Abstract
AIM: To compare gene expression profiles of pancreatic adenocarcinoma tissue specimens, human pancreatic and colon adenocarcinoma and leukemia cell lines and normal pancreas samples in order to distinguish differentially expressed genes and to validate the differential expression of a subset of genes by quantitative real-time RT-PCR (RT-QPCR) in endoscopic ultrasound-guided fine needle aspiration (EUS-guided FNA) specimens.
METHODS: Commercially dedicated cancer cDNA macroarrays (Atlas Human Cancer 1.2) containing 1176 genes were used. Different statistical approaches (hierarchical clustering, principal component analysis (PCA) and SAM) were used to analyze the expression data. RT-QPCR and immunohistochemical studies were used for validation of results.
RESULTS: RT-QPCR validated the increased expression of LCN2 (lipocalin 2) and for the first time PLAT (tissue-type plasminogen activator or tPA) in malignant pancreas as compared with normal pancreas. Immunohistochemical analysis confirmed the increased expression of LCN2 protein localized in epithelial cells of ducts invaded by carcinoma. The analysis of PLAT and LCN2 transcripts in 12 samples obtained through EUS-guided FNA from patients with pancreatic adenocarcinoma showed significantly increased expression levels in comparison with those found in normal tissues, indicating that a sufficient amount of high quality RNA can be obtained with this technique.
CONCLUSION: Expression profiling is a useful method to identify biomarkers and potential target genes. Molecular analysis of EUS-guided FNA samples in pancreatic cancer appears as a valuable strategy for the diagnosis of pancreatic adenocarcinomas.
Keywords: Pancreas, Colon, Adenocarcinoma, Gene expression profiling, Endoscopic ultrasonography, Ultrasound, Fine needle aspiration
INTRODUCTION
Pancreatic cancer remains one of the most deadly tumor types. The 5-year survival rate after diagnosis is less than 3.5%[1]. So far, the difficulties persist to diagnose pancreatic cancer early. Indeed, the molecular mechanisms underlying pancreatic oncogenesis remain partially understood. Several fundamental studies suggest the implication of a number of molecules involved in cell cycling, apoptosis or signal transduction. Unfortunately, the clinical relevance of these molecules is still pending. For example, the screening of the activating mutations of the proto-oncogene K-ras, which are the most frequent alterations observed to date in pancreatic cancer, is not thought to be sufficiently robust as a diagnostic or prognostic marker[2]. The combination of several genetic alterations such as those found in tumor suppressor genes p16, DPC4, p53 did not improve the sensitivity and specificity of K-ras mutation test for the diagnosis[3]. The development of new diagnostic tools is hence crucial for the detection of pancreatic cancer at an early stage.
Large-scale analysis of gene expression has been widely proposed as a powerful method for malignancy diagnosis, predicting invasion and metastasis through the identification of biomarkers. Pancreatic cancer has previously been the focus for such studies[4-8]. However, a rather low concordance between different studies was found in a meta-analysis of several of these studies[9]. There are several possible explanations; one of them being differences in probe design (oligonucleotides or cDNA), support (nylon membranes or glass slides) or the underlying detection technology (fluorescence or radioactivity). No matter what the reason is, there are controversies about the need of multiple well-defined and validated approaches to find a transcriptional “consensus” for a given tissue or cell type.
In this study, we used commercially dedicated macroarrays containing 1176 genes, selected on their functional implication in cancer biology, to study the expression profiles in pancreatic adenocarcinomas (surgical specimens and cell lines). We compared these profiles with those of normal pancreas, other adenocarcinoma cell lines of colon origin as well as with a non-adenocarcinoma leukemia cancer cell line. This array has never been used in expression profiling studies of pancreatic adenocarcinomas.
An important issue is the limited access to pancreatic tissue specimens. Being a minimally invasive technique for patient exploration, endoscopic ultrasound-guided fine needle aspiration (EUS-guided FNA) is now largely used for pancreatic tumor diagnosis[10]. Therefore, to support the clinical relevance of these studies, we determined whether quantification of these markers would be feasible in EUS-guided FNA specimens for prognostic or molecular diagnosis procedures.
