Abstract
Bioreporter bacteria, i.e., strains engineered to respond to chemical exposure by production of reporter proteins, have attracted wide interest because of their potential to offer cheap and simple alternative analytics for specified compounds or conditions. Bioreporter construction has mostly exploited the natural variation of sensory proteins, but it has been proposed that computational design of new substrate binding properties could lead to completely novel detection specificities at very low affinities. Here we reconstruct a bioreporter system based on the native Escherichia coli ribose binding protein RbsB and one of its computationally designed variants, reported to be capable of binding 2,4,6-trinitrotoluene (TNT). Our results show in vivo reporter induction at 50 nM ribose, and a 125 nM affinity constant for in vitro ribose binding to RbsB. In contrast, the purified published TNT-binding variant did not bind TNT nor did TNT cause induction of the E. coli reporter system.
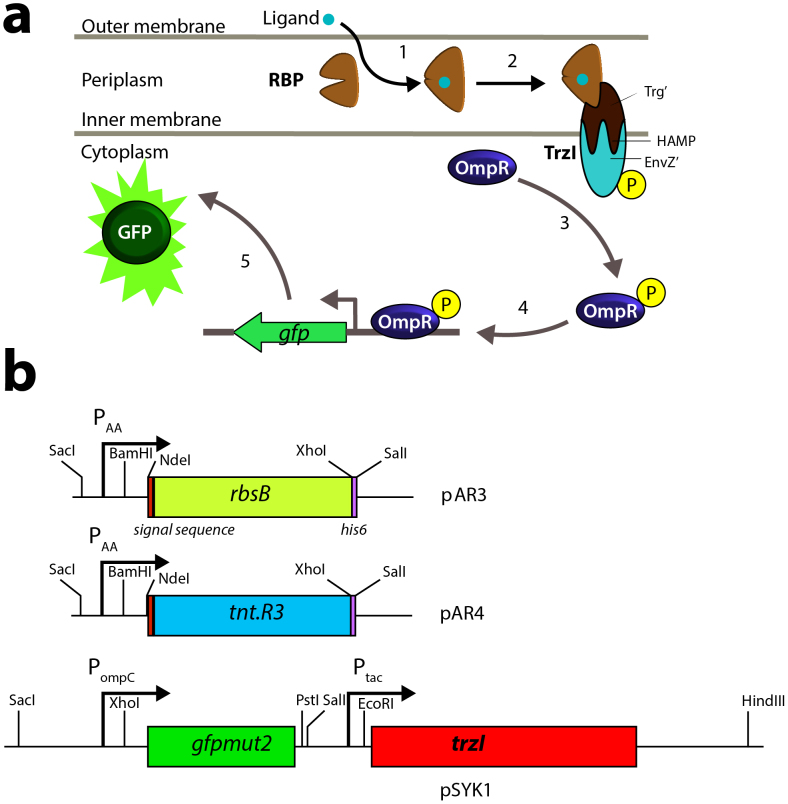
Construction of bioreporter bacteria typically starts with identifying a sensory protein that controls expression of a target gene promoter in dependence on one or more chemical inducers1,2,3,4,5,6,7,8. As an example, the ArsR protein represses its cognate promoter Pars, but when cells are exposed to arsenite (AsIII), this oxyanion will interact with ArsR causing it to lose affinity for the operator site close to Pars, thus increasing the rate of transcription from Pars6. The expression of reporter genes, such as those for luciferase, autofluorescent proteins or beta-galactosidase, when coupled to Pars, will consequently increase in the presence of arsenite; it is this increase in reporter protein signal or activity that is quantified in the bioreporter assay9. Despite the interest in and potential applicability of bioreporter assays, the weak part in their design is the availability of suitable sensory proteins for recognition of the target chemicals. Most of the “low-hanging fruits” in form of bacterial transcription regulators for e.g., heavy metals and metalloids, organic compounds or global stress responses, have been exploited7,10,11,12,13,14. Although it has been shown to be possible to somewhat expand substrate recognition properties of known transcription regulators through mutagenesis and selection, this is a rather cumbersome approach15,16,17,18,19,20. In this context, a landmark study over ten years ago suggested a completely different framework for the construction of bioreporter systems, based on computerized design of de novo substrate binding properties of periplasmic binding proteins (PBPs)8. Substrate binding to the redesigned PBP would lead to an interaction with a hybrid membrane receptor, thereby triggering reporter gene expression, as will be explained more in detail below (Fig. 1).
Figure 1. Schematic outline of the ribose-binding protein based reporter signaling chain.
(a) Ribose-binding protein (RBP) captures its ligand, leading to a conformational change. Ribose-RBP binds the TrzI hybrid transmembrane receptor, which causes a phosphorylation cascade leading to OmpR ~ P binding the ompC-promoter and consecutive gfpmut2 expression. (b) Relevant plasmid constructions. rbsB or tnt.R3 with original rbsB periplasmic export signal sequence and 3′-hexahistidine tag under transcriptional control of the weak constitutive PAA promoter29. Plasmid pSYK1 with gfpmut2 under the ompC promoter control and trzI under control of Ptac (note that pSYK1 carries the lacIq gene).
PBPs consist of a broad class of proteins that carry a conserved protein structure, the bilobal structural fold21. PBPs scavenge molecules for the cell, which they can present to specific transporter channels, and/or link compound binding to chemotactic movement. As an example, the galactose- (GBP) and ribose-binding proteins (RBP) of Escherichia coli enable the cell to sense galactose and ribose, respectively22. The sugars are bound by their respective PBP, and a fraction of sugar-bound GBP and RBP binds to the Trg chemoreceptor; the other fraction is presented to the transport channels MglAC (for GBP-galactose) or RbsAC (for RBP-ribose)23.
The binding of the PBP to a chemoreceptor can be transformed into de novo gene expression by using a hybrid membrane chemoreceptor-histidine kinase. This was shown almost 20 years ago by the group of Hazelbauer, who linked the EnvZ histidine kinase of the E. coli osmoregulation system to the Trg receptor via the so-called HAMP domain (Fig. 1a)24. The HAMP domain is a conserved domain among histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins and phosphatases, and plays a crucial role in signal transduction25. The resulting hybrid receptor kinase (named TrzI) combines the 265 N-terminal amino acids of Trg with the 230 C-terminal amino acids of EnvZ24. Galactose-GBP and/or ribose-RBP binding to TrzI will trigger histidine kinase activity of the cytoplasmic EnvZ-domain, leading to phosphorylation of the cognate DNA-binding response regulator OmpR. Phosphorylated OmpR (OmpR ~ P) binds the low affinity sites within the OmpC promoter (PompC) and increases transcription rate from this promoter (Fig. 1a)26. A proof of concept was presented by fusing PompC with the gene coding for β-galactosidase (lacZ) in E. coli, demonstrating that trzI expression yielded enhanced β-galactosidase activity when exposed to increasing ribose concentrations24.
