with previous studies[83], the apparent molecular masses of the intracellular as well as secreted tE2 proteins are larger than that calculated from the amino acid sequence and the secreted tE2 proteins contain species of even higher molecular mass than the intracellular tE2 protein, suggesting that intracellular and extracellular E2 proteins may have been modified by different glycosylation.
Intradermal injection of this E2 DNA vaccine elicited strong E2-specific antibody responses in mice as demonstrated by high IgG levels (Figure 2). IgG isotyping, IFN-γ ELISPOT, lymphocyte proliferation, and CTL assays (Figures 2-4) indicated that both Th1 lymphocyte and cytotoxic T lymphocyte responses were induced by E2 DNA vaccine. Interestingly, DNA vaccine alone or DNA vaccine followed by an E2 protein formulated with CpG ODN and 10% Emulsigen induced comparable E2-specific IgG2a to IgG1 ratios (Figure 2B) and CTL responses (Figure 4). This is in agreement with the proven roles of immuno-stimulatory CpG ODN in promoting Th1 immune responses[41,89-91]. However, although DNA prime and protein boost strategy induced a relatively strong IFN-γ response, the number of IFN-γ secreting cells in the splenocytes derived from mice immunized with DNA prime and protein boost was significantly lower than that of DNA vaccine group (Figure 3B).
DNA vaccines are often less effective in large model animals and in humans than in mice[31,92]. Therefore, it is critical to test the vaccine efficacy in an out-bred, large animal model. Our piglet trial indicated that E2 DNA vaccine induced E2-specific IgG titers (Figure 5A) that were about 100-fold lower than those detected in mice (Figure 2A). As for the type of vaccine-induced immune response, E2 DNA vaccine induced a more balanced response in piglets (Figure 5B), in contrast to a strongly Th1-biased response in mice (Figure 2B). The effect of protein boost injection in modulating the type of the immune response was also different: while protein boost had little effect in mice (Figure 2B), a shift towards Th2 direction was detected in piglets (Figure 5B). Taken together, while these results demonstrate that E2 vaccines were also effective in inducing antigen-specific immune responses in piglets, inter-species difference in immune responses to different vaccine strategies requires further investigation.
In conclusion, our results demonstrate that a DNA vaccine expressing the secreted form of HCV E2 protein induces E2-specific immune responses in mice and piglets. An E2 protein vaccine formulated with CpG ODN and 10% Emulsigen further increases the antibody responses. In addition, our results highlight the importance of testing the magnitude and type of vaccine-induced immune responses in multiple model species before primate or human trials are initiated.
ACKNOWLEDGMENTS
We would like to thank Sylvia van Drunen Littel-van den Hurk, Philip Griebel, and Lou Qualtiere for providing reagents and/or helpful discussions. We thank Lucy Liu, Shirley Lam, Candice Jackel, Marlene Snider, Laura Latimer, Ponn Benjamin, and Satya Viswanathan for their contributions to this work. We thank VIDO Animal Care staff for doing the animal trials. This work is published as VIDO Journal series #427. LAB is a recipient of a Canada research chair in vaccinology.
COMMENTS
Background
Hepatitis C is a devastating liver disease worldwide. Current interferon and ribavirin therapy is far from satisfactory. An effective vaccine for hepatitis C is urgently needed.
Research frontiers
To develop an effective vaccine for hepatitis C, a rational design based on the understanding of the magnitude and the quality of the immune responses is critical for a success. The application of appropriate adjuvants and vaccine delivery means also play a major role.
Innovations and breakthroughs
The novelty of this article lies in the design of the HCV E2 vaccine, namely the hypervariable region-1 was replaced by the signal peptide sequence of the tissue plasminogen activator and the hydrophobic region at the carboxyl-terminus was removed.
Applications
The HCV E2 DNA vaccine developed in this study should be further tested in primate models with hepatitis C virus challenge to demonstrate protective activity of the vaccine.
Peer review
HCV infection is a big burden worldwide. Numerous studies have evaluated the potential of using E2 protein as a hepatitis C vaccine candidate through different strategies, but the efficacy of E2 vaccines is less satisfactory in inducing sterilizing immunity. It is interesting to investigate further improvement of E2 vaccines by evaluating additional vaccination strategies. The authors characterized the immunogenicity of a HCV E2 DNA vaccine alone or with a protein vaccine boost in murine and porcine animal models. They found that this E2 DNA vaccine elicited E2-specific immune responses in mice and piglets and recombinant E2 protein vaccination boosting significantly increased the antibody response in piglets. The study is well designed and results are convincing. The presentation and readability of the manuscript is satisfactory.