MATERIALS AND METHODS
Cell and tissue samples
All cell lines were of human origin and were grown at 37°C, 50 mL/L CO2 in the presence of 10 mL/L fetal calf serum, penicillin/streptomycin (Invitrogen Inc. Carlsbad, CA, USA) and 2 mmol/L L-glutamine (Invitrogen). ASPC-1 (ATCC: CRL-1682), Capan-1 (ATCC: HTB-79), Capan-2 (ATCC: HTB-80), NP29 (kindly provided by Dr. Gabriel Capellá, Barcelona, Spain), HCT 116 (ATCC: CCL-247) and K562 (ATCC: CLL-243) cells were grown in RPMI 1640 medium (Invitrogen). PANC-1 (ATCC: CRL-1469), B × PC-3 (ATCC: CRL-1687), SW480 (ATCC: CLL-228), SW620 (ATCC: CLL-227) and MIAPaCa-2 (ATCC: CRL-1420) cells were grown in Dulbecco’s modified Eagle medium (DMEM), 1.0 g/L glucose. HT-29 cells (ATCC: HTB-38) were maintained in DMEM with 4.5 g/L glucose. Caco-2 cells (ATCC: HTB-37) were grown in RPMI 1640 in the presence of 10 g/L non-essential amino acids. Pancreatic ductal adenocarcinoma specimens were obtained from patients undergoing pancreaticoduodenectomy, after written consent and in accordance with French ethical guidelines. Pancreatic cancer samples were obtained through EUS-guided FNA in patients who have given their written consent. The protocol was approved by the Ethical Committee from Midi-Pyrénées CPPRB-1. Briefly, FNA was performed using GF-UC 30p ultrasound endoscope (Olympus, Rungis, France). Samples of pancreatic cancer tissue were obtained from each patient. The core biopsies were then transferred in Dubosq-Brazil medium and the cellular material remaining in the needle was immediately put in RNA later (Ambion, Woodward Austin, TX, USA). All cases of pancreatic cancer were diagnosed based on histological features. One of the normal pancreatic specimens was obtained from an organ donor. The other two normal pancreatic RNA samples were from Clontech (Palo Alto, CA, USA), each a pool from two individuals.
RNA extraction, cDNA labeling and membrane hybridization
Cells were rinsed in PBS and total RNA was extracted with the RNeasy mini-kit (QIAGEN, Valencia CA, USA). Adenocarcinoma tissues were first grinded mechanically in liquid nitrogen with a pestle in a mortar. Cellular samples obtained by EUS-guided FNA were temporarily stored at -25°C in RNA later (Ambion), and total RNA purified using the RNeasy micro-kit (QIAGEN). The quality and the quantity of the RNA were systematically determined with an Agilent Bioanalyzer 2100. Atlas Pure Total RNA Labeling System (Clontech) and α33P-dATP (-92.5 TBq/mmol) (GE-Amersham, Saclay, France) were used for cDNA target synthesis of 25-40 μg of total RNA. Only targets with a total activity superior to 1 million cpm were used. The membranes (Atlas Human Cancer 1.2, Clontech) were hybridized overnight, washed according to the manufacturer’s recommendations and exposed on phosphor storage screens for generally three days. The screens were then scanned in a phosphorimager. All cell lines were analyzed at least twice, i.e. independent RNA extraction, labeling and hybridization procedures.
Expression data analysis
The scanned images were analyzed with the ImaGene software (V.4.0) (Biodiscovery Inc. Los Angeles, CA). Spots having an intensity inferior to the negative spots (foreign DNA) on the membrane (mean + 2SD) in more than 50% of the experiments in each category were eliminated. By this procedure, 871 genes were retained and used for further analysis. The gene names used are respecting the nomenclature proposed by HUGO. Functions and tissue expression distribution came from the database SOURCE available at http://source.stanford.edu or from the relevant references as indicated. The Genesis software (V1.5.0)[11] was used for clustering analysis. For principal component analysis (PCA) with biplot representation [12] the R software (V.1.9) was used (http://www.r-project.org/). FDR analysis was performed with the Significance Analysis of Microarrays (SAM) software (V1.21): http://www-stat.stanford.edu/~tibs/SAM/.
Real-time RT-QPCR
For quantitative real-time RT-PCR (RT-QPCR) analysis, 3 μg (tumor samples, cell lines, and normal pancreas) or 5-10 ng (cell aspiration) of total RNA were reversely transcribed using Superscript II RNase H- reverse transcriptase (Invitrogen) and random hexamers using standard conditions. Generally, 5% of the RT reaction was used as template for the subsequent RT-QPCR reactions using the SYBR Green technology (Applied Biosystems Foster City, CA, USA) in a GeneAmp 5700 sequence detector system (Applied Biosystems). The expression levels were normalized to 18S ribosomal RNA levels. The sequence, orientation and corresponding exon localization of the primers used are shown in Table 1 as well as the length of the amplicon.
Table 1.