A major conceptual advancement was made when it was proposed that by molecular dynamics modeling, on the basis of the resolved crystal structure of RBP, with and without ligand, it would be possible to predict the amino acid changes in RBP necessary for binding with new ligands27. This would create a possibly universal scaffold for engineering of new ligand-binding specificities, which could all be hosted in the same signaling “chassis” presented by the hybrid TrzI-OmpR system. To provide proof of principle, the binding pockets of glucose-binding protein (GBP), ribose-binding protein (RBP), arabinose-binding protein (ABP), glutamine-binding protein (QBP) and histidine-binding protein (HBP) were redesigned by computational simulation in order to bind toxic and non-natural molecules, such as serotonin, dinitrotoluene and TNT8. Simulation results suggested that nM binding affinities could be obtained, and experimental data were presented showing that expressing the mutant RBPs in an E. coli TrzI-OmpR background with ompCp-lacZ reporter led to nM detection specificity of TNT by measuring β-galactosidase activity8.
Motivated by the potential importance and implications of a universal scaffold for the engineering of bioreporter ligand specificity, we have set out to repeat the construction of one of the developed bioreporter strains with reported nM affinity for recognition of TNT8. A wide variety of methods is available for detection of TNT (see, for example, ref. 28), but bioreporter-based assays could be interesting for field application. Genes for both wild-type (rbsB) and mutant RBP (tnt.R3) were produced by DNA synthesis and cloned in an E. coli TrzI-OmpR background expressing the GFPmut2 protein from the ompC-promoter, to measure sensitivity of the reporter strains for ribose and TNT, respectively. We examined expression of wild-type and mutant proteins in the reporter strains, and have investigated ligand binding of the purified proteins by iso-thermal calorimetry (ITC). Whereas wild-type RBP produced an excellent and sensitive ribose sensor in E. coli, the published TNT-binding RBP variant did not show any significant binding to TNT, and the bioreporter cells harbouring it did not display any response to TNT or ribose.
Results
In vitro characterization of RbsB and TNT.R3 substrate binding
To test substrate binding by RbsB and TNT.R3 we purified both proteins from E. coli and determined the heat released by substrate addition to the purified protein fractions by isothermal microcalorimetry (ITC). Both proteins were overexpressed from the T7 promoter as C-terminal hexahistidine tagged variants in E. coli BL21 (DE3) pLysS using IPTG induction of the T7 RNA polymerase. Proteins were purified from culture-cleared lysates using Ni-NTA affinity chromatography. Figure 2 shows the different affinity binding steps and the purity of the final protein fraction (Elution II, at 250 mM imidazol). Concentrations of RbsB-His6 and TNT.R3-His6 after elution were between 0.2 and 0.8 mg/mL (Table 1).
Figure 2. Purification and expression in E. coli of RbsB-His6 and TNT-His6.
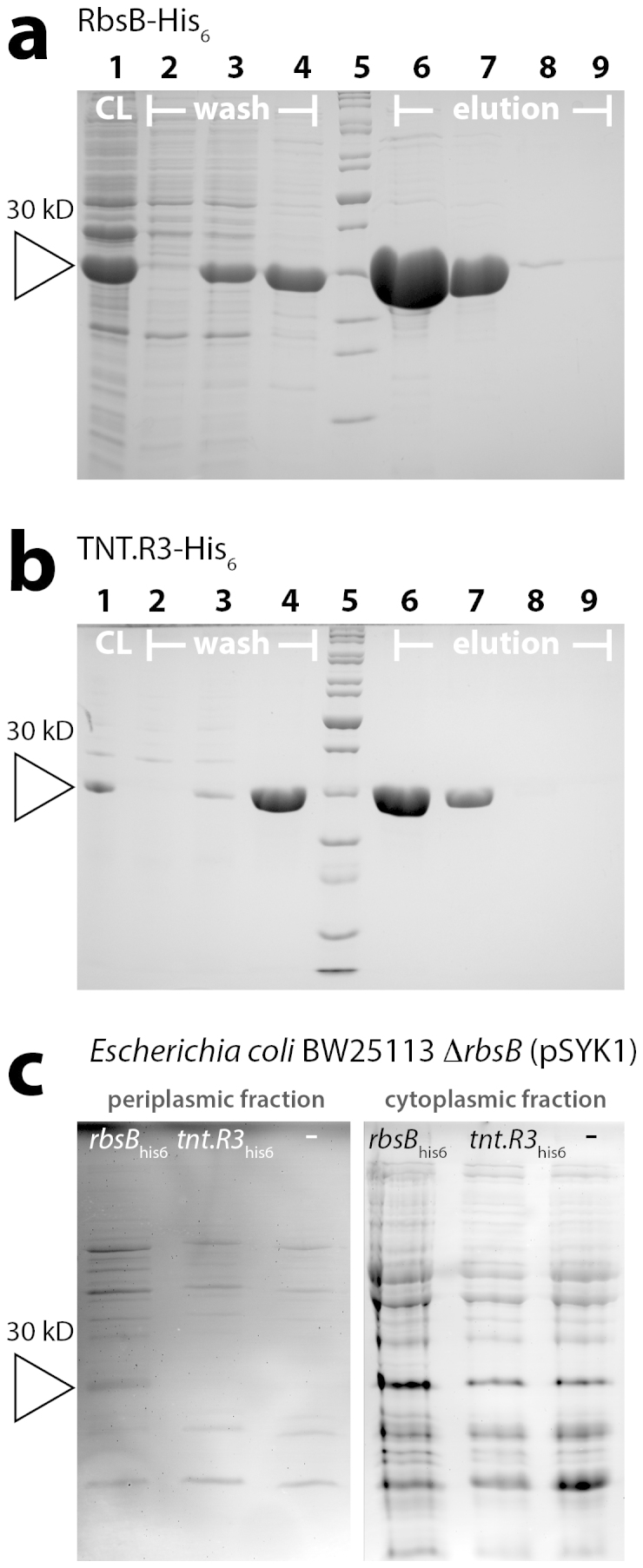
(a) Coomassie-stained SDS-PAGE gel of purified fractions of RbsB-His6 from E. coli BL21 (pAR1, strain 3725). Lanes: 1, cleared lysate; 2, flowthrough; 3, Washing step I; 4, Washing step II; 5, Peqgold protein marker 2; 6, Elution I; 7, Elution II; 8, Elution III; 9, Elution IV. (b) As A, but for E. coli BL21 (pAR2, strain 3325). Lanes: as for panel (A). (c) InVision His6-tag stained SDS-PAGE gel of cell extracts from E. coli BW25113 ΔrbsB (pSYK1) expressing rbsB-his6 (from plasmid pAR3, strain 4175), tnt.R3-his6 (from plasmid pAR4, strain 4176), or with empty pSTV plasmid (strain 4497). Left panel, periplasmic protein fraction. Right panel, whole soluble protein fraction. Open triangle indicates the expected position (30 kDa) of RbsB-His6 and TNT.R3-His6.
