Abstract
AIM: To study the association between Crohn’s disease (CD), Mycobacterium avium subspecies paratuberculosis (MAP), and genetic factors by examining the role of natural resistance-associated macrophage protein 1 (NRAMP1) gene polymorphisms (now SLC11A1) in Sardinian patients with CD and controls.
METHODS: Thirty-seven CD patients and 34 controls with no inflammatory bowel disease (IBD) were recruited at the University of Sassari after giving written consent. Six SCL11A1 polymorphisms previously reported to be the most significantly associated with IBD were searched. M. paratuberculosis was identified by IS900 PCR and sequencing. Logistic regression was used to calculate odds ratios (OR) for the associations among CD, presence of MAP, and 6 loci described above.
RESULTS: For the first time, a strong association was observed between polymorphisms at NRAMP1 locus 823C/T and CD. While CD was strongly associated with both NRAMP1 and MAP, NRAMP1 polymorphisms and MAP themselves were not correlated.
CONCLUSION: Combined with previous work on the NOD2/CARD15 gene, it is clear that the interplay of genetic, infectious, and immunologic factors in the etiology of CD is complex.
Keywords: Mycobacterium avium subspecies paratuberculosis, Crohn’s disease, SCL11A1 polymorphisms
INTRODUCTION
Natural resistance-associated macrophage protein 1 (NRAMP1), now strictly referred to as SLC11A1 (Solute carrier 11a1) and the gene which encodes for it is recognized as having a role in the susceptibility of men and animals to a number of mycobacterial infections. In human beings, the NRAMP1 gene is located on human chromosome region 2q35. It is composed of 15 exons and covers at least 16 kb of DNA. It encodes an integral membrane protein of 550 amino acids that is expressed exclusively in the lysosomal compartment of monocytes and macrophages[1]. The promoter region of the NRAMP1 gene possesses a polymorphism within a possible enhancer element containing a Z-DNA-forming dinucleotide repeat[2,3].
It has been proposed that NRAMP1 polymorphisms play a role in susceptibility to mycobacterial infections[4,5]. To date, studies have yielded contradictory results[2]. In a West African population, NRAMP1 variants have been associated with susceptibility to Mycobacterium tuberculosis[6]. Malik et al[7] reported that variants of NRAMP1 are associated with tuberculosis (TB) in children. NRAMP1 gene polymorphisms have been linked with genetic susceptibility to infection with M. tuberculosis and progression of TB into severe clinical forms in eastern China[8]. Abe et al[9] reported that genetic variation in the NRAMP1 gene is associated with tuberculous cavitation of the lungs in Japanese patients. Kim et al[10] found that NRAMP1 polymorphisms are associated with tuberculous pleurisy.
Various studies have been carried out to establish a connection between NRAMP1 variants and diseases caused by mycobacteria other than the TB bacilli, such as Mycobacterium ulcerans causing Buruli ulcers[11] and Mycobacterium leprae[12]. A recent study[5] showed that the NRAMP1 promoter region polymorphism are positively associated with leprosy but not with the Mitsuda reaction (intradermal injection of lepromin), and that variants of the NRAMP1 gene favour microbial survival inside the macrophages by blocking the efficient transport of iron. It was reported that the NRAMP1 gene may have a role in some autoimmune diseases, including rheumatoid arthritis (RA)[13]. It was also reported that NRAMP1 823 C/C prevents the development of rheumatoid nodules in RA patients[14], type 1 diabetes[15] and multiple sclerosis[16]. Variants of the NRAMP1 gene have been associated with improved response to Bacillus Calmette-Guerin immunotherapy for superficial bladder cancer[17]. Hofmeister et al showed that NRAMP1 variants are specifically associated with Crohn’s disease (CD)[18].
In a previous study, we found that CD is associated with polymorphisms of the NOD2 gene and the pathogen MAP in Sardinian patients[19]. NOD2 protein is an intracellular protein that activates NFkB upon binding to microbial peptidoglycan. MAP is the etiological agent of Johne’s disease, a granulomatous enteritis of ruminants and other monogastric animals[20]. Although the association between MAP and CD (a human equivalent of paratuberculosis) has been postulated for a long time, only recent studies have made improvements in isolation and genomic techniques have allowed this link to be firmly established in a number of different populations[21-24].