Sense (S) and antisense (AS) primers used in quantitative RT-PCR analyses
| Primer | Exon | Sequence (5’-3’) | Amplicon |
| LCN2 (Lipocalin2) S | 1 | TGATCCCAGCCCCACCT | 74 bp |
| LCN2 (Lipocalin2) AS | 2 | CCACTTCCCCTGAATTGGT | |
| PLAT (tPA) S | 2 | TGGAGAGAAAACCTCTGCGAG | 72 bp |
| PLAT (tPA) AS | 3 | CCATGATTGCTTCACAGCGT | |
| KRT7 (Keratin7) S | 7 | CTCTGTGATGAATTCCACTGGTG | 72 bp |
| KRT7 (Keratin7) AS | 8 | CCCATGGTTCCCCCGA | |
| 18S S | AAACGGCTACCACATCCAAG | 155 bp | |
| 18S AS | CCTCCAATGGATCCTCGTTA |
Immunohistochemical analysis
Paraffin-embedded tissue sections were dewaxed and permeabilized with citrate buffer, pH 6, for 3 min ×5 min in a microwave oven. The slides were rinsed 3 min× 5 min in PBS at room temperature and treated for 10 min with Dako protein block solution (Dako, Glostrup, Denmark). The sections were then incubated with a monoclonal mouse anti-human lipocalin 2 antibody (clone 211-01) (The Antibody Shop, Denmark), (1:100 in PBS, 10 g/L BSA) over night at 4°C. After rinsing (3 min × 5 min in PBS), the sections were treated with 30 mL/L H2O2, 100 mL/L methanol in PBS for 15 min followed by two rinses (2 min × 5 min) in PBS, 10 g/L BSA, and the incubation with an HRP-coupled rabbit anti-mouse IgG antibody (P0161; Dako) (1:50 in PBS, 10 g/L BSA) for 1 h at 20°C. The slides were then rinsed 3 min × 5 min in PBS + 10 g/L BSA and incubated 1 h (20°C) with an HRP-coupled goat anti-rabbit IgG antibody (P0448; Dako) (1:50 in PBS, 10 g/L BSA) and finally rinsed 2 min × 5 min in PBS + 1% BSA prior to development with one drop of AEC + substrate chromogen solution (Dako) for 15 min. The slides were washed in running cold water for 10 min and counterstained with Mayer’s hemalun solution (Merck, Darmstadt, Germany) and mounted with Dako glycerol mounting medium. Pictures were taken with the Visiolab 2000 software using a Nikon eclipse E400 microscope.
RESULTS
Expression profiling
Using Clontech’s Human Cancer 1.2 macroarrays, the expression profile of 1176 genes associated with cancer was studied in 7 human pancreatic adenocarcinoma cell lines (PCL), PANC-1, MIA Paca-2, ASPC-1, BxPC-3, Capan-1, Capan-2, NP29, and 6 tumor samples, PT1-PT6, obtained through surgical resection of pancreatic adenocarcinomas. In order to assess the specificity of the expression of pancreatic tumor cells, 5 human colon cancer cell lines (CCL), SW480, HT-29, HCT116, SW620, Caco-2, 1 leukemia cell line (LCL), K562 and 3 normal human pancreatic tissue samples, NormPanc1-3, were included in the study. After screening of the expression data as described in the material and methods section, 871 genes were retained for further analysis. The expression data are available at the following URL: http://ifr31w3.toulouse.inserm.fr/microArrayPancreas/DataSummary.xls. In a hierarchical cluster analysis of samples based on centered mean expression data, two main clusters could be identified, corresponding to cell lines (up) and tissue samples (down), respectively (Figure 1). Among the clusters identified within the cell lines, one was purely pancreatic (BxPC-3, Capan-1, Capan-2 and NP29) and one comprised only colon cancer cell lines (HT-29, HCT116 and SW620). At the top of the dendrogram, a colon cancer cell line SW480 clustered with two pancreatic cancer cell lines, PANC-1 and MIA PaCa-2. In a hierarchical clustering analysis of the 871 genes retained after filtering, three subclusters (A, B and C) contained genes with an elevated expression in pancreatic tumor samples (PT) and a lower expression in normal pancreas and non-pancreatic cancer cell lines (NormPanc, CCL and LCL) (Figure 2). Subcluster A containing four genes, IFITM1, KRT7, PLAT and LCN2, exhibited an expression profile particularly specific for neoplastic pancreas. These four genes were overexpressed in the majority of PCL and in PT1 to 6, but not in non-pancreatic cell lines and in normal pancreas. Subclusters B and C showed genes overexpressed in all PT, in some pancreatic cancer cell lines but not in normal pancreas.
Figure 1.
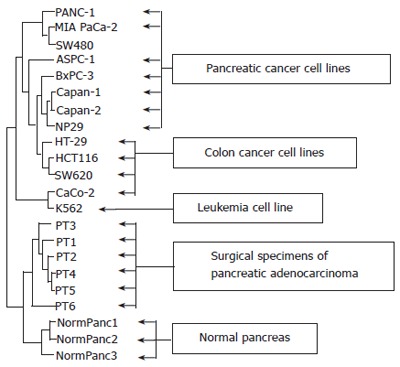
Hierarchical clustering analysis of sample expression profiles. Dendrogram of centered mean expression data using euclidian distance with average linkage clustering, showing relationships in gene expression profiles of samples. Closely related samples are found in the same branch of the tree and a reduced branch height represents a closer relationship between groups.
Figure 2.
