Abstract
AIM: To investigate the role of transglutaminase 3 (TGM3) gene in human esophageal squamous cell carcinoma (ESCC), and analyze its relationship with clinicopathological parameters.
METHODS: Gene expression of TGM3 in fresh esophageal cancer tissues and their corresponding normal mucosas was detected immunohistochemically(IHC) by means of tissue microarray(TMA). Its correlation with clinical characteristics was evaluated and analyzed by univariate analysis. All statistical analyses were performed by SPSS version 10.0.
RESULTS: Expression rate of TGM3 was reduced to 81.8% in ESCC. Expression of TGM3 was significantly inversely correlated with histological grade of esophageal carcinoma (in grade I, II and III tumors, the reduced expression was 4/7, 57/71, and 20/21, respectively, P < 0.05), whereas it had no obvious correlations with lymph node metastasis and depth of invasion.
CONCLUSION: Reduced expression of TGM3 may play an important role in esophageal carcinogenesis.
Keywords: Esophageal squamous cell carcinoma, Immunohistochemically, Tissue microarray
INTRODUCTION
Esophageal cancer ranks among the 10 most frequent cancers in the world, with a predominant distribution in developing countries. Our previous study showed that genetic susceptibility to esophageal cancer was one of the important causes for the high prevalence and familial aggregation of this disease in some areas of northern China[1]. Chen et al observed the down-regulation of TGM3 gene in human ESCC using polymerase chain reaction (RT-PCR) and Northern blot techniques[2]. To our best knowledge, this is the first time to investigate the protein expression of TGM3 in ESCC. The expression levels of TGM3 gene decreased in ESCC tissues compared with their normal counterparts. The results are consistent with that from RT-PCR and Northern blot. Therefore, TGM3 gene might be related to human ESCC and further study may provide insight into the mechanisms of carcinogenesis of esophagus. TGM3 is a member of a family of Ca2+-dependent enzymes, which can catalyze the substrates to form a complex that is related to the differentiation of the stratified squamous epithelia[3]. The gene is localized on chromosome 20q11.2[4]. The full-length cDNA of TGM3 is about 2.6 kb, encoding 692 amino acids. TGM3 as a proenzyme, whose molecular weight is about 77 ku, becomes highly active upon proteolytic cleavage into 50 ku (amino-terminal) and 27 ku species[2].
Tissue microarray was first introduced in 1998[5], which allows simultaneous analysis of many tumors using small-diameter cores sampled from larger blocks of tissue, but may be limited by tumor heterogeneity[6]. This study used TMA for the study of four molecules of interest as prognostic factors in ESCC, including methods for assessing immunocytochemical scoring of microarrays. TMA blocks were constructed from 99 cases of ESCC with corresponding normal tissues, taking four cores from different areas of each tumor and adjacent esophageal epithelia. Immunocytochemical labelling was performed for TGM3. The extent and intensity of scoring was determined for each core and the degree of agreement was determined for results from the assessment of two, three or four cores for each carcinoma. In a subset of 99 representative cases, the labelling in the TMA was compared with that in whole-tissue sections of the same carcinomas. An adequate sample of carcinoma was obtained in more than 90% of the 396 cores. TMA is a reliable tool to demonstrate cellular and molecular alterations in ESCC. We recommend using the mean results from four cores for biological studies, with analysis of categorical data based on quartile groups.
MATERIALS AND METHODS
Materials
Specimens of cancer tissues and matched adjacent normal mucosa were taken from 99 consecutive patients with squamous cell carcinoma of the thoracic esophagus who had undergone esophagectomy with regional lymph node dissected from Oct 2004 to Sept 2005 at the Department of Thoracic Surgery, the First Affiliated Hospital of Anhui Medical University. None of them received irradiation or chemotherapy preoperatively. The patients included 87 men and 12 women with a mean age of 54 (range 36-79) years. Six tumors were located in the upper thorax, 61 in the middle thorax and 32 in the lower thorax (Table 1). The removed specimens were examined histologically with hematoxylin and eosin staining, and then the clinicopathologic stage was determined according to TNM classification.
Table 1.