Table 1. Protein concentrations during purification of RbsB-His6 and TNT.R3-His6 from E. coli BL21 (DE3).
| Total protein concentration (mg/ml) | ||
|---|---|---|
| Sample | RbsB-His6 | TNT.R3-His6 |
| Column flow through | 43.9 | 27.1 |
| Washing step I | 22.9 | 14.4 |
| Washing step II | 2.4 | 1.2 |
| Elution I | 5.0 | 0.7 |
| Elution II | 0.8 | 0.2 |
| Elution III | 0.12 | 0.01 |
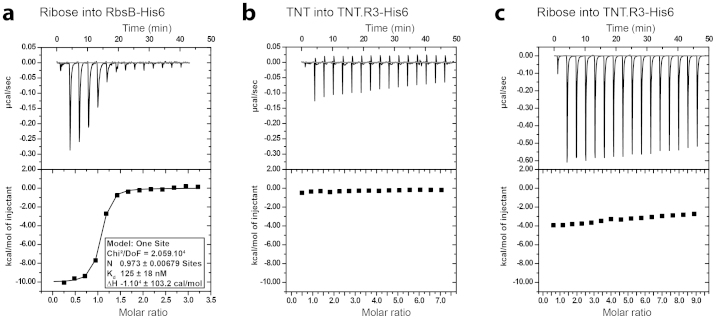
Addition of 250 μM ribose to 34 μM RbsB-His6 solution produced clear evidence for ribose binding with successively decreased heat release upon accumulated ribose additions (Fig. 3a). Assuming a single binding site for ribose per RbsB, an affinity constant (Kd) of 125 nM was calculated from the fitted data sets. Titration of buffer into RbsB-His6 solution, or of ribose solution into buffer produced no heat release (Figure S1). In contrast, neither 1 mM TNT nor 1 mM ribose titration into 20 μM TNT.R3-His6 solution released heat that was different from the addition of buffer alone (Figure 3b, c). Titration of TNT alone into buffer did not produce any consistent heat release as well (Figure S1).
Figure 3. In vitro substrate binding using isothermal microcalorimetry.
(a) Injections of 250 nM ribose into 34 μM purified RbsB-His6 solution. (b) Injections of 1 μM TNT into 50 μM purified TNT.R3-His6 solution. (c) Injections of 1 μM ribose into 50 μM purified TNT.R3-His6 solution. Graphs display immediate heat release in μcal/s (upper panels) and calculated heat release per mol of injectant (lower panels). Note the expected binding of ribose to RbsB (A), but the absence of any detectable binding of TNT or ribose by TNT.R3. For further controls see SI Figure 1.
Because the maximum aqueous solubility of TNT, 140 mg/L at 20°C, may have been limiting for saturation of TNT.R3 binding sites, we also tested the opposite titration in ITC (i.e., protein into substrate solution). In this manner, the TNT concentration in the measurement cell can be maintained below aqueous solubility. However, titration of 50 μM TNT.R3-His6 solution into 150 μM TNT also did not produce consistent heat release (Figure S1A, B). In contrast, titration of 100 μM RbsB-His6 solution into 4 μM ribose did produce heat release (Figure S1C), although in comparison to the binding curve in Figure 3a the injection peaks were not as clear. This may have been due to secondary effects caused by the dissolution of protein agglomerates during the injection. We thus concluded from the in vitro experiments that purified RbsB-His6 is indeed capable of binding ribose, but that TNT.R3-His6 neither binds TNT nor ribose, although the protein can be purified and is detectable on SDS-PAGE without any apparent degradation (Fig. 2b).
RBP-based bioreporter assays
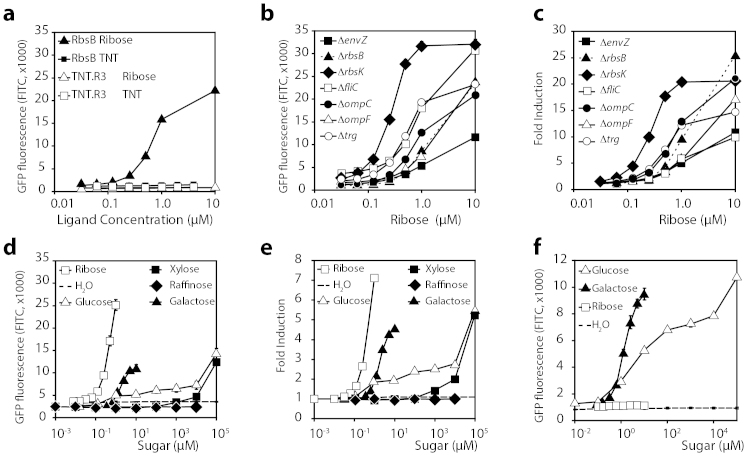
To verify that E. coli expressing the TrzI-hybrid-OmpR signaling chain is indeed a good chassis for an RbsB-based bioreporter, we reconstructed a reporter strain using gfpmut2 instead of the original lacZ under control of the ompC promoter (Fig. 1b, plasmid pSYK1), and measured induction of GFPmut2 over time in the presence of different sugars. This E. coli (strain 4175) lacks the chromosomal rbsB but constitutively expresses rbsB-his6 from a moderately constitutive promoter (PAA, plasmid pAR3). In addition, this strain expresses trzI from the lac promoter and carries the ompCp-gfpmut2 fusion (both on plasmid pSYK1). Background fluorescence of E. coli BW25113 strain 4175 grown on MM with fumarate as sole carbon and energy source was very low, and the lowest measured ribose concentration that resulted in statistically significant GFPmut2 induction compared to the medium-only control within 2 h incubation time was 50 nM (Fig. 4a). Maximum induction reached 25-fold at a ribose concentration of 10 μM (Fig. 4c). To determine the reporter strain's specificity we examined GFPmut2 production upon addition of a variety of other sugars, the majority of which (xylose, arabinose, sucrose, fructose, maltose, mannose, lactose) did not elicit any GFPmut2 induction from the E. coli reporter strain at concentrations below 1 mM. In contrast, significant GFPmut2 production was observed upon incubation with galactose and glucose between 300 nM and 10 μM (Fig. 4d, e). This is likely caused by interference from GBP, which can bind either galactose or glucose, and interacts in its closed (substrate-bound) configuration with the hybrid receptor TrzI (Fig. 4f). The reaction to glucose is lower than to galactose at the same concentration, possibly due to the more rapid metabolism of this sugar.
Figure 4. Reporter gene expression from the hybrid TrzI-OmpR-ompCp::gfpmut2 signaling chain.
(a) Average GFPmut2 fluorescence in flow cytometry (detected using the FITC channel, arbitrary units), as a function of ribose or TNT concentration after 2 h induction time, for E. coli BW25113 ΔrbsB expressing rbsB-his6 (strain 4175) or tnt.r3-his6 (strain 4176). (b) As A, but as a function of ribose exposure only and for a range of E. coli BW25113 backgrounds (Table 2). (c) As B, but expressed as -fold induction compared to the blank. (d) As A, but as a function of exposure to different sugars for E. coli BW25113 ΔrbsB expressing rbsB-his6 (strain 4175). (e) As D, but expressed as -fold induction compared to the blank. (f) Average GFPmut2 fluorescence of E. coli BW25113 ΔrbsB (pSYK1, pSTV28, strain 4497) exposed to glucose, galactose or ribose. Data points show means of GFPmut2 fluorescence from biological triplicate assays, each sampling 10,000 cells. Error bars indicate calculated standard deviations from the mean (when not visible, inside symbol size).