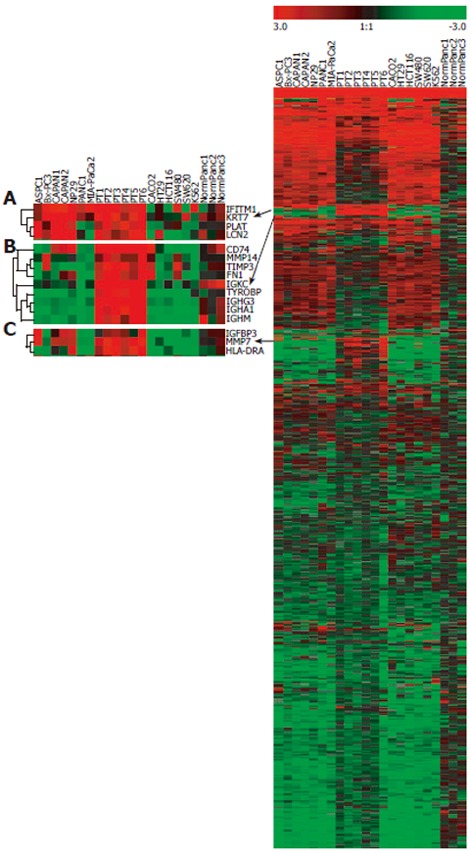
Hierarchical clustering analysis of gene expression profiles. Right: Expression profile of the 871 genes retained after data-filtering. Left: Three magnified subclusters (A-C) showing the genes overexpressed in most of pancreatic tumors. Red color indicates high expression levels; green color indicates low expression levels.
In order to estimate the variability of the experimental procedures, several biological and technical replicates were performed. In a hierarchical clustering analysis, these replicates either clustered directly together or regrouped within clusters of the respective cell line validating our experimental protocols and data acquisition procedures (data not shown). We used the SAM software to detect the genes that were significantly over-expressed in malignant pancreas, referring to the mean expression in pancreatic tumors (PT) and pancreatic cancer cell lines (PCL) as compared with the other groups normal pancreas (NormPanc), colon cancer cell lines (CCL) and leukemia cell line (LCL). Among the genes selected by SAM (FDR < 2%), the ones with an at least 2-fold higher expression level in malignant pancreas are shown in Table 2. In the same table are also represented other expression fold ratios, i.e. the comparison between pancreatic and colon cancer cell lines (column PCL/CCL) as well as the fold ratio between pancreatic tumors and normal pancreas (column PT/NormPanc). The five genes with the highest expression ratios were PLAT, KRT7, CD74, MMP7, and LCN2. The ratio of mean expression level of these genes in PT as compared with that in NormPanc, was found to be equal to or above 3.36, indicating that they discriminate well between adenocarcinoma and normal tissues. The ratio of the gene mean expression level in PCL as compared with that in CCL discriminates between pancreatic and colon cancer cells, which is important to differential diagnosis.
Table 2.
Significantly overexpressed genes (over 2-fold) in malignant pancreas
| Gene symbol | Gene description |
Mean fold expression ratios |
||
| Malignant | PCL/ | PT/ | ||
| Panc/others | CCL | NormPanc | ||
| PLAT | Plasminogen activator, tissue-type | 6.22 | 4.36 | 6.74 |
| KRT7 | Keratin 7 | 5.72 | 4.92 | 3.97 |
| CD74 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | 5.44 | 1.61 | 8.00 |
| MMP7 | Matrix metalloproteinase 7 (matrilysin, uterine) | 5.36 | 3.73 | 5.70 |
| LCN2 | Lipocalin 2 (oncogene 24p3), NGAL | 5.25 | 8.32 | 3.36 |
| HLA-G | HLA-G histocompatibility antigen, class I, G | 4.04 | 1.57 | 4.32 |
| IGHG3 | Immunoglobulin heavy constant gamma 3 | 3.95 | 0.94 | 11.81 |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | 3.70 | 1.57 | 7.01 |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) | 3.50 | 1.61 | 9.20 |
| ITGA3 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | 3.48 | 3.00 | 2.40 |
| ITGB4 | Integrin, beta 4 | 3.36 | 1.16 | 4.34 |
| KRT19 | Keratin 19 | 3.27 | 1.34 | 2.98 |
| CTSD | Cathepsin D (lysosomal aspartyl protease) | 3.21 | 2.75 | 2.89 |
| CASP4 | Caspase 4, apoptosis-related cysteine protease | 3.10 | 2.43 | 1.80 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | 3.08 | 3.53 | 1.87 |
| PLAU | Plasminogen activator, urokinase | 3.08 | 2.83 | 2.01 |
| MMP11 | Matrix metalloproteinase 11 (stromelysin 3) | 2.96 | 1.11 | 7.22 |
| DTR | Diphtheria toxin receptor (heparin-binding epidermal growth factor-like growth factor) | 2.84 | 1.74 | 3.33 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 2.76 | 1.65 | 3.17 |
| LAMA4 | Laminin, alpha 4 | 2.74 | 2.14 | 3.30 |
| ITGB8 | Integrin, beta 8 | 2.59 | 2.31 | 4.44 |
| IFITM1 | Interferon induced transmembrane protein 1 (9-27) | 2.41 | 5.47 | 7.10 |
| AXL | AXL receptor tyrosine kinase | 2.35 | 1.63 | 2.91 |
| ITGAE | Integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) | 2.31 | 1.58 | 3.56 |
| CD59 | CD59 antigen p18-20 (antigen identified by monoclonal antibodies 16.3A5, EJ16, EJ30, EL32 and G344) | 2.26 | 2.11 | 2.95 |
| KRT10 | Keratin 10 (epidermolytic hyperkeratosis; keratosis palmaris et plantaris) | 2.09 | 2.18 | 1.60 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | 2.03 | 2.41 | 1.22 |
| PTGES | Prostaglandin E synthase | 2.03 | 2.84 | 1.11 |
| HIF1A | Hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 2.01 | 1.24 | 2.87 |
Although clustering methods are the most widely used technique to group genes based on expression patterns, PCA is rapidly gaining acceptance in the field of transcriptome analysis. PCA permits data visualization of complex multidimensional datasets by projecting data into a sub-space with 2 or 3 dimensions. Moreover, a biplot representation of PCA results (Figure 3) provides a powerful visual overview of the relationships between genes and samples. The sub-space determined by PCA captures the highest amount of the total variability. In our line and column centered data matrix, the first (PC1) and second (PC2) principal components captured 39% and 13% of the total variability, respectively (Figure 3). In the biplot, samples are shown as arrows, and the genes are shown by their HUGO ID symbols. Arrows of similar length, pointing in the same direction display a higher degree of correlation. As observed in the hierarchical clustering analysis (Figure 1), SW480 was more associated to pancreatic cell lines than to other colon cell lines. Also in agreement with figure 1, the leukemia cell line K562 was correlated with the colon cell line Caco-2. An analysis of the PC1 (horizontal x-axis) revealed a clear distinction between tissue samples and cell lines, with a positive x-coordinate value for all tissue samples and negative for all cell lines. More interestingly, the same analysis of PC2 (vertical y-axis) discriminated malignant pancreas (pancreatic tumors and pancreatic cell lines) from the other samples categories studied, with the exception of SW480.