Clinical and histopathological characteristics of patients
| Characteristics | n=99 (%) |
| Sex | |
| Male | 87 (87.9) |
| Female | 12 (12.1) |
| Location of tumor | |
| Upper thoracic | 6 (6.1) |
| Middle thoracic | 61 (61.6) |
| Lower thoracic | 32 (32.3) |
| Histological grade | |
| I | 7 (7.1) |
| II | 71 (71.7) |
| III | 21 (21.2) |
| Depth of invasion | |
| T1 | 5 (5.1) |
| T2 | 35 (35.4) |
| T3 | 59 (59.6) |
| T4 | 0 (0) |
| Lymph node metastasis | |
| Positive | 32 (32.3) |
| Negative | 67 (67.7) |
Construction of TMA
Archival paraffin-embedded, formalin-fixed tissues are collected. Two pathologists selected representative areas from each donor tumor block, and then punched cores 0.6 mm in diameter, from the donor block. Sections of the resultant tissue microarray were sliced and transferred to glass slides for processing of TGM3 by immunohistochemistry (Pictured is a 120-specimen TMA with 0.6 mm spots).
Immunohistochemical staining
Immunohistochemical analysis was done retrospectively. Resected esophageal specimens, including both tumor and normal mucosae, were fixed in a 40 g/L formaldehyde solution and embedded in paraffin. The following antibody was used in this study: goat polyclonal anti-human TGM3 antibody (Santa Cruz Biotech Co, USA) diluted 1:100 in PBS. Four μm thick sections of formalin-fixed paraffin-embedded tissue blocks of esophageal tumors were sliced. TMAs were deparaffinized, dehydrated and blocked to remove endogenous peroxidase activity by 3 mL/L H2O2 in methanol for 30 min. TMAs were treated with microwave in 0.1 mol/L citrate buffer pH 6.0 at 96°C for 15 min. After incubation with 100 mL/L normal goat serum to block non-specific binding, they were then incubated with the primary antibodies overnight at 4°C. After antibody was washed with PBS, TMAs were incubated with the secondary antibody and the third antibody (Streptavidin/HRP) according to the manufacturer’s instructions, and finally DAB was visualized. Tissues were counterstained with hematoxylin. Negative control was designed by using PBS instead of primary antibody.
Evaluation of staining
The cytoplasmic expression of TGM3 in the ESCC was evaluated as described previously and scored as follows: (a) 0, < 5% of the epithelial cells in the respective lesions; (b) 1, 5%-25% of the epithelial cells in the respective lesions; (c) 2, 26%-50% of the epithelial cells in the respective lesions; (d) 3, 51%-75% of the epithelial cells in the respective lesions; and (e) 4, >75% of the epithelial cells in the respective lesions. The intensity was graded as follows: (a) 0, negative; (b) 1+, weak; (c) 2+, moderate; and (d) 3+, strong (as intense as normal mucosa). A final score between 0 and 12 was achieved by multiplication of the extent of positivity and intensity[7,8]. Scores of 8-12 were defined as “strong expression”, scores of 3-7 were defined as “reduced expression”, and scores of 0-3 were defined as “markedly reduced expression”. Positive staining more than 5% in cell cytoplasm was defined as positive staining; less than 50% in cell cytoplasm was defined as reduced expression; more than 50% in cell cytoplasm was defined as preserved expression[9].
Statistical analysis
χ2 test or Fisher’s exact probability test and Spearman rank correlation coefficient analysis were used to assess the association between immunohistochemical features and clinicopathological characteristics. A P value less than 0.05 was considered statistically significant. All the statistical analyses were performed using the SPSS 10.0 V for Windows.
RESULTS
Expression of TGM3 in esophageal squamous cell carcinoma
TGM3 positive expression showed brown stained signals in the normal mucosa cytoplasm and reduced or positive expression in ESCC, with only a small number of expressions in cell membranes. No nuclear expression of TGM3 was observed in cells. The positive expression rate of TGM3 in 99 esophageal cancer patients was 60.6% (60/99). Significant positive correlation was found in TGM3 expression of the cases between paired normal and cancerous tissue of esophageal carcinoma (P < 0.05). The reduced expression rate of TGM3 was 81.8% (81/99) (Figure 1).
Figure 1.