Next, we examined the possible influence of a number of key proteins in the chemotaxis or osmolarity sensing pathways on reporter gene induction in the trzI-ompR ompCp-gfpmut2 bioreporter strain. Figure 4b displays the characteristic GFPmut2 induction profiles of E. coli BW25113 carrying individual gene deletions as a function of ribose concentration, whereas Figure 4c shows the fold induction levels. In comparison to BW25113 lacking native rbsB, the isogenic strain with envZ interruption showed a loss of responsiveness to ribose (Fig. 4b). In contrast, interruption of rbsK increased the sensitivity of the reporter to ribose (Fig. 4b). A less drastic increase in sensitivity was obtained when deleting trg or fliC. Deleting ompC or ompF had very little effect on the sensitivity of the reporter strain for ribose (Fig. 4b).
Interestingly, depending on the host, on ribose concentration and on the deleted gene, the heterogeneity of GFPmut2 production among cells in the population varied significantly (Fig. 5). In general, the per-cell variability in GFPmut2 expression decreased at higher inducer concentrations (Fig. 5). Compared to the host without functional chromosomal rbsB, deletion of fliC, ompC, or rbsK led to a more homogenously reacting population at lower ribose concentrations (i.e., lower coefficient of variation, Fig. 5c, d, f). Conversely, deleting ompF or envZ resulted in higher cellular variability of GFPmut2 production, almost irrespective of ribose concentrations (Fig. 5a, e). Deleting the chromosomal trg chemoreceptor did not result in any difference of GFPmut2 heterogeneity compared to the ΔrbsB strain (Fig. 5b, g).
Figure 5. Coefficient of variation of per-cell GFPmut2 expression as a function of ribose concentration for different E. coli BW25113 backgrounds (A–G, Table 2) expressing rbsB-his6 from pAR3 and the TrzI-OmpR-ompCp::gfpmut2 hybrid signaling chain of pSYK1.
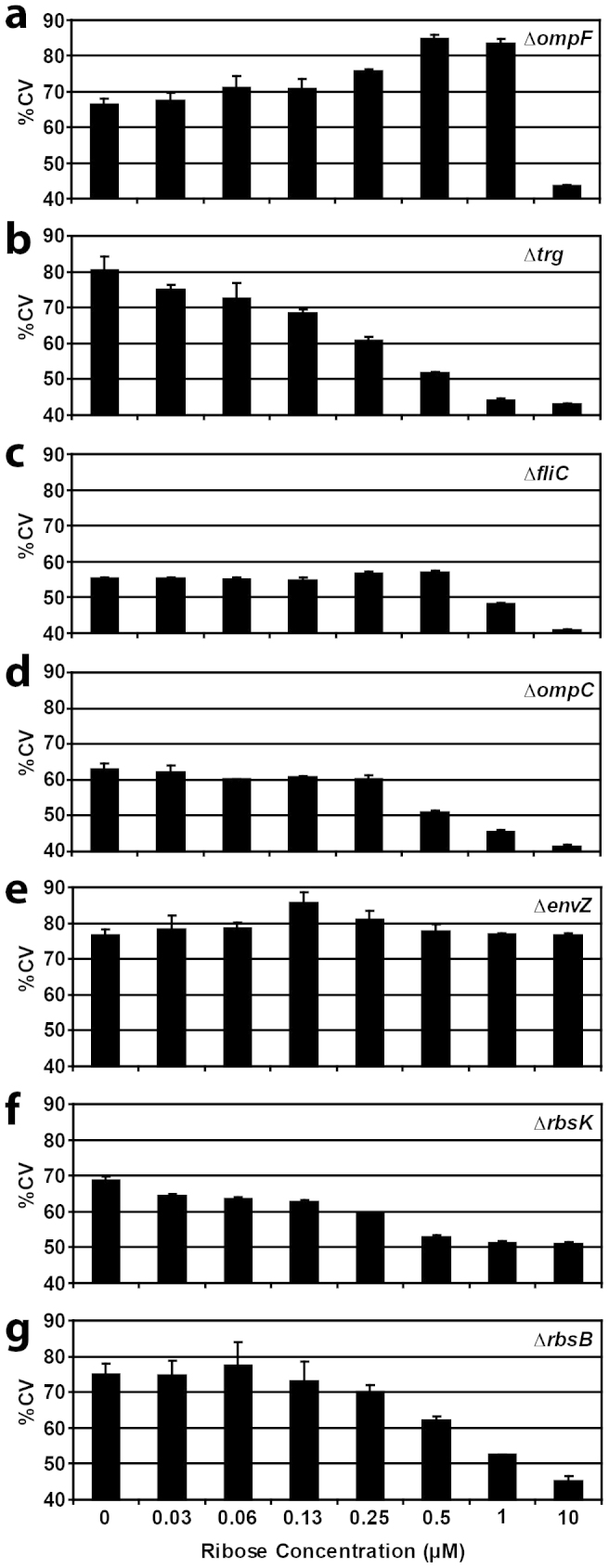
Bars indicate the mean coefficient of variation from biological triplicate assays (each measuring GFPmut2 expression in 10,000 cells), plus one calculated standard deviation (error bar).
A non-functional TNT bioreporter
In contrast to the wild-type rbsB-based reporter, replacing rbsB by tnt.R3 in an isogenic host background (E. coli BW25113 ΔrbsB, strain 4176, Table 2) led to complete loss of sensitivity to ribose (Fig. 4a). More importantly, the tnt.R3-based bioreporter was completely unresponsive to TNT over a broad concentration range, 0.06 to 4 μM (Fig. 4a). Since we did not observe any reporter signal from the reconstructed TNT.R3 bioreporter, but excellent sensitivity from the wild-type RbsB bioreporter for ribose, we also examined whether E. coli BW25113 expressed the TNT.R3 protein from the PAA-promoter and periplasmic transport signal sequence at the same level as the RbsB protein (Fig. 1b). Expression of the RbsB-His6 and TNT.R3-His6 proteins in the BW25113 bioreporter strain is under control of the same low constitutive PAA-promoter29 in the same plasmid background. Unfortunately, anti-His6-antibodies did not produce sufficient sensitivity and selectivity to detect both proteins in Western blots of cytoplasmic cell extracts from the BW25113 strains (not shown). In-gel staining of the His6-tag revealed fluorescent bands with an apparent size of around 30 kDa in the cytoplasmic protein fraction, which were not completely specific for the expressed RbsB-His6 and TNT.R3-His6 in extracts from E. coli BW25113 (Fig. 2c). Analysis of the periplasmic fraction indicated presence of a detectable RbsB-His6 but not TNT.R3-His6 (Fig. 2c). Gel-extracted and trypsin-digested protein fractions in the size range of 28–36 kDa were further analyzed by nano-liquid chromatography followed by direct peptide mass identification (Table 3). This analysis confirmed that both RbsB-His6 and TNT.R3-His6 are produced by E. coli BW25113 ΔrbsB from the PAA-promoter. However, whereas we identified RbsB-His6 in the whole soluble and in the periplasmic protein extracts, TNT.R3-His6 was only detectable in the whole soluble but not in the periplasmic protein extract (Table 3). We conclude from this part that whereas both RbsB-His6 and TNT.R3-His6 are produced from the same plasmid type and promoter in E. coli, TNT.R3-His6 is less abundant and does not seem to be transported into the periplasmic space.