Footnotes
Supported by the Canadian Network for Vaccines and Immuno-therapeutics
S- Editor Pan BR L- Editor Zhu LH E- Editor Ma WH
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 3.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 4.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 5.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 991–1041. [Google Scholar]
- 7.Major ME, Rehermann B, Feinstone SM. Hepatitis C Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1127–1161. [Google Scholar]
- 8.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 9.Petit MA, Lièvre M, Peyrol S, De Sequeira S, Berthillon P, Ruigrok RW, Trépo C. Enveloped particles in the serum of chronic hepatitis C patients. Virology. 2005;336:144–153. doi: 10.1016/j.virol.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Schofield DJ, Bartosch B, Shimizu YK, Allander T, Alter HJ, Emerson SU, Cosset FL, Purcell RH. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology. 2005;42:1055–1062. doi: 10.1002/hep.20906. [DOI] [PubMed] [Google Scholar]
- 11.Yuki N, Hayashi N, Kasahara A, Hagiwara H, Mita E, Ohkawa K, Katayama K, Fusamoto H, Kamada T. Quantitative analysis of antibody to hepatitis C virus envelope 2 glycoprotein in patients with chronic hepatitis C virus infection. Hepatology. 1996;23:947–952. doi: 10.1053/jhep.1996.v23.pm0008621173. [DOI] [PubMed] [Google Scholar]
- 12.Netski DM, Mosbruger T, Depla E, Maertens G, Ray SC, Hamilton RG, Roundtree S, Thomas DL, McKeating J, Cox A. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis. 2005;41:667–675. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- 13.Chen M, Sällberg M, Sönnerborg A, Weiland O, Mattsson L, Jin L, Birkett A, Peterson D, Milich DR. Limited humoral immunity in hepatitis C virus infection. Gastroenterology. 1999;116:135–143. doi: 10.1016/s0016-5085(99)70237-4. [DOI] [PubMed] [Google Scholar]
- 14.Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter HJ, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu YK, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell RH, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 17.Ishii K, Rosa D, Watanabe Y, Katayama T, Harada H, Wyatt C, Kiyosawa K, Aizaki H, Matsuura Y, Houghton M, et al. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology. 1998;28:1117–1120. doi: 10.1002/hep.510280429. [DOI] [PubMed] [Google Scholar]
- 18.Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 19.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 20.Neumann-Haefelin C, Blum HE, Chisari FV, Thimme R. T cell response in hepatitis C virus infection. J Clin Virol. 2005;32:75–85. doi: 10.1016/j.jcv.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Koziel MJ. Cellular immune responses against hepatitis C virus. Clin Infect Dis. 2005;41 Suppl 1:S25–S31. doi: 10.1086/429492. [DOI] [PubMed] [Google Scholar]
- 22.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 23.Ward S, Lauer G, Isba R, Walker B, Klenerman P. Cellular immune responses against hepatitis C virus: the evidence base 2002. Clin Exp Immunol. 2002;128:195–203. doi: 10.1046/j.1365-2249.2002.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leroux-Roels G, Esquivel CA, DeLeys R, Stuyver L, Elewaut A, Philippé J, Desombere I, Paradijs J, Maertens G. Lymphoproliferative responses to hepatitis C virus core, E1, E2, and NS3 in patients with chronic hepatitis C infection treated with interferon alfa. Hepatology. 1996;23:8–16. doi: 10.1002/hep.510230102. [DOI] [PubMed] [Google Scholar]
- 25.Sarobe P, Lasarte JJ, García N, Civeira MP, Borrás-Cuesta F, Prieto J. Characterization of T-cell responses against immunodominant epitopes from hepatitis C virus E2 and NS4a proteins. J Viral Hepat. 2006;13:47–55. doi: 10.1111/j.1365-2893.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 26.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 27.Berzofsky JA, Ahlers JD, Belyakov IM. Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol. 2001;1:209–219. doi: 10.1038/35105075. [DOI] [PubMed] [Google Scholar]