As a part of our ongoing studies with NOD2/CARD15, we also looked at the role of other genes, especially NRAMP1, which may be associated with the survival of intracellular pathogens such as MAP. The role of NRAMP1 in regulating microbial survival inside phagosomes is related to iron transport, although the mechanism has not yet been completely elucidated[5,25].
MATERIALS AND METHODS
Patients were recruited at the University of Sassari after giving written consent. Using a case-control design, we analyzed 37 CD patients and 34 controls with no inflammatory bowel disease (IBD).
We searched for polymorphisms in 6 loci previously reported to be the most significantly associated with IBD[13]. The SCL11A1 polymorphisms that we analyzed included a (GT)n microsatellite in the promoter region, (-)237 C/T; 469 + 14G/C, INT4; a non-conservative base substitution at codon 543 (D543N); 823 C/T; and a 4-bp TGTG deletion locating 55 nucleotides downstream of the last codon in exon 15 (1729 + del55del4).
Amplification was performed as previously reported[13] using 100 ng of template genomic DNA previously extracted from intestinal tissues. The primer sequences that we used were previously reported[13]. Detection of MAP also was performed as previously reported[24].
Logistic regression was used to calculate odds ratios (OR) for the associations among CD, presence of MAP, and the 6 loci described above. Saturated and reduced models were computed and their comparability was assessed using the likelihood ratio test. The reduced model was assessed for goodness of fit using the covariate patterns as groups. In addition, the actual and model-predicted probabilities of CD according to levels of the independent variables in the final model were calculated.
RESULTS
Table 1 shows the full and reduced logistic regression models computed for this study. The full model showed significant associations between CD and the presence of MAP, the 1729 + 55del4 deletion polymorphism and the 823 C/T CT polymorphism. No significant associations were found between CD and the INT4, D534R, GT (n) and (-) 237 C/T loci. These non-significant loci were dropped in the reduced model and likelihood ratio testing showed no difference between the full and reduced models (likelihood ratio test: P = 0.436). Furthermore, the reduced model showed good fit (Pearson goodness-of-fit test: P = 0.407) and predictive power. We tested for significant interactions between MAP infection and each of the genetic loci, none of which was statistically significant. We therefore selected the reduced main effect model as our final model. No association was found between MAP and the 823 C/T or 1729 + 55del4 polymorphisms.
Table 1.
Full and reduced logistic regression main effects models for the association between Crohn's disease, MAP infection, and the presence of a number of genetic polymorphisms of the NRAMP1 gene
|
Full model |
Reduced model2 |
||||
| Odds Ratio | P | Odds Ratio | P | ||
| MAP | Negative | 1.01 | 1.01 | ||
| Positive | 45.5 | < 0.001 | 42.4 | < 0.001 | |
| 1729 + 55del4 | +/+ | 1.01 | 1.01 | ||
| +/DEL, DEL/DEL | 5.6 | 0.014 | 5.6 | 0.011 | |
| 823 C/T | CC | 1.01 | 1.01 | ||
| CT | 75.6 | 0.001 | 50.8 | 0.001 | |
| D534R | GG | 1.01 | |||
| GC | 1.1 | 0.913 | |||
| (-)237C/T | CC | 1.01 | |||
| CT, TT | 0.5 | 0.350 | |||
| INT4 | GG | 1.01 | |||
| GC, CC | 2.5 | 0.224 | |||
| GT (n) | Allele1 | 1.01 | |||
| Allele2 | 1.2 | 0.807 | |||
| Allele3 | 0.6 | 0.673 | |||
Reference category;
Model statistics: Likelihood ratio test for equivalence with full model: P = 0.436; Pearson goodness-of-fit test: P = 0.407; Area under Receiver-Operating Characteristic curve: 0.886.
Figure 1 shows the actual and final model-predicted probabilities of CD by MAP infection and the two statistically significant loci, 823 C/T and 1729 + 55del4. The close similarity between the actual and predicted probabilities reflected the good fit of our final model to the data. Strong independent effects of MAP infection and each of the loci on the probability of CD were observed (Figure 1). Even with no mutant polymorphism, MAP infection was highly predictive of CD. Having the CT polymorphism at the 823 locus, the probability of CD was greatly increased both among patients infected with MAP and among patients not infected with MAP. The effect of deletions at the 1729 locus was more moderate. It should be noted that these probabilities were sensitive to the ratio of cases to controls in the study, which were shown here to illustrate the particularly strong association among CD, MAP infection and the 823 C/T locus as well as the close fit between the experimental data and our chosen statistical model.