Table 2. List of strains used in this study with their relevant characteristics.
| Strain No | Host | Plasmid(s) | Relevant characteristics | Reference |
|---|---|---|---|---|
| 97 | E. coli BL21 (DE3) | pLysS | Host strain for overexpression from the T7 promoter | 40 |
| 3325 | E. coli BL21 (DE3) pLysS | pAR2 | Host 97, cytoplasmic overexpression of His6-tagged TNT.R3 | This study |
| 3725 | E. coli BL21 (DE3) pLysS | pAR1 | Host 97, cytoplasmic overexpression of His6-tagged RbsB | This study |
| 4076 | Escherichia coli BW25113 ΔrbsB | rbsB | 42 | |
| 4175 | E. coli BW25113 ΔrbsB | pAR3 pSYK1 | expression of His6-tagged RbsB with signal sequence from PAA expression of TrzI, gfpmut2 fusion to ompC promoter | This study |
| 4176 | E. coli BW25113 ΔrbsB | pAR4, pSYK1 | as 4175, but expressing the TNT.R3 mutant protein | This study |
| 4497 | E. coli BW25113 ΔrbsB | pSTV28, pSYK1 | as 4175, but with empty vector. | This study |
| 4500 | E. coli BW25113 ΔompF | ompF | 42 | |
| 4501 | E. coli BW25113 Δtrg | trg | 42 | |
| 4502 | E. coli BW25113 ΔfliC | fliC | 42 | |
| 4503 | E. coli BW25113 ΔompC | ompC | 42 | |
| 4504 | E. coli BW25113 ΔenvZ | envZ | 42 | |
| 4505 | E. coli BW25113 ΔrbsK | rbsK | 42 | |
| 4515 | E. coli BW25113 ΔompF | pAR3, pSYK1 | as 4175 in host 4500 | This study |
| 4516 | E. coli BW25113 Δtrg | pAR3, pSYK1 | as 4175 in host 4501 | This study |
| 4517 | E. coli BW25113 ΔfliC | pAR3, pSYK1 | as 4175 in host 4502 | This study |
| 4518 | E. coli BW25113 ΔompC | pAR3, pSYK1 | as 4175 in host 4503 | This study |
| 4519 | E. coli BW25113 ΔenvZ | pAR3, pSYK1 | as 4175 in host 4504 | This study |
| 4520 | E. coli BW25113 ΔrbsK | pAR3, pSYK1 | as 4175 in host 4505 | This study |
Table 3. Occurrence of RbsB-His6 and TNT.R3-His6 in E. coli BW25113 ΔrbsB (pSYK1) protein fractions.
| Periplasmic protein fraction1 | Whole soluble protein fraction1 | |||||
|---|---|---|---|---|---|---|
| Identified protein(s) | Strain 4175 RbsB-His6 | Strain 4176 TNT.R3-His6 | Strain 4497 Ctrl | Strain 4175 RbsB-His6 | Strain 4176 TNT.R3-His6 | Strain 4497 Ctrl |
| RbsB-His6 | 18 (8)2 | 5 (5) | 4 (4) | 14 (5) | 1 (1) | 1 (1) |
| TNT.R3-His6 | ND3 | ND | ND | ND | 2 (2) | ND |
| RbsB-His6 + TNT.R3-His64 | 18 (5) | 6 (6) | ND | 10 (5) | 5 (3) | ND |
| Total RbsB-His6 + TNT.R3-His65 | 36 (13) | 11 (11) | 4 (4) | 24 (10) | 8 (6) | 1 (1) |
| Total proteins | 116 (387)6 | 86 (277)6 | 104 (349)6 | 259 (1273)6 | 234 (971)6 | 254 (1061)6 |
1) 28–36 kD fragments on SDS-PAGE gel from extracts as shown in Fig. 2C, originating from E. coli BW25113 ΔrbsB (pSYK1) expressing rbsB-his6 (from plasmid pAR3, strain 4175), E. coli BW25113 ΔrbsB (pSYK1) expressing tnt.R3-his6 (from plasmid pAR4, strain 4176), or E. coli BW25113 ΔrbsB (pSYK1) carrying empty pSTV (strain 4497, Ctrl).
2) Normalized number of peptide mass fragments identifying the protein(s) as total number of specific peptide fragments by the averaged total peptide count in the sample; between brackets, number of unique peptide mass fragments identifying the protein(s).
3) ND, not detected.
4) Peptides covering regions common to both RbsB-His6 and TNT.R3-His6.
5) All peptides identifying both RbsB-His6 and TNT.R3-His6.
6) Total number of identified proteins within the 27–32 kDa purified gel region; within brackets: total number of identified peptide mass fragments.
Discussion
We revisit here the use of a PBP-based microbial biosensor, proposed over a decade ago as a general scaffold for computational design of new binding specificities8,30. We show through independent de novo synthesis that a wild-type RbsB-based signaling cascade is fully functional in detecting low concentrations of ribose, but one of the most interesting computationally designed variants for detecting TNT8 is not. We conclude this from three different experimental lines of evidence. First, we demonstrated that the purified RbsB bound ribose, as expected, whereas purified TNT.R3 protein bound neither TNT nor ribose at detectable levels (Fig. 3). Second, constitutive expression of rbsB-his6 under a moderate promoter in an E. coli harboring the TrzI hybrid receptor and ompCp-gfpmut2 fusion led to a ribose-dependent production of GFPmut2 fluorescence already at 50 nM ribose (Fig. 4a, c). In contrast, assays in the same genetic background and plasmid constructions with a tnt.R3-his6 gene produced no detectable GFPmut2 in presence of either TNT or ribose (Fig. 4a). Finally, we showed that both RbsB-His6 and TNT.R3-His6 are produced in such E. coli background, but whereas RbsB-His6 appears to reach the periplasmic space we did not find evidence that this is also true for TNT.R3-His6 (Fig. 2c, Table 3). The absence of any reporter signal in strains carrying the tnt.r3-his6 gene to TNT is, therefore, unlikely the result of poor expression of TNT.R3-His6. Possibly, the introduced mutations in the TNT.R3 variant that were predicted to turn RbsB into a TNT-binder, made the protein less stable in the cytoplasmic environment (Table 3), and hindered the folding and transport process. In addition, it is possible that the intramolecular movement upon ligand binding, or even the binding to the TrzI receptor, are affected. But since we have no evidence that TNT.R3-His6 reaches the periplasmic space (Table 3), this is a less likely explanation for the absence of GFPmut2 fluorescence in tnt.R3-his6 reporter strains. Misfolding of the protein was not specifically examined in our assays, but had been suggested as a reason for concern in other studies performed in order to structurally characterize the computational designs of the receptors31,32,33.