- 28.Sharma AK, Khuller GK. DNA vaccines: future strategies and relevance to intracellular pathogens. Immunol Cell Biol. 2001;79:537–546. doi: 10.1046/j.1440-1711.2001.01044.x. [DOI] [PubMed] [Google Scholar]
- 29.Lemieux P. Technological advances to increase immunogenicity of DNA vaccines. Expert Rev Vaccines. 2002;1:85–93. doi: 10.1586/14760584.1.1.85. [DOI] [PubMed] [Google Scholar]
- 30.Doria-Rose NA, Haigwood NL. DNA vaccine strategies: candidates for immune modulation and immunization regimens. Methods. 2003;31:207–216. doi: 10.1016/s1046-2023(03)00135-x. [DOI] [PubMed] [Google Scholar]
- 31.Manoj S, Babiuk LA, van Drunen Littel-van den Hurk S. Approaches to enhance the efficacy of DNA vaccines. Crit Rev Clin Lab Sci. 2004;41:1–39. doi: 10.1080/10408360490269251. [DOI] [PubMed] [Google Scholar]
- 32.Rath A, Choudhury S, Batra D, Kapre SV, Rupprecht CE, Gupta SK. DNA vaccine for rabies: relevance of the trans-membrane domain of the glycoprotein in generating an antibody response. Virus Res. 2005;113:143–152. doi: 10.1016/j.virusres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Barnett SW, Rajasekar S, Legg H, Doe B, Fuller DH, Haynes JR, Walker CM, Steimer KS. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine. 1997;15:869–873. doi: 10.1016/s0264-410x(96)00264-2. [DOI] [PubMed] [Google Scholar]
- 34.Letvin NL, Montefiori DC, Yasutomi Y, Perry HC, Davies ME, Lekutis C, Alroy M, Freed DC, Lord CI, Handt LK, et al. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc Natl Acad Sci USA. 1997;94:9378–9383. doi: 10.1073/pnas.94.17.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung L, Srivastava IK, Kan E, Legg H, Sun Y, Greer C, Montefiori DC, zur Megede J, Barnett SW. Immunogenicity of HIV-1 Env and Gag in baboons using a DNA prime/protein boost regimen. AIDS. 2004;18:991–1001. doi: 10.1097/00002030-200404300-00006. [DOI] [PubMed] [Google Scholar]
- 36.Song MK, Lee SW, Suh YS, Lee KJ, Sung YC. Enhancement of immunoglobulin G2a and cytotoxic T-lymphocyte responses by a booster immunization with recombinant hepatitis C virus E2 protein in E2 DNA-primed mice. J Virol. 2000;74:2920–2925. doi: 10.1128/jvi.74.6.2920-2925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yosida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 38.van Drunen Littel-van den Hurk S, Braun RP, Karvonen BC, King T, Yoo D, Babiuk LA. Immune responses and protection induced by DNA vaccines encoding bovine parainfluenza virus type 3 glycoproteins. Virology. 1999;260:35–46. doi: 10.1006/viro.1999.9793. [DOI] [PubMed] [Google Scholar]
- 39.Yu H, Babiuk LA, van Drunen Littel-van den Hurk S. Priming with CpG-enriched plasmid and boosting with protein formulated with CpG oligodeoxynucleotides and Quil A induces strong cellular and humoral immune responses to hepatitis C virus NS3. J Gen Virol. 2004;85:1533–1543. doi: 10.1099/vir.0.79821-0. [DOI] [PubMed] [Google Scholar]
- 40.Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 41.Ioannou XP, Gomis SM, Karvonen B, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. CpG-containing oligodeoxynucleotides, in combination with conventional adjuvants, enhance the magnitude and change the bias of the immune responses to a herpesvirus glycoprotein. Vaccine. 2002;21:127–137. doi: 10.1016/s0264-410x(02)00378-x. [DOI] [PubMed] [Google Scholar]
- 42.Rankin R, Pontarollo R, Ioannou X, Krieg AM, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. CpG motif identification for veterinary and laboratory species demonstrates that sequence recognition is highly conserved. Antisense Nucleic Acid Drug Dev. 2001;11:333–340. doi: 10.1089/108729001753231713. [DOI] [PubMed] [Google Scholar]
- 43.Van Pel A, De Plaen E, Boon T. Selection of highly transfectable variant from mouse mastocytoma P815. Somat Cell Mol Genet. 1985;11:467–475. doi: 10.1007/BF01534840. [DOI] [PubMed] [Google Scholar]