Figure 3.
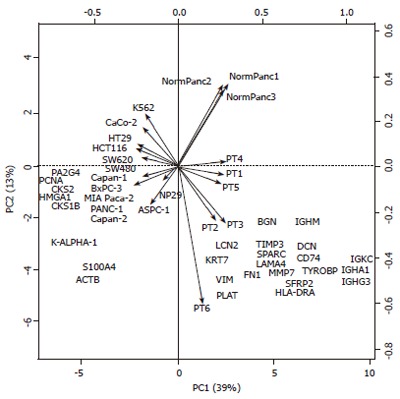
Principal component analysis (PCA) of gene expression data. Biplot resulting from a PCA of the line and column centered data containing 871 genes (individuals) in lines and 22 samples in columns (variables). The 28 genes contributing the most to the total variability are shown. The two principal components (PC1 and PC2) contribute to more than 50% (39% and 13%) of the total variability and resolve four biological sample categories: PC1 on the horizontal x-axis distinguish between cell lines (left) and tissue samples (right) whereas PC2 on the vertical y-axis distinguish between malignant pancreas samples (bottom) and other sample categories (top).
To improve the interpretation, only the 28 genes contributing most to the total variability of the data set are shown in the biplot. The co-localization between a gene and a sample type signifies an overexpression in the given sample type. Accordingly, genes such as PCNA, HMGA1, PA2G4, CKS2 and CKS1B were specifically overexpressed in cell line samples, whereas IGHM, IGKC, IGHA1 and IGHG3, all immunoglobulins, were highly associated with tumor samples. The extreme position of these genes on the horizontal axis in the biplot indicates they are involved in the distinction between cells and tissue samples. The overexpression of genes of the immune response is a characteristic of cancer tissue samples versus cell lines. Furthermore, a set of genes, including PLAT, VIM, HLA-DRA, IGHG3, ACTB, S100A4, and others located in the lower half of the biplot, was involved in the discrimination between malignant pancreas and all other samples. Among them, LCN2, KRT7, VIM and PLAT, appeared to be highly correlated with both pancreatic tumors and pancreatic cell lines. However, in the third principal component (PC3), VIM was positioned far from the three other genes (data not shown). Interestingly, CTRL (chymotrypsin-like), was found to be specifically expressed in normal pancreas and absent in PT or PCL. CTRL is a poorly characterized serine protease with chymotrypsin- and elastase-2-like activities, which is expressed as a pro-protease in normal pancreatic tissue[13]. The absence of CTRL expression in adenocarcinoma samples is in agreement with the absence of other serine protease digestive enzyme, such as chymotrypsin and trypsin, present in normal pancreatic tissue and known to be down-regulated in pancreatic cancer.
Differentially expressed genes in neoplastic pancreas
Taken together, the three statistical methods used in our study (hierarchical clustering, Figure 2, the SAM method, Table 2, and PCA, Figure 3) retained seven genes as differentially overexpressed in malignant pancreas. Among these genes, 3 are involved in the immune response: CD74, HLADRA, and IGHG3, and 4 genes previously suggested as key proteins in pancreatic cell biology or oncogenesis: KRT7, MMP7, and PLAT and LCN2. Among them, three genes LCN2, KRT7 and PLAT were selected for further validation by real-time RT-QPCR. This choice was based on the highly specific expression profile in malignant pancreas (Figure 2; subcluster A) and elevated mRNA levels in the corresponding samples (Table 2) as well as on their specific positioning in the PCA analysis, that is in the interface between the pancreatic tumors and pancreatic cancer cell lines. These studies confirmed significantly elevated levels of LCN2 and PLAT mRNA in malignant pancreas samples, tumors and cancer cell lines (Mann-Whitney test: aP < 0.05) (Table 3). However, they were unable to confirm the macroarray results concerning KRT7 expression levels.