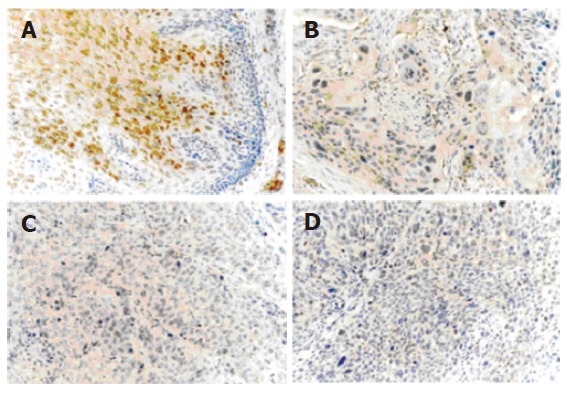
Immunohistochemical analysis of TGM3 in paired ESCC tissue samples using anti- TGM3 antibody (1:100). Diffuse and strong staining was detected in the cytoplasm of the normal epithelial cells (A), while sporadic and weak staining was observed in the cytoplasm of matched esophageal cancer epithelial cells (B: well-differentiated, C: moderately-differentiated, D: poorly-differentiated) (original magnification × 200).
Relationship between TGM3 expression and clinicopathologic variables in esophageal squamous cell carcinoma
Expression of TGM3 correlated significantly only with histological grade of esophageal squamous cell carcinoma. Significant inverse correlation existed between the intensity of TGM3 expression and histological grade (P < 0.05). No significant correlation was found between abnormal expression of TGM3 and lymph node metastasis and depth of invasion (Table 2).
Table 2.
Relationship between clinicopathological parameters and expression of TGM3
| Type | Cases |
TGM3 |
P | |
| Preserved | Reduced | |||
| Histological grade | ||||
| I | 7 | 3 | 4 | < 0.05 |
| II | 71 | 14 | 57 | |
| III | 21 | 1 | 20 | |
| Depth of invasion | ||||
| Mucous layer | 5 | 0 | 5 | > 0.05 |
| Muscular layer | 35 | 5 | 30 | |
| Full-thickness | 59 | 13 | 46 | |
| Lymph node metastases | ||||
| Positive | 32 | 6 | 26 | > 0.05 |
| Negative | 67 | 12 | 55 | |
DISCUSSION
Transglutaminase(TGM) enzymes are widespread in both plants and animals[10,11]. They catalyze the formation of anisodipeptide cross-linking between the ε-NH2 side chain of a protein-bound lysine residue and the γ-amide side chain of aprotein-bound glutamine residue, thereby forming an insoluble macromolecular aggregate that is used for a variety of cellular functions. To date, there are nine known TGM enzymes encoded in the human genome[12], and interestingly, three of them are active in the epidermis and its appendages. These include: the TGM1 enzyme[13] which can function as membrane-associated[14], soluble full-length,and soluble proteolytically activated processed forms in the epidermis[15]; the soluble, “tissue” TGM2 enzyme[16], and the soluble TGM3 proenzyme, which also requires proteolytic activation[17,18]. The TGM3 enzyme is expressed during the late stages of the terminal differentiation of the epidermis and in certain cell types of the hair follicle[19]. The enzyme is thought to be critically involved in the cross-linking of structural proteins and in the formation of the cornified cell envelope, thereby contributing to rigid structures that play vital roles in shape determination and/or barrier functions[20-22]. Although TGM3 mRNA represents less than 2% of the TGM transcripts, the activated TGM3 accounts for up to 75% of the total TGM activity in mammalian epidermis[15].
The understanding of the molecular basis of tumor development has progressed dramatically in the last two decades. It is well known that tumor is essentially a genetic disease. Therefore it is important to demonstrate what these oncogenes are and how they work in carcinogenesis. Identifying the genetic differences between normal and tumor cells or tissues will help discover the genes that directly cause tumor or are associated with tumorigenesis and provide novel markers for early detection and appropriate therapy.