- 44.Liu Q, Tikoo SK, Babiuk LA. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology. 2001;285:91–99. doi: 10.1006/viro.2001.0922. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Zaiss AK, Colarusso P, Patel K, Haljan G, Wickham TJ, Muruve DA. The role of capsid-endothelial interactions in the innate immune response to adenovirus vectors. Hum Gene Ther. 2003;14:627–643. doi: 10.1089/104303403321618146. [DOI] [PubMed] [Google Scholar]
- 46.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 47.Braun R, Babiuk LA. Compatibility of plasmids expressing different antigens in a single DNA vaccine formulation. J Gen Virol. 1998;79(Pt 12):2965–2970. doi: 10.1099/0022-1317-79-12-2965. [DOI] [PubMed] [Google Scholar]
- 48.Rothschild MF. Porcine genomics delivers new tools and results: this little piggy did more than just go to market. Genet Res. 2004;83:1–6. doi: 10.1017/s0016672303006621. [DOI] [PubMed] [Google Scholar]
- 49.Fournillier A, Nakano I, Vitvitski L, Depla E, Vidalin O, Maertens G, Trépo C, Inchauspé G. Modulation of immune responses to hepatitis C virus envelope E2 protein following injection of plasmid DNA using single or combined delivery routes. Hepatology. 1998;28:237–244. doi: 10.1002/hep.510280131. [DOI] [PubMed] [Google Scholar]
- 50.Pancholi P, Liu Q, Tricoche N, Zhang P, Perkus ME, Prince AM. DNA prime-canarypox boost with polycistronic hepatitis C virus (HCV) genes generates potent immune responses to HCV structural and nonstructural proteins. J Infect Dis. 2000;182:18–27. doi: 10.1086/315646. [DOI] [PubMed] [Google Scholar]
- 51.Pancholi P, Perkus M, Tricoche N, Liu Q, Prince AM. DNA immunization with hepatitis C virus (HCV) polycistronic genes or immunization by HCV DNA priming-recombinant canarypox virus boosting induces immune responses and protection from recombinant HCV-vaccinia virus infection in HLA-A2.1-transgenic mice. J Virol. 2003;77:382–390. doi: 10.1128/JVI.77.1.382-390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tedeschi V, Akatsuka T, Shih JW, Battegay M, Feinstone SM. A specific antibody response to HCV E2 elicited in mice by intramuscular inoculation of plasmid DNA containing coding sequences for E2. Hepatology. 1997;25:459–462. doi: 10.1002/hep.510250234. [DOI] [PubMed] [Google Scholar]
- 53.Ha SJ, Chang J, Song MK, Suh YS, Jin HT, Lee CH, Nam GH, Choi G, Choi KY, Lee SH, et al. Engineering N-glycosylation mutations in IL-12 enhances sustained cytotoxic T lymphocyte responses for DNA immunization. Nat Biotechnol. 2002;20:381–386. doi: 10.1038/nbt0402-381. [DOI] [PubMed] [Google Scholar]
- 54.Ha SJ, Kim DJ, Baek KH, Yun YD, Sung YC. IL-23 induces stronger sustained CTL and Th1 immune responses than IL-12 in hepatitis C virus envelope protein 2 DNA immunization. J Immunol. 2004;172:525–531. doi: 10.4049/jimmunol.172.1.525. [DOI] [PubMed] [Google Scholar]
- 55.Puig M, Major ME, Mihalik K, Feinstone SM. Immunization of chimpanzees with an envelope protein-based vaccine enhances specific humoral and cellular immune responses that delay hepatitis C virus infection. Vaccine. 2004;22:991–1000. doi: 10.1016/j.vaccine.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Vidalin O, Fournillier A, Renard N, Chen M, Depla E, Boucreux D, Brinster C, Baumert T, Nakano I, Fukuda Y, et al. Use of conventional or replicating nucleic acid-based vaccines and recombinant Semliki forest virus-derived particles for the induction of immune responses against hepatitis C virus core and E2 antigens. Virology. 2000;276:259–270. doi: 10.1006/viro.2000.0566. [DOI] [PubMed] [Google Scholar]
- 57.Saito T, Sherman GJ, Kurokohchi K, Guo ZP, Donets M, Yu MY, Berzofsky JA, Akatsuka T, Feinstone SM. Plasmid DNA-based immunization for hepatitis C virus structural proteins: immune responses in mice. Gastroenterology. 1997;112:1321–1330. doi: 10.1016/s0016-5085(97)70146-x. [DOI] [PubMed] [Google Scholar]
- 58.Lee SW, Cho JH, Lee KJ, Sung YC. Hepatitis C virus envelope DNA-based immunization elicits humoral and cellular immune responses. Mol Cells. 1998;8:444–451. [PubMed] [Google Scholar]