Figure 1.
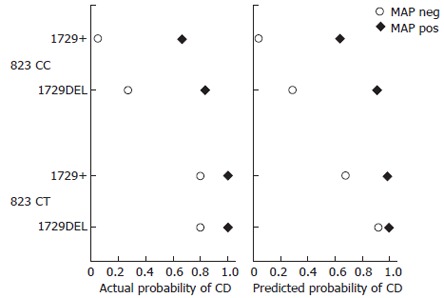
Actual probability of CD and predicted probability of CD showing strong independent effects of MAP infection and each of the loci on the probability of CD.
DISCUSSION
For the first time, a strong association has been observed between CD and polymorphisms at the 823C/T and 1729 + 55del4 loci in the NRAMP1 gene in Sardinians.
Although the combination of MAP infection and 823 CT mutation could perfectly predict CD, generalizations were limited by the small sample size in our study. Only 8 patients in this study simultaneously had MAP infection, 823 CT mutation, and CD. It would be worthwhile to follow up this finding in a larger study to determine if these two factors are a useful prognostic index for the development of CD.
It was reported that polymorphisms in the NOD2 gene and the presence of MAP are strongly associated with CD[19]. Both NOD2 polymorphisms and MAP infection had a strong independent association with CD, and are also associated with one another, suggesting that susceptibility to MAP infection may be influenced by NOD2 polymorphisms[26]. NOD2 protein is involved in the activation of NFkB factor and failure of its priming signal causes failure of pathogen clearance, possibly explaining the abnormal adaptive immune response to pathogens[27].
Our findings agree and disagree with some previous genetic studies of various NRAMP1 loci and CD. Our finding which is lacking of association between GT (n) alleles and CD is consistent with the failure to find an association between any of these alleles and IBD in Americans[28]. Stokkers et al[29] found that mutations at the 823C/T or (-) 274C/T loci mutations are not associated with CD, which is in agreement with our findings at (-) 274C/T locus in this study. However, we have found a very strong association between CD and the 823CT polymorphism.
Although previous studies have suggested that NRAMP1 mutations may favour microbial survival, the fact that this study failed to find any association between NRAMP1 polymorphisms and MAP infection does not support that viewpoint. Interestingly, although a previous study[6] showed allele 2 of this polymorphism is associated with mycobacterial infections, we did not find such an association between this allele and MAP infection.
The lack of association between NRAMP1 polymorphisms and MAP infection is particularly curious because NRAMP1 is thought to directly impact microbial survival inside of macrophages. NRAMP1 is thought to either deny needed iron in intraphagosomal microbes, or to increase transphagosomal iron needed to create bactericidal hydroxyl radicals via the Haber-Weiss/Fenton reaction[25].
The 823CT and 1729 + 55del4 polymorphisms have been associated with autoimmune disease[14], but not with another mycobacterial disease, tuberculosis[30], which probably does not have an autoimmune etiology. In the present study, these polymorphisms were found to be associated with CD which probably does have an autoimmune component, but not associated with MAP infection. These findings, combined with the strong association between MAP infection and CD, suggest that NRAMP1 polymorphisms may not cause CD by affecting the survival of MAP in intestinal tissue, but rather work together with MAP infection to cause CD via an autoimmune mechanism. In other words, one possible interpretation may be that these data support the aetiology of CD with both infectious and autoimmune components. At least, one study has elucidated a mechanism by which NRAMP1 affects the survival of intracellular pathogens and is simultaneously involved in autoimmune responses[25].
A further step of the study is to test the derived model on a large cohort of patients and controls.
In conclusion, CD may be strongly influenced by both NRAMP1 and MAP, although NRAMP1 polymorphisms and MAP infection are not themselves correlated. Combined with previous work on the NOD2/CARD15 gene, a complex interplay of genetic, immunologic and infectious factors may play a role in the aetiology of CD.