Table 3.
Relative mRNA levels presented as 2[Ct(18S)-Ct(gene of interest)] (mean x 10-2±SEM)
| Gene | n | KRT7 | LCN2 | PLAT |
| Pancreatic tumors | 6 | 1.14. ± 0.47 | 3.58 ± 0.65a | 2.34 ± 0.89a |
| Pancreatic cell lines | 6 | 79.62 ± 90.97 | 188.99 ± 81.13a | 13.51 ± 4.67a |
| Normal pancreas | 3 | 0.42 ± 0.23 | 1.05 ± 0.85 | 0.14 ± 0.08 |
P < 0.05, vs normal pancreas.
The PLAT protein is known to be overexpressed in pancreatic cancer tissues[14]. However, even though several studies have reported elevated LCN2 mRNA levels in pancreatic cancer[15], and LCN2 protein in human pancreatic juice in patients with pancreatic cancer[16], the presence of the LCN2 protein in pancreatic adenocarcinoma is less well characterized. Therefore, in order to evaluate the expression of the lipocalin 2 protein in pancreatic tumors, we performed immunohistochemical analysis using a monoclonal anti-lipocalin 2 antibody. As shown in Figure 4A, little or no labeling was observed in normal pancreas (n = 3). However, in sections of pancreatic adenocarcinomas (n = 5), a strong apical labeling was detected in ducts invaded by carcinoma (Figure 4B and C).
Figure 4.
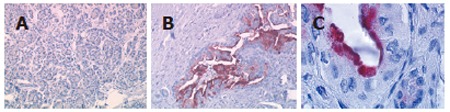
Lipocalin 2 protein expression in pancreatic cancer tissue Immunohistochemical analysis by AEC + substrate chromogen staining. A (x 40): normal pancreas; B (x 200), C (x 1000): pancreatic adenocarcinomas.
LCN2, KRT7 and PLAT transcripts in EUS-guided FNA samples
We investigated the possibility to use EUS-guided FNA as a source of pancreatic sample to study the expression of KRT7, LCN2 and PLAT genes as diagnostic biomarkers. The presence of these mRNAs was therefore examined by RT-QPCR analysis in 12 samples, originating from different individuals, obtained through EUS-guided FNA from patients with pancreatic adenocarcinoma. Compared with normal pancreas, the expression level of the KRT7, LCN2 and PLAT transcripts were 26.7 ± 18.16, 20.6 ± 7.6 and 70.8 ± 35.4 (mean ± SE), respectively. In each case, 18S ribosomal RNA was used for normalization. Even though 10 out of 12 patients presented elevated levels of expression for KRT7 and LCN2 and 11 for PLAT, only the levels of PLAT and LCN2 expression were significantly different from normal pancreas (unilateral Student t test: P < 0.05). These results were thus perfectly concordant with those obtained from pancreatic cancer cell lines and tumors.
DISCUSSION
Large scale analysis of gene expression has been widely proposed as a powerful method for identification of molecular markers of neoplasia and for the generation of novel taxonomies for cancer. Accordingly, several studies, have been conducted to characterize the expression profiles in human pancreatic adenocarcinomas[15,17]. However, to the best of our knowledge, none of these studies report the comparison between pancreatic and colon cancer cell lines. Moreover, this study reports, for the first time, about the use of the dedicated Atlas Human Cancer 1.2 cDNA macroarray containing 1176 cancer-associated genes to study expression profile in pancreatic carcinoma. We report the expression profiles of pancreatic adenocarcinoma cell lines and tumors and compared with that of normal pancreas specimens, colon cancer cell lines and one hematopoietic cell line K562. This was done to differentiate pancreatic and non-pancreatic cancer gene profile in view of differential diagnosis. The fact that several groups have previously reported elevated expression levels of LCN2 mRNA in pancreatic cancer[18-21], was reassuring for the validation of our study design. However, reports validating this overexpression at the protein level are very scarce[22]. Lipocalins are small extracellular proteins with important role in cell proliferation and differentiation, possessing protease inhibitory properties and/or carrying lipophilic ligands such as retinoids and fatty acids into the cells[23]. The localization of LCN2 protein at the apical side of cells from ducts invaded by the tumoral process might be of pathophysiological importance and supports further analysis for a better understanding of the role of this protein in cancer.
Interestingly, the overexpression of PLAT in pancreatic adenocarcinoma has not been reported in previous expression profiling studies. PLAT was recently shown to play a critical role in tumor angiogenesis and in the development of exocrine pancreatic cancer[24], contributing to an invasive phenotype[14,25]. The vast majority of pancreatic adenocarcinomas express KRT7[26,27] and it has previously been indicated as overexpressed in pancreatic cancer by gene expression profiling studies[28]. However, the KRT7 protein is suggested as a marker of normal pancreatic duct epithelial cells playing a role in cell differentiation[29]. The reported expression of KRT7 in both normal and malignant pancreas could, at least in part, explain why we were unable to observe a significant difference in expression levels between both sample types. Among the seven genes retained on our study, three are highly implicated in the immune response: CD74, HLADRA, IGHG3. IGHG3 was only overexpressed in tumor samples suggesting an immune cell origin. In contrast, elevated levels of CD74 and HLADRA were found in tumor samples and also in some pancreatic cancer cell lines. Elevated levels of CD74, and HLADRA were observed in several types of cancers, including gastric cancer and renal epithelial neoplasms[30,31].