- 59.Esumi M, Rikihisa T, Nishimura S, Goto J, Mizuno K, Zhou YH, Shikata T. Experimental vaccine activities of recombinant E1 and E2 glycoproteins and hypervariable region 1 peptides of hepatitis C virus in chimpanzees. Arch Virol. 1999;144:973–980. doi: 10.1007/s007050050559. [DOI] [PubMed] [Google Scholar]
- 60.Abraham JD, Himoudi N, Kien F, Berland JL, Codran A, Bartosch B, Baumert T, Paranhos-Baccala G, Schuster C, Inchauspé G, et al. Comparative immunogenicity analysis of modified vaccinia Ankara vectors expressing native or modified forms of hepatitis C virus E1 and E2 glycoproteins. Vaccine. 2004;22:3917–3928. doi: 10.1016/j.vaccine.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Forns X, Payette PJ, Ma X, Satterfield W, Eder G, Mushahwar IK, Govindarajan S, Davis HL, Emerson SU, Purcell RH, et al. Vaccination of chimpanzees with plasmid DNA encoding the hepatitis C virus (HCV) envelope E2 protein modified the infection after challenge with homologous monoclonal HCV. Hepatology. 2000;32:618–625. doi: 10.1053/jhep.2000.9877. [DOI] [PubMed] [Google Scholar]
- 62.Youn JW, Park SH, Lavillette D, Cosset FL, Yang SH, Lee CG, Jin HT, Kim CM, Shata MT, Lee DH, et al. Sustained E2 antibody response correlates with reduced peak viremia after hepatitis C virus infection in the chimpanzee. Hepatology. 2005;42:1429–1436. doi: 10.1002/hep.20934. [DOI] [PubMed] [Google Scholar]
- 63.Forns X, Emerson SU, Tobin GJ, Mushahwar IK, Purcell RH, Bukh J. DNA immunization of mice and macaques with plasmids encoding hepatitis C virus envelope E2 protein expressed intracellularly and on the cell surface. Vaccine. 1999;17:1992–2002. doi: 10.1016/s0264-410x(98)00448-4. [DOI] [PubMed] [Google Scholar]
- 64.Rollier C, Depla E, Drexhage JA, Verschoor EJ, Verstrepen BE, Fatmi A, Brinster C, Fournillier A, Whelan JA, Whelan M, et al. Control of heterologous hepatitis C virus infection in chimpanzees is associated with the quality of vaccine-induced peripheral T-helper immune response. J Virol. 2004;78:187–196. doi: 10.1128/JVI.78.1.187-196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inchauspé G, Major ME, Nakano I, Vitvitski L, Trépo C. DNA vaccination for the induction of immune responses against hepatitis C virus proteins. Vaccine. 1997;15:853–856. doi: 10.1016/s0264-410x(96)00275-7. [DOI] [PubMed] [Google Scholar]
- 66.Zhu LX, Liu J, Ye Y, Xie YH, Kong YY, Li GD, Wang Y. A candidate DNA vaccine elicits HCV specific humoral and cellular immune responses. World J Gastroenterol. 2004;10:2488–2492. doi: 10.3748/wjg.v10.i17.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, Forns X, Gutierrez R, Mushahwar IK, Wu T, Payette PJ, Bukh J, Purcell RH, Davis HL. DNA-based vaccination against hepatitis C virus (HCV): effect of expressing different forms of HCV E2 protein and use of CpG-optimized vectors in mice. Vaccine. 2002;20:3263–3271. doi: 10.1016/s0264-410x(02)00304-3. [DOI] [PubMed] [Google Scholar]
- 68.Zucchelli S, Capone S, Fattori E, Folgori A, Di Marco A, Casimiro D, Simon AJ, Laufer R, La Monica N, Cortese R, et al. Enhancing B- and T-cell immune response to a hepatitis C virus E2 DNA vaccine by intramuscular electrical gene transfer. J Virol. 2000;74:11598–11607. doi: 10.1128/jvi.74.24.11598-11607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youn JW, Park SH, Cho JH, Sung YC. Optimal induction of T-cell responses against hepatitis C virus E2 by antigen engineering in DNA immunization. J Virol. 2003;77:11596–11602. doi: 10.1128/JVI.77.21.11596-11602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park SH, Yang SH, Lee CG, Youn JW, Chang J, Sung YC. Efficient induction of T helper 1 CD4+ T-cell responses to hepatitis C virus core and E2 by a DNA prime-adenovirus boost. Vaccine. 2003;21:4555–4564. doi: 10.1016/s0264-410x(03)00499-7. [DOI] [PubMed] [Google Scholar]