ACKNOWLEDGMENTS
The authors are thankful to Professor Giovanni Fadda for his guidance and support. The authors are also thankful to the International Society for Genomic and Evolutionary Microbiology (ISOGEM) for supporting and endorsing the study.
References
- 1.Canonne-Hergaux F, Gruenheid S, Govoni G, Gros P. The Nramp1 protein and its role in resistance to infection and macrophage function. Proc Assoc Am Physicians. 1999;111:283–289. doi: 10.1046/j.1525-1381.1999.99236.x. [DOI] [PubMed] [Google Scholar]
- 2.Blackwell JM, Barton CH, White JK, Searle S, Baker AM, Williams H, Shaw MA. Genomic organization and sequence of the human NRAMP gene: identification and mapping of a promoter region polymorphism. Mol Med. 1995;1:194–205. [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell JM, Searle S. Genetic regulation of macrophage activation: understanding the function of Nramp1 (=Ity/Lsh/Bcg) Immunol Lett. 1999;65:73–80. doi: 10.1016/s0165-2478(98)00127-8. [DOI] [PubMed] [Google Scholar]
- 4.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. 2003;4:4–11. doi: 10.1038/sj.gene.6363915. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira FR, Goulart LR, Silva HD, Goulart IM. Susceptibility to leprosy may be conditioned by an interaction between the NRAMP1 promoter polymorphisms and the lepromin response. Int J Lepr Other Mycobact Dis. 2004;72:457–467. doi: 10.1489/1544-581X(2004)72<457:STLMBC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 6.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 7.Malik S, Abel L, Tooker H, Poon A, Simkin L, Girard M, Adams GJ, Starke JR, Smith KC, Graviss EA, et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc Natl Acad Sci USA. 2005;102:12183–12188. doi: 10.1073/pnas.0503368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Shao L, Weng X, Hu Z, Jin A, Chen S, Pang M, Chen ZW. Variants of the natural resistance-associated macrophage protein 1 gene (NRAMP1) are associated with severe forms of pulmonary tuberculosis. Clin Infect Dis. 2005;40:1232–1236. doi: 10.1086/428726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe T, Iinuma Y, Ando M, Yokoyama T, Yamamoto T, Nakashima K, Takagi N, Baba H, Hasegawa Y, Shimokata K. NRAMP1 polymorphisms, susceptibility and clinical features of tuberculosis. J Infect. 2003;46:215–220. doi: 10.1053/jinf.2002.1064. [DOI] [PubMed] [Google Scholar]
- 10.Kim JH, Lee SY, Lee SH, Sin C, Shim JJ, In KH, Yoo SH, Kang KH. NRAMP1 genetic polymorphisms as a risk factor of tuberculous pleurisy. Int J Tuberc Lung Dis. 2003;7:370–375. [PubMed] [Google Scholar]
- 11.Stienstra Y, van der Werf TS, Oosterom E, Nolte IM, van der Graaf WT, Etuaful S, Raghunathan PL, Whitney EA, Ampadu EO, Asamoa K, et al. Susceptibility to Buruli ulcer is associated with the SLC11A1 (NRAMP1) D543N polymorphism. Genes Immun. 2006;7:185–189. doi: 10.1038/sj.gene.6364281. [DOI] [PubMed] [Google Scholar]
- 12.Abel L, Sánchez FO, Oberti J, Thuc NV, Hoa LV, Lap VD, Skamene E, Lagrange PH, Schurr E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 13.Yang YS, Kim SJ, Kim JW, Koh EM. NRAMP1 gene polymorphisms in patients with rheumatoid arthritis in Koreans. J Korean Med Sci. 2000;15:83–87. doi: 10.3346/jkms.2000.15.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CH, Cho CL, Tsai WC, Ou TT, Wu CC, Yen JH, Liu HW. Inhibitors of kB-like gene polymorphisms in rheumatoid arthritis. Immunol Lett. 2006;105:193–197. doi: 10.1016/j.imlet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Satoh J, Kojima Y, Negoro K, Hirai M, Hinokio Y, Kinouchi Y, Suzuki S, Matsuura N, Shimosegawa T, et al. Promoter polymorphism of SLC11A1 (formerly NRAMP1) confers susceptibility to autoimmune type 1 diabetes mellitus in Japanese. Tissue Antigens. 2004;63:231–236. doi: 10.1111/j.1399-0039.2004.000172.x. [DOI] [PubMed] [Google Scholar]