We acknowledge that the original study designed a number of periplasmic-binding-protein variants, of which we independently reconstructed only the one proposed as the most prominent8. We find that the published TNT.R3 mutant cannot be used as a sensor for the detection of TNT, contrary to the reported lowest detection limit of between 10−4 and 10−3 μM and a Kd of 2 nM8, a value 65 times lower than the affinity we measured for the wild type RbsB protein towards ribose (125 nM). These findings, however, do not null the notion that a platform using biosensors based on periplasmic binding proteins could be a powerful tool. Indeed, E. coli expressing RbsB in combination with the TrzI-hybrid-OmpR ompCp-gfpmut2 signaling chain turned out to be an excellent reporter for ribose (method detection limit of ~50 nM), with a good selectivity (no reaction to multiple sugars and 10-fold lower detection threshold of ribose than galactose, Fig. 4). We showed that the response from the hybrid signaling chain could even be further optimized by using an rbsK-mutant of E. coli rather than rbsB (Fig. 4C, D). The higher response of this strain to ribose may be explained by the fact that deleting rbsK interrupts ribose metabolism, leaving on average more ribose to activate the signaling chain through RbsB-TrzI-OmpR rather than going through the ribose transport system. Also, a trg host mutant background produced a more sensitive response, but with overall lower fold-induction (Fig. 4c), which may be due to less internal competition for ribose-bound RbsB by the natural Trg chemotaxis receptor. Deletion of envZ, ompF, and, surprisingly, fliC, resulted in a much poorer response to ribose (Fig. 4b, c). Disruption of native envZ may result, in spite of the presence of TrzI, in a lower overall amount of OmpR ~ P in the cell, causing less frequent binding to the weak-affinity OmpR-sites in the PompC promoter, and thus to a lower level of gfpmut2 transcription. These results show that the host chassis for a ribose-binding protein based reporter may be further improved and would prove useful, once the limitations in computational design are overcome.
Along with techniques such as directed evolution, computational protein design has been successful in the design of enzyme catalysts, new protein folds, or antigens, but its predictions still fall short especially in the protein-small molecule domain34,35,36. Many methods adopted in the current context, including the technique of dead-end elimination along with a semi-empirical potential energy function, used in designing TNT.R3 and other PBP-variant receptors8, do not take into consideration the flexibility of the protein backbone. This concerns specifically the substrate binding pocket flexibility, dynamics of the structure upon binding, and calculation of the protein stability or consistency of the 3D fold of the designs. Because of the magnitude of the combinatorial search problem to be tackled in protein design, the chemical accuracy of the calculations is significantly reduced. For a receptor like RBP, the energetic or entropic cost of the conformational re-organization or domain re-orientation during binding can be very important but difficult to calculate. This also holds for other relevant steps in the PBP signalling cascade, such as the binding of the ligand-bound PBP with the transmembrane receptor. So far, therefore, these steps have not been included in energy calculations.
Very recently, it has been demonstrated that in-silico design of small-molecule binding proteins can drive the binding affinity down to picomolar level37. The computational method was modified so that the designs have binding pockets similar to the naturally occurring ones, in terms of the favourable hydrogen-bonding or van der Waals interactions with the ligand, and high shape complementarity to the ligand. These advances may be highly beneficial for the design of sensory proteins and extremely advantageous to the biosensing field, given that the set of known characterized transcription factors is small and in view of the difficulties involved in the identification of suitable transcription factors for the detection of new compounds of interest.
Methods
Strains and growth conditions
All E. coli strains used for this work are listed in Table 2. For cloning purposes, E. coli strains were cultured at 37°C on Luria Bertani (LB) medium38, supplemented with appropriate antibiotics to select for plasmid maintenance. In case of ampicillin (Amp), a concentration of 100 μg/ml was used; for chloramphenicol (Cm), we used 30 μg/ml. Culturing conditions for protein overexpression and for reporter assays are specified below.
Plasmid constructions
The mutant tnt.R3 gene was produced by DNA synthesis (DNA2.0, CA, USA) on the basis of the mutant sequence provided in Looger et al8. The gene sequence further encoded a C-terminal hexahistidine (His6) tag, restriction sites for NdeI (N-terminal), XhoI and BamHI (C-terminal), and for NcoI at the end of the signal sequence (Figure S2). The wild type rbsB gene was amplified from pAI12 (a kind gift of Hazelbauer's lab, Pullman, Washington) using primers with restriction sites for NdeI and XhoI. The PCR product was digested and placed into pET22b(+) in order to attach the vector-located hexahistidine tag to the C-terminus of the rbsB gene. The resulting rbsB-His6 gene was again amplified with primers containing NdeI and SalI restriction sites. The PAA promoter29 used to drive constitutive transcription of both tnt.R3 and rbsB was synthesized (DNA2.0) under inclusion of BamHI and SacI restriction sites. The promoter fragment was first cloned into pUC18 and recovered by SacI and XbaI digestion. This PAA-fragment was ligated with a recovered DNA fragment containing the multiple cloning site of pET22(+) (using XbaI and SalI), and with pSTV28 (Takara, Japan), digested with SacI and SalI. After transformation in E. coli, this resulted in plasmid pSTV28PAAmcs. Finally, rbsB-His6 and tnt.R3-His6 were placed into pSTV28PAAmcs using NdeI and SalI or NdeI and XhoI, respectively, resulting in pAR3 and pAR4 (Fig. 1b, Table 2).
For overexpression the synthesized tnt.R3-His6 fragment was digested with NcoI and BamHI and placed into pET3d39 resulting in pAR2. This removes the signal sequence for transport in the periplasmic space. RbsB-His6 was amplified from pAI12, now without the signal sequence using primers containing NdeI and XhoI restriction sites. The NdeI-XhoI fragment was purified and placed into pET22b(+) digested with the same enzymes, directly in front of the hexahistidine tag. After transformation this resulted in plasmid pAR1. All final constructs were verified by sequencing. Plasmids for overexpression were transformed into E. coli BL21 (DE3) carrying pLysS40.
Plasmid pSYK1 carries the gene for the hybrid receptor trzI under the control of Ptac and gfpmut2 under the control of PompC (Fig. 1b). The ompC promoter fused to the gfpmut2 reporter gene was amplified from plasmid pUA66-ompC::GFPmut241 (kindly provided by Prof. Uri Alon from the Department of Molecular Cell Biology at the Weizmann Institute of Science, Rehovot, Israel) using primers that introduced BamHI restriction sites on either end. This fragment was ligated into the unique BamHI site of plasmid pRB020, which carries a lacIq gene and places trzI under control of the tac promoter24. After transformation this resulted in plasmid pSYK1. Reporter constructs were cotransformed into E. coli BW25113 background carrying either rbsB deletion or deletions in other genes of the hybrid signaling pathway (Table 2).