Although the role of TGM3 has been well established in skin keratinocytes, little information is available concerning its involvement in esophageal epithelia. In previous study, TGM3 gene showed down-regulation in human ESCC tissues[2,23]. To verify this differential expression, we first investigated immunohistochemically the expression of TGM3 protein in paired ESCC by means of TMA. The results of IHC revealed that TGM3 reduced expression in 81.8% (81/99) examined tumor tissues relative to the corresponding normal tissues. Among 99 esophageal tumors examined, which were histologically squamous-cell carcinomas, seven tumors were in grade I, 71 were in grade II and 21 were in grade III. Our data showed that decreased TGM3 expression was uncommon in 3 phases of primary esophageal cancer. This is in agreement with previous studies on a variety of cancers, such as laryngeal carcinoma[24], head and neck squamous cell carcinoma[25], oral squamous cell carcinoma from leukoplakia[26]. In all these studies, reduction or loss of TGM3 expression was significantly associated with dedifferentiation, increased invasiveness, and high incidence of lymph node metastasis, hematogenous recurrence and poor prognosis in a number of human carcinomas. In our investigation, 81.8% of ESCC showed reduced expression of TGM3. As a marker associated with squamous cell histological grade, the level of TGM3 expression had an inverse correlation with histological grade, but not with lymphatic metastases and depth of tumor invasion. TGM3, therefore, is one of the most important adhesion molecules expressed by epithelial cells and is regarded as an invasion suppressor molecule. Further investigations are needed to predict the prognostic value of TGM3 for esophageal tumor patients.
ACKNOWLEDGMENTS
We thank Hao Li for paraffin-embedded, formalin-fixed tissues used in this work. We are grateful to Min Zhao for construction of TMA.
Footnotes
Supported by National Key Program on Basic Research, No. 2004CB518604
S- Editor Wang J L- Editor Zhu LH E- Editor Bi L
References
- 1.Zhang W, Bailey-Wilson JE, Li W, Wang X, Zhang C, Mao X, Liu Z, Zhou C, Wu M. Segregation analysis of esophageal cancer in a moderately high-incidence area of northern China. Am J Hum Genet. 2000;67:110–119. doi: 10.1086/302970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen BS, Wang MR, Xu X, Cai Y, Xu ZX, Han YL, Wu M. Transglutaminase-3, an esophageal cancer-related gene. Int J Cancer. 2000;88:862–865. doi: 10.1002/1097-0215(20001215)88:6<862::aid-ijc4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Ahvazi B, Boeshans KM, Idler W, Baxa U, Steinert PM, Rastinejad F. Structural basis for the coordinated regulation of transglutaminase 3 by guanine nucleotides and calcium/magnesium. J Biol Chem. 2004;279:7180–7192. doi: 10.1074/jbc.M312310200. [DOI] [PubMed] [Google Scholar]
- 4.Wang M, Kim IG, Steinert PM, McBride OW. Assignment of the human transglutaminase 2 (TGM2) and transglutaminase 3 (TGM3) genes to chromosome 20q11.2. Genomics. 1994;23:721–722. doi: 10.1006/geno.1994.1571. [DOI] [PubMed] [Google Scholar]
- 5.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 6.Gomaa W, Ke Y, Fujii H, Helliwell T. Tissue microarray of head and neck squamous carcinoma: validation of the methodology for the study of cutaneous fatty acid-binding protein, vascular endothelial growth factor, involucrin and Ki-67. Virchows Arch. 2005;447:701–709. doi: 10.1007/s00428-005-0002-7. [DOI] [PubMed] [Google Scholar]
- 7.Sarbia M, Loberg C, Wolter M, Arjumand J, Heep H, Reifenberger G, Gabbert HE. Expression of Bcl-2 and amplification of c-myc are frequent in basaloid squamous cell carcinomas of the esophagus. Am J Pathol. 1999;155:1027–1032. doi: 10.1016/S0002-9440(10)65203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao XP, Pretlow TG, Rao JS, Pretlow TP. Beta-catenin expression is altered in human colonic aberrant crypt foci. Cancer Res. 2001;61:8085–8088. [PubMed] [Google Scholar]