- 16.Comabella M, Altet L, Peris F, Villoslada P, Sánchez A, Montalban X. Genetic analysis of SLC11A1 polymorphisms in multiple sclerosis patients. Mult Scler. 2004;10:618–620. doi: 10.1191/1352458504ms1097oa. [DOI] [PubMed] [Google Scholar]
- 17.Decobert M, Larue H, Bergeron A, Harel F, Pfister C, Rousseau F, Lacombe L, Fradet Y. Polymorphisms of the human NRAMP1 gene are associated with response to bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J Urol. 2006;175:1506–1511. doi: 10.1016/S0022-5347(05)00653-1. [DOI] [PubMed] [Google Scholar]
- 18.Hofmeister A, Neibergs HL, Pokorny RM, Galandiuk S. The natural resistance-associated macrophage protein gene is associated with Crohn's disease. Surgery. 1997;122:173–18; discussion 173-18;. doi: 10.1016/s0039-6060(97)90006-4. [DOI] [PubMed] [Google Scholar]
- 19.Sechi LA, Gazouli M, Ikonomopoulos J, Lukas JC, Scanu AM, Ahmed N, Fadda G, Zanetti S. Mycobacterium avium subsp. paratuberculosis, genetic susceptibility to Crohn's disease, and Sardinians: the way ahead. J Clin Microbiol. 2005;43:5275–5277. doi: 10.1128/JCM.43.10.5275-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chacon O, Bermudez LE, Barletta RG. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu Rev Microbiol. 2004;58:329–363. doi: 10.1146/annurev.micro.58.030603.123726. [DOI] [PubMed] [Google Scholar]
- 21.Bull TJ, McMinn EJ, Sidi-Boumedine K, Skull A, Durkin D, Neild P, Rhodes G, Pickup R, Hermon-Taylor J. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J Clin Microbiol. 2003;41:2915–2923. doi: 10.1128/JCM.41.7.2915-2923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci USA. 2005;102:12344–12349. doi: 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet. 2004;364:1039–1044. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- 24.Sechi LA, Scanu AM, Molicotti P, Cannas S, Mura M, Dettori G, Fadda G, Zanetti S. Detection and Isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn's disease in Sardinia. Am J Gastroenterol. 2005;100:1529–1536. doi: 10.1111/j.1572-0241.2005.41415.x. [DOI] [PubMed] [Google Scholar]
- 25.Alter-Koltunoff M, Ehrlich S, Dror N, Azriel A, Eilers M, Hauser H, Bowen H, Barton CH, Tamura T, Ozato K, et al. Nramp1-mediated innate resistance to intraphagosomal pathogens is regulated by IRF-8, PU.1, and Miz-1. J Biol Chem. 2003;278:44025–44032. doi: 10.1074/jbc.M307954200. [DOI] [PubMed] [Google Scholar]
- 26.Sieswerda LE, Bannatyne RM. Mapping the effects of genetic susceptibility and Mycobacterium avium subsp. paratuberculosis infection on Crohn's disease: strong but independent. J Clin Microbiol. 2006;44:1204–1205. doi: 10.1128/JCM.44.3.1204-1205.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, Ahmad T, McGovern DP, Onnie C, Negoro K, Goldthorpe S, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 28.Stokkers PC, de Heer K, Leegwater AC, Reitsma PH, Tytgat GN, van Deventer SJ. Inflammatory bowel disease and the genes for the natural resistance-associated macrophage protein-1 and the interferon-gamma receptor 1. Int J Colorectal Dis. 1999;14:13–17. doi: 10.1007/s003840050177. [DOI] [PubMed] [Google Scholar]
- 29.Crawford NP, Eichenberger MR, Colliver DW, Lewis RK, Cobbs GA, Petras RE, Galandiuk S. Evaluation of SLC11A1 as an inflammatory bowel disease candidate gene. BMC Med Genet. 2005;6:10. doi: 10.1186/1471-2350-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvaraj P, Chandra G, Kurian SM, Reetha AM, Charles N, Narayanan PR. NRAMP1 gene polymorphism in pulmonary and spinal tuberculosis. Current Science. 2002;82:451–454. [Google Scholar]