Analysis of expression of RbsB-His6 and TNT.R3-His6 in E. coli BW25113
Ten mL LB medium containing Amp and Cm were inoculated with a single colony from a freshly grown LB agar plate with both antibiotics, and incubated at 37°C. The cells were harvested by centrifugation at 3200 x g at mid-exponential phase (OD600 = 0.7) and resuspended in 400 μL of 30 mM Tris-HCl containing 20% sucrose at a pH of 8.0. Half of the suspension (for the preparation of the total soluble protein fraction or cell extract) was transferred to 1.5 mL screw-capped microtubes containing 0.1 g acid-washed glass beads (<106 μm, Sigma, USA). Suspensions were homogenized in a bead beater (Fastprep FP120, Thermo Electron, USA) for 30 s at a speed of 4.0. EDTA was added to the other half to a final concentration of 1 mM for the preparation of the soluble periplasmic protein fraction. This suspension was incubated on ice for 10 min with gentle agitation. After centrifugation at 16,000 x g for 20 min at 4°C the supernatant was removed and the pellet was resuspended in 200 μL of ice-cold solution of 5 mM MgSO4. During the following incubation on ice for 10 min the tubes were inverted 10 times every minute for 10 s. Both whole soluble and periplasmic fractions were centrifuged for 30 min at 16,000 x g and 4°C, after which the supernatant was transferred to a clean Eppendorf tube and placed on ice. Samples were then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), by mixing various protein amounts with SDS loading buffer, according to Sambrook38. Samples for SDS-PAGE were incubated for 5 min on a thermomixer (Eppendorf, Schweiz) at 99°C, and centrifuged for 1 min at 16,000 x g immediately before loading. The samples were loaded onto 13% acrylamide gel and proteins were separated for 1.5 h at 150 V, according to standard procedures38.
After electrophoresis the gels were stained using InVision colorant (Life technologies, USA) in order to detect His6-tagged proteins. The gel was placed into a fixing solution (40% v/v ethanol and 10% v/v acetic acid in ultrapure water) for 1 h and then washed with water for 20 min. Subsequently, the gel was washed with buffer A (20 mM imidazole, 50 mM NaH2PO4 and 500 mM NaCl, pH 8.0) for 10 min, and incubated with InVision stain for 1 h. The gel was destained with 20 mM phosphate buffer (pH 7.8) for 20 min, and subsequently scanned on a Typhoon imager (Amersham Biosciences, United Kingdom) at a resolution of 50 μm using a 532 nm laser. Subsequently, gels were washed with water for 10 min, and restained using Coomassie blue for 1 h according to standard procedures38. After destaining the gel was photographed with a Nikon D5100 camera equipped with a 18–55 mm objective (Nikon, Schweiz) under white light illumination.
Expression and export of RbsB-His6 and TNT.R3-His6 from the PAA-promoter was separately analyzed using direct peptide mass identification. E. coli BW25113 cultures were grown and periplasmic or whole soluble protein fractions were prepared as described above. Proteins were separated by SDS-PAGE and proteins in a size region of 28-36 kD were excised. Proteins were subsequently digested with trypsin and peptides were separated on an Ultimate 3000 Nano LC System (Dionex), followed by detection in a Thermo Scientific LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Mass spectra were analyzed using Scaffold Viewer (http://www.proteomesoftware.com/), using protein and peptide identification thresholds of 99.9% and 99.99%, respectively. The minimum number of peptides for identification was 2.
RbsB-His6 and TNT.R3-His6 overexpression and purification
In order to analyze in vitro substrate binding, we first overexpressed and purified RbsB-His6 and TNT.R3-His6. We used the C-terminal added hexahistidine tag in combination with Ni-NTA affinity column chromatography (Qiagen, Germany). Proteins were overexpressed in E. coli BL21(DE3)pLysS carrying the appropriate plasmid (pAR1 for RbsB-His6 or pAR2 for TNT.R3-His6). Cultures were launched at 37°C in 200 mL LB medium containing 100 μg/mL Amp and 30 μg/mL Cm and inoculated with a single colony from a freshly grown agar plate. At a culture turbidity of OD600 = 0.3 overexpression was induced through addition of IPTG to 1 mM final concentration. Cultures were incubated further for 16 h at 20°C, after which the cells were harvested by centrifugation at 3,200 x g in four 50 mL Falcon tubes. Cell pellets were stored at −80°C until protein isolation.
For purification of the His6-tagged proteins, one cell pellet was resuspended in 4 mL of buffer A (see above). The suspension was transferred into 1.5 mL screw-capped plastic tubes containing 0.1 g glass beads (see above), and homogenized in a bead beater (Fastprep) for 3 times at 20 s and a speed of 4.0, with intermittent cooling on ice. After centrifugation for 30 min at 16,000 x g and 4°C the supernatant was transferred to a clean tube, and mixed with 750 μL of Ni-NTA resin (Qiagen, Germany) for one hour at 4°C using a multi axle rotator (A257, Denley Instruments LTD, United Kingdom). Subsequently, the protein-Ni-NTA suspension was poured onto a 1 mL polypropylene column (Qiagen, Germany) to collect the protein-bound-resin. After consecutive washing with 1 mL of buffer A containing first 40 mM and then 80 mM imidazole, the proteins were eluted in fractions of 600 μL using buffer A containing 250 mM imidazole. Flow-through from the column was collected and analysed by SDS-PAGE and Coomassie blue staining (see above). Protein concentrations were determined by using the Bradford assay and by NanoDrop spectrophotometry (Thermo Scientific, USA), using the “Protein A280” mode with calculated theoretical molar extinction coefficient and molecular weight as parameters. Purified protein was stored on ice and used within 5 h for the substrate-binding assay, without further dialysis.
Analysis of substrate binding using isothermal microcalorimetry (ITC)
Quantified amounts of purified protein, typically 280 μl of between 0.6 and 1 μg/μl, were pipetted into the measurement cell of an isothermal titration calorimetry instrument (MicroCal iTC200, GE Healthcare Life Sciences, USA). A volume of 280 μl of elution buffer (buffer A with 250 mM imidazole) was used as a reference. An appropriate concentration of the test ligand (of between 0.25 and 1 mM in buffer A containing 250 mM imidazole) was filled into the injection syringe. In other experiments we tested injecting purified protein solution (of between 1.4 and 2.8 μg/μl) into substrate solution in the measurement cell. The substrates tested were ribose and TNT. Measurements were taken at 20°C, with a reference power of 11 μcal/s, a stirring velocity of 1000 rpm and a “low feedback” mode. Raw data were recorded in μcal/s over time and integrated to kcal/mol over molar ratio. Wherever possible regression curves were calculated based on a one-binding site model.