- 9.Lin YC, Wu MY, Li DR, Wu XY, Zheng RM. Prognostic and clinicopathological features of E-cadherin, alpha-catenin, beta-catenin, gamma-catenin and cyclin D1 expression in human esophageal squamous cell carcinoma. World J Gastroenterol. 2004;10:3235–3239. doi: 10.3748/wjg.v10.i22.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 11.Haroon ZA, Hettasch JM, Lai TS, Dewhirst MW, Greenberg CS. Tissue transglutaminase is expressed, active, and directly involved in rat dermal wound healing and angiogenesis. FASEB J. 1999;13:1787–1795. doi: 10.1096/fasebj.13.13.1787. [DOI] [PubMed] [Google Scholar]
- 12.Grenard P, Bates MK, Aeschlimann D. Evolution of transglutaminase genes: identification of a transglutaminase gene cluster on human chromosome 15q15. Structure of the gene encoding transglutaminase X and a novel gene family member, transglutaminase Z. J Biol Chem. 2001;276:33066–33078. doi: 10.1074/jbc.M102553200. [DOI] [PubMed] [Google Scholar]
- 13.Kim IG, McBride OW, Wang M, Kim SY, Idler WW, Steinert PM. Structure and organization of the human transglutaminase 1 gene. J Biol Chem. 1992;267:7710–7717. [PubMed] [Google Scholar]
- 14.Chakravarty R, Rice RH. Acylation of keratinocyte transglutaminase by palmitic and myristic acids in the membrane Anchorage region. J Biol Chem. 1989;264:625–629. [PubMed] [Google Scholar]
- 15.Kim SY, Chung SI, Steinert PM. Highly active soluble processed forms of the transglutaminase 1 enzyme in epidermal keratinocytes. J Biol Chem. 1995;270:18026–18035. doi: 10.1074/jbc.270.30.18026. [DOI] [PubMed] [Google Scholar]
- 16.Gentile V, Saydak M, Chiocca EA, Akande O, Birckbichler PJ, Lee KN, Stein JP, Davies PJ. Isolation and characterization of cDNA clones to mouse macrophage and human endothelial cell tissue transglutaminases. J Biol Chem. 1991;266:478–483. [PubMed] [Google Scholar]
- 17.Kim IG, Lee SC, Lee JH, Yang JM, Chung SI, Steinert PM. Structure and organization of the human transglutaminase 3 gene: evolutionary relationship to the transglutaminase family. J Invest Dermatol. 1994;103:137–142. doi: 10.1111/1523-1747.ep12392470. [DOI] [PubMed] [Google Scholar]
- 18.Lichti U, Ben T, Yuspa SH. Retinoic acid-induced transglutaminase in mouse epidermal cells is distinct from epidermal transglutaminase. J Biol Chem. 1985;260:1422–1426. [PubMed] [Google Scholar]
- 19.Kim IG, Gorman JJ, Park SC, Chung SI, Steinert PM. The deduced sequence of the novel protransglutaminase E (TGase3) of human and mouse. J Biol Chem. 1993;268:12682–12690. [PubMed] [Google Scholar]
- 20.Marvin KW, George MD, Fujimoto W, Saunders NA, Bernacki SH, Jetten AM. Cornifin, a cross-linked envelope precursor in keratinocytes that is down-regulated by retinoids. Proc Natl Acad Sci U S A. 1992;89:11026–11030. doi: 10.1073/pnas.89.22.11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steven AC, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107(Pt 2):693–700. [PubMed] [Google Scholar]
- 22.Steinert PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–17711. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- 23.Luo A, Kong J, Hu G, Liew CC, Xiong M, Wang X, Ji J, Wang T, Zhi H, Wu M, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene. 2004;23:1291–1299. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 24.He G, Zhao Z, Fu W, Sun X, Xu Z, Sun K. [Study on the loss of heterozygosity and expression of transglutaminase 3 gene in laryngeal carcinoma] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:120–123. [PubMed] [Google Scholar]
- 25.Gonzalez HE, Gujrati M, Frederick M, Henderson Y, Arumugam J, Spring PW, Mitsudo K, Kim HW, Clayman GL. Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129:754–759. doi: 10.1001/archotol.129.7.754. [DOI] [PubMed] [Google Scholar]
- 26.Ohkura S, Kondoh N, Hada A, Arai M, Yamazaki Y, Sindoh M, Takahashi M, Matsumoto I, Yamamoto M. Differential expression of the keratin-4, -13, -14, -17 and transglutaminase 3 genes during the development of oral squamous cell carcinoma from leukoplakia. Oral Oncol. 2005;41:607–613. doi: 10.1016/j.oraloncology.2005.01.011. [DOI] [PubMed] [Google Scholar]