- 71.Jin J, Yang JY, Liu J, Kong YY, Wang Y, Li GD. DNA immunization with fusion genes encoding different regions of hepatitis C virus E2 fused to the gene for hepatitis B surface antigen elicits immune responses to both HCV and HBV. World J Gastroenterol. 2002;8:505–510. doi: 10.3748/wjg.v8.i3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heile JM, Fong YL, Rosa D, Berger K, Saletti G, Campagnoli S, Bensi G, Capo S, Coates S, Crawford K, et al. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J Virol. 2000;74:6885–6892. doi: 10.1128/jvi.74.15.6885-6892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee AY, Polakos NK, Otten GR, Ulmer JB, Houghton M, Paliard X. Quantification of the number of cytotoxic T cells specific for an immunodominant HCV-specific CTL epitope primed by DNA immunization. Vaccine. 2000;18:1962–1968. doi: 10.1016/s0264-410x(99)00486-7. [DOI] [PubMed] [Google Scholar]
- 74.Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dubuisson J. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr Top Microbiol Immunol. 2000;242:135–148. doi: 10.1007/978-3-642-59605-6_7. [DOI] [PubMed] [Google Scholar]
- 77.Goffard A, Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85:295–301. doi: 10.1016/s0300-9084(03)00004-x. [DOI] [PubMed] [Google Scholar]
- 78.Inudoh M, Nyunoya H, Tanaka T, Hijikata M, Kato N, Shimotohno K. Antigenicity of hepatitis C virus envelope proteins expressed in Chinese hamster ovary cells. Vaccine. 1996;14:1590–1596. doi: 10.1016/s0264-410x(96)00154-5. [DOI] [PubMed] [Google Scholar]
- 79.Sato K, Okamoto H, Aihara S, Hoshi Y, Tanaka T, Mishiro S. Demonstration of sugar moiety on the surface of hepatitis C virions recovered from the circulation of infected humans. Virology. 1993;196:354–357. doi: 10.1006/viro.1993.1488. [DOI] [PubMed] [Google Scholar]
- 80.Cocquerel L, Meunier JC, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cocquerel L, Op de Beeck A, Lambot M, Roussel J, Delgrange D, Pillez A, Wychowski C, Penin F, Dubuisson J. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 2002;21:2893–2902. doi: 10.1093/emboj/cdf295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spaete RR, Alexander D, Rugroden ME, Choo QL, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 83.Michalak JP, Wychowski C, Choukhi A, Meunier JC, Ung S, Rice CM, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78(Pt 9):2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 84.Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 85.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 87.Scottà C, Tuosto L, Masci AM, Racioppi L, Piccolella E, Frasca L. Hypervariable region 1 variant acting as TCR antagonist affects hepatitis C virus-specific CD4+ T cell repertoire by favoring CD95-mediated apoptosis. J Leukoc Biol. 2005;78:372–382. doi: 10.1189/jlb.0804456. [DOI] [PubMed] [Google Scholar]
- 88.Pennica D, Holmes WE, Kohr WJ, Harkins RN, Vehar GA, Ward CA, Bennett WF, Yelverton E, Seeburg PH, Heyneker HL, et al. Cloning and expression of human tissue-type plasminogen activator cDNA in E. coli. Nature. 1983;301:214–221. doi: 10.1038/301214a0. [DOI] [PubMed] [Google Scholar]
- 89.Brazolot Millan CL, Weeratna R, Krieg AM, Siegrist CA, Davis HL. CpG DNA can induce strong Th1 humoral and cell-mediated immune responses against hepatitis B surface antigen in young mice. Proc Natl Acad Sci USA. 1998;95:15553–15558. doi: 10.1073/pnas.95.26.15553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weeratna RD, Brazolot Millan CL, McCluskie MJ, Davis HL. CpG ODN can re-direct the Th bias of established Th2 immune responses in adult and young mice. FEMS Immunol Med Microbiol. 2001;32:65–71. doi: 10.1111/j.1574-695X.2001.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 91.Davis HL, Weeratna R, Waldschmidt TJ, Tygrett L, Schorr J, Krieg AM. CpG DNA is a potent enhancer of specific immunity in mice immunized with recombinant hepatitis B surface antigen. J Immunol. 1998;160:870–876. [PubMed] [Google Scholar]
- 92.Donnelly JJ, Wahren B, Liu MA. DNA vaccines: progress and challenges. J Immunol. 2005;175:633–639. doi: 10.4049/jimmunol.175.2.633. [DOI] [PubMed] [Google Scholar]