The quantitative RT-PCR on EUS-guided FNA samples of PLAT and LCN2, validated the overexpression found by the macroarray analysis. Thus, the quality and the amount of cellular sampling using pancreatic EUS-guided FNA allow the extraction of sufficient quantities of RNA to perform RT-QPCR analysis as a new tool for early diagnosis, as described recently for lymph node metastasis[32]. Furthermore, when performed directly on resected tumor pieces, fine needle aspiration has been shown to produce a relative enrichment of cancer cells, in comparison to tumor samples. This enrichment has been attributed to the capability of epithelial cancer cells to be aspirated more easily than stromal cells[33]. To the best of our knowledge, no gene expression had been evaluated with EUS-guided FNA biopsies from patients with pancreatic adenocarcinoma. EUS-guided FNA is a safe method for patient exploration. Morbidity rate ranges from 1% to 3% when performed by experienced endosonographers[34]. Therefore, identification and quantification of potential molecular markers for pancreatic cancer on cellular samples obtained by EUS-guided FNA could be a promising approach for the diagnosis of solid pancreatic masses. Future studies are needed to prospectively evaluate new molecular biomarkers using this procedure, in order to increase the accuracy of current standard histological and cytological analyses, that was only 80 to 85% in pilot studies[35,36].
ACKNOWLEDGMENTS
The authors would thank J-J Maoret, IFR31, Toulouse, France, for his technical assistance.
Footnotes
Supported by Contrat Université Paul Sabatier, Toulouse, France, ASUPS 2000 (N. Vaysse); AOL DRC Hôpitaux de Toulouse 2001, (L. Buscail); Région Midi-Pyrénées (L. Buscail). H. Laurell was supported by a grant from European Community Plan 99 ECC QLG3-CT-1999-0908 (C. Susini). The Agilent 2100 Bioanalyzer and the phosphoimager (Molecular Dynamics, Sunnyvale, CA, USA) were at the Transcriptome Platform, Toulouse Génopole, and at the molecular biology platform at the Institute Louis Bugnard, IFR31, Toulouse, France, respectively
S- Editor Pan BR L- Editor Ma JY E- Editor Bai SH
References
- 1.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2003;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis M, Van Laethem JL, Parma J, De Maertelaer V, Delhaye M, Devière J. Predictive factors for pancreatic cancer in patients with chronic pancreatitis in association with K-ras gene mutation. Endoscopy. 2004;36:535–542. doi: 10.1055/s-2004-814401. [DOI] [PubMed] [Google Scholar]
- 3.Costentin L, Pagès P, Bouisson M, Berthelémy P, Buscail L, Escourrou J, Pradayrol L, Vaysse N. Frequent deletions of tumor suppressor genes in pure pancreatic juice from patients with tumoral or nontumoral pancreatic diseases. Pancreatology. 2002;2:17–25. doi: 10.1159/000049443. [DOI] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan ZJ, Hu XG, Cao GS, Tang Y. Analysis of gene expression profile of pancreatic carcinoma using cDNA microarray. World J Gastroenterol. 2003;9:818–823. doi: 10.3748/wjg.v9.i4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 8.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 9.Grützmann R, Boriss H, Ammerpohl O, Lüttges J, Kalthoff H, Schackert HK, Klöppel G, Saeger HD, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 10.Kahl S, Malfertheiner P. Role of endoscopic ultrasound in the diagnosis of patients with solid pancreatic masses. Dig Dis. 2004;22:26–31. doi: 10.1159/000078732. [DOI] [PubMed] [Google Scholar]
- 11.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 12.Chapman S, Schenk P, Kazan K, Manners J. Using biplots to interpret gene expression patterns in plants. Bioinformatics. 2002;18:202–204. doi: 10.1093/bioinformatics/18.1.202. [DOI] [PubMed] [Google Scholar]
- 13.Reseland JE, Larsen F, Solheim J, Eriksen JA, Hanssen LE, Prydz H. A novel human chymotrypsin-like digestive enzyme. J Biol Chem. 1997;272:8099–8104. doi: 10.1074/jbc.272.12.8099. [DOI] [PubMed] [Google Scholar]
- 14.Paciucci R, Torà M, Díaz VM, Real FX. The plasminogen activator system in pancreas cancer: role of t-PA in the invasive potential in vitro. Oncogene. 1998;16:625–633. doi: 10.1038/sj.onc.1201564. [DOI] [PubMed] [Google Scholar]
- 15.Brandt R, Grützmann R, Bauer A, Jesnowski R, Ringel J, Löhr M, Pilarsky C, Hoheisel JD. DNA microarray analysis of pancreatic malignancies. Pancreatology. 2004;4:587–597. doi: 10.1159/000082241. [DOI] [PubMed] [Google Scholar]
- 16.Grønborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 17.Grützmann R, Saeger HD, Lüttges J, Schackert HK, Kalthoff H, Klöppel G, Pilarsky C. Microarray-based gene expression profiling in pancreatic ductal carcinoma: status quo and perspectives. Int J Colorectal Dis. 2004;19:401–413. doi: 10.1007/s00384-003-0563-3. [DOI] [PubMed] [Google Scholar]
- 18.Furutani M, Arii S, Mizumoto M, Kato M, Imamura M. Identification of a neutrophil gelatinase-associated lipocalin mRNA in human pancreatic cancers using a modified signal sequence trap method. Cancer Lett. 1998;122:209–214. doi: 10.1016/s0304-3835(97)00391-1. [DOI] [PubMed] [Google Scholar]
- 19.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, et al. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4324. [PubMed] [Google Scholar]
- 20.Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missiaglia E, Blaveri E, Terris B, Wang YH, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Lemoine NR. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 22.Friedl A, Stoesz SP, Buckley P, Gould MN. Neutrophil gelatinase-associated lipocalin in normal and neoplastic human tissues. Cell type-specific pattern of expression. Histochem J. 1999;31:433–441. doi: 10.1023/a:1003708808934. [DOI] [PubMed] [Google Scholar]
- 23.Bratt T. Lipocalins and cancer. Biochim Biophys Acta. 2000;1482:318–326. doi: 10.1016/s0167-4838(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 24.Díaz VM, Planaguma J, Thomson TM, Reventós J, Paciucci R. Tissue plasminogen activator is required for the growth, invasion, and angiogenesis of pancreatic tumor cells. Gastroenterology. 2002;122:806–819. doi: 10.1053/gast.2002.31885. [DOI] [PubMed] [Google Scholar]
- 25.Díaz VM, Hurtado M, Thomson TM, Reventós J, Paciucci R. Specific interaction of tissue-type plasminogen activator (t-PA) with annexin II on the membrane of pancreatic cancer cells activates plasminogen and promotes invasion in vitro. Gut. 2004;53:993–1000. doi: 10.1136/gut.2003.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein NS, Bassi D. Cytokeratins 7, 17, and 20 reactivity in pancreatic and ampulla of vater adenocarcinomas. Percentage of positivity and distribution is affected by the cut-point threshold. Am J Clin Pathol. 2001;115:695–702. doi: 10.1309/1NCM-46QX-3B5T-7XHR. [DOI] [PubMed] [Google Scholar]
- 27.Tot T. Cytokeratins 20 and 7 as biomarkers: usefulness in discriminating primary from metastatic adenocarcinoma. Eur J Cancer. 2002;38:758–763. doi: 10.1016/s0959-8049(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 28.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, et al. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 29.Bouwens L. Cytokeratins and cell differentiation in the pancreas. J Pathol. 1998;184:234–239. doi: 10.1002/(SICI)1096-9896(199803)184:3<234::AID-PATH28>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 30.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Iwashige H, Aridome K, Hokita S, Aikou T. Invariant chain expression in gastric cancer. Cancer Lett. 2001;168:87–91. doi: 10.1016/s0304-3835(01)00503-1. [DOI] [PubMed] [Google Scholar]
- 31.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellisé M, Castells A, Ginès A, Agrelo R, Solé M, Castellví-Bel S, Fernández-Esparrach G, Llach J, Esteller M, Bordas JM, et al. Detection of lymph node micrometastases by gene promoter hypermethylation in samples obtained by endosonography- guided fine-needle aspiration biopsy. Clin Cancer Res. 2004;10:4444–4449. doi: 10.1158/1078-0432.CCR-03-0600. [DOI] [PubMed] [Google Scholar]
- 33.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, et al. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7446. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole D, Palazzo L, Arotçarena R, Dancour A, Aubert A, Hammel P, Amaris J, Ruszniewski P. Assessment of complications of EUS-guided fine-needle aspiration. Gastrointest Endosc. 2001;53:470–474. doi: 10.1067/mge.2001.112839. [DOI] [PubMed] [Google Scholar]
- 35.Voss M, Hammel P, Molas G, Palazzo L, Dancour A, O'Toole D, Terris B, Degott C, Bernades P, Ruszniewski P. Value of endoscopic ultrasound guided fine needle aspiration biopsy in the diagnosis of solid pancreatic masses. Gut. 2000;46:244–249. doi: 10.1136/gut.46.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raut CP, Grau AM, Staerkel GA, Kaw M, Tamm EP, Wolff RA, Vauthey JN, Lee JE, Pisters PW, Evans DB. Diagnostic accuracy of endoscopic ultrasound-guided fine-needle aspiration in patients with presumed pancreatic cancer. J Gastrointest Surg. 2003;7:118–126; discussion 127-128. doi: 10.1016/S1091-255X(02)00150-6. [DOI] [PubMed] [Google Scholar]