RBP-based bioreporter assays using the TrzI-OmpR hybrid signaling chain
In order to measure the capacity of RbsB or TNT.R3 to induce the TrzI-hybrid-OmpR ompCp-gfpmut2 signaling chain in the presence of appropriate inducer, we used E. coli BW25113ΔrbsB cotransformed with pSTV-based plasmids (pAR3, to express rbsB, or pAR4 for tnt.R3) and plasmid pSYK1 (to provide the hybrid signaling chain, see Table 2). Upon induction, these strains produce GFPmut2, the fluorescence intensity of which was measured using flow cytometry. The bioreporter assay was optimized for minimal background GFPmut2 expression and medium fluorescence. Hereto, 5 mL of minimal medium with Amp and Cm (SI Table 1) with 20 mM fumarate as sole carbon and energy source were inoculated with a single colony from a freshly grown LB plate containing the same antibiotics. Cultures were incubated overnight at 37°C with rotary shaking at 180 rpm. The next morning, 2 μL culture aliquots were transferred into 5 mL of fresh minimal medium (SI Table 1), and incubated for 4 h at 37°C with rotary shaking at 180 rpm. Samples (180 μL) of this culture were then introduced into the wells of a 96-well plate (F96 Cert.Maxisorp, Nunc, Denmark) and mixed with 20 μL of the appropriate ligand in a range of concentrations to start induction. Notably, we used ribose (D-(-)-ribose, Aldrich, USA) at concentrations between 7.8 nM and 250 μM. 2,4,6-Trinitrotoluene (TNT) was purchased from Dr. Ehrenstorfer GmbH, Germany and used at concentrations between 62 nM and 4 μM, by dissolving the TNT in water. Different other gene deletion variants of BW2511342 were tested with the same plasmids, in order to discern the effect of background host genes on the induction from the hybrid signalling chain (Table 2). Induction was allowed to proceed for 2 h at 37°C, after which aliquots of 150 μl of each of the wells were auto-sampled, and values of individual cell forward scatter (FSC) and GFPmut2 fluorescence (FITC-channel) were recorded by a Becton Dickinson Fortessa flow cytometer (LRS FortessaTM, Becton Dickinson, USA). The flow rate was set to 3 μl/s and the cell density was between 100-1000 cells/μL. Sensitivities for the FSC and the FITC channels were set at 350 V and 676 V, respectively. Recorded data were gated to remove background particles. The mean fluorescence values of the gated uninduced or induced populations were calculated. All experiments were carried out in triplicate, and the standard deviation and coefficient of variation were calculated from the mean fluorescence.
Author Contributions
A.R. and S.Y.K. performed experiments. A.R. and J.R.M. prepared Figures 1–5. S.B. and S.Y.K. contributed strains and experimental advice. A.R., S.R. and J.R.M. wrote the main manuscript. All authors reviewed the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grant 244405 (Biomonar) from the European Seventh Framework Programme (FP7). The pAI12 plasmid was kindly provided by Hazelbauer's lab (Washington State University, USA). We thank Sebastian Ritzmann for help in optimizing RbsB protein purification and Karine Lapouge for instructions in ITC measurements. Patrice Waridel and Manfredo Quadroni from the University of Lausanne Protein Analysis Facility are thanked for their help in peptide analysis. Work in the Belkin lab was partially supported by the Minerva Center for Bio-hybrid Complex Systems.
References
- Siegfried K. et al. Field testing of arsenic in groundwater samples of Bangladesh using a test kit based on lyophilized bioreporter bacteria. Environ Sci Technol 46, 3281–3287 (2012). [DOI] [PubMed] [Google Scholar]
- Pedahzur R., Polyak B., Marks R. S. & Belkin S. Water toxicity detection by a panel of stress-responsive luminescent bacteria. J Appl Toxicol 24, 343–348 (2004). [DOI] [PubMed] [Google Scholar]
- Branco R., Cristovao A. & Morais P. V. Highly sensitive, highly specific whole-cell bioreporters for the detection of chromate in environmental samples. PLoS One 8, e54005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan Ul Haque M., Nadalig T., Bringel F., Schaller H. & Vuilleumier S. Fluorescence-based bacterial bioreporter for specific detection of methyl halide emissions in the environment. Appl Environ Microbiol 79, 6561–6567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Martin M. A., Mateo P., Leganes F. & Fernandez-Pinas F. A battery of bioreporters of nitrogen bioavailability in aquatic ecosystems based on cyanobacteria. Sci Total Environ 47, 2050–2064 (2013). [DOI] [PubMed] [Google Scholar]
- Stocker J. et al. Development of a set of simple bacterial biosensors for quantitative and rapid field measurements of arsenite and arsenate in potable water. Environ Sci Technol 37, 4743–4750 (2003). [DOI] [PubMed] [Google Scholar]
- van der Meer J. R. & Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol 8, 511–522 (2010). [DOI] [PubMed] [Google Scholar]
- Looger L. L., Dwyer M. A., Smith J. J. & Hellinga H. W. Computational design of receptor and sensor proteins with novel functions. Nature 423, 185–189 (2003). [DOI] [PubMed] [Google Scholar]
- Merulla D. et al. Bioreporters and biosensors for arsenic detection. Biotechnological solutions for a world-wide pollution problem. Curr Opin Biotechnol 24, 534–541 (2013). [DOI] [PubMed] [Google Scholar]
- Hynninen A. & Virta M. Whole-cell bioreporters for the detection of bioavailable metals. Adv Biochem Eng Biotechnol 118, 31–63 (2010). [DOI] [PubMed] [Google Scholar]
- Ivask A., Rolova T. & Kahru A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol 9, 41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher P., Jaspers M., Harms H., Zehnder A. J. B. & van der Meer J. R. Development and characterization of a whole cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol 63, 4053–4060 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin S. A panel of stress-responsive luminous bacteria for monitoring wastewater toxicity. Methods Mol Biol 102, 247–258 (1998). [DOI] [PubMed] [Google Scholar]
- de las Heras A. & de Lorenzo V. In situ detection of aromatic compounds with biosensor Pseudomonas putida cells preserved and delivered to soil in water-soluble gelatin capsules. Anal Bioanal Chem 400, 1093–1104 (2011). [DOI] [PubMed] [Google Scholar]
- Galvao T. C. & de Lorenzo V. Transcriptional regulators a la carte: engineering new effector specificities in bacterial regulatory proteins. Curr Opin Biotechnol 17, 34–42 (2006). [DOI] [PubMed] [Google Scholar]
- Garmendia J., Devos D., Valencia A. & de Lorenzo V. A la carte transcriptional regulators: unlocking responses of the prokaryotic enhancer-binding protein XylR to non-natural effectors. Mol Microbiol 42, 47–59 (2001). [DOI] [PubMed] [Google Scholar]
- Reed B., Blazeck J. & Alper H. Evolution of an alkane-inducible biosensor for increased responsiveness to short-chain alkanes. J Biotechnol 158, 75–79 (2012). [DOI] [PubMed] [Google Scholar]
- Beggah S., Vogne C., Zenaro E. & van der Meer J. R. Mutant transcription activator isolation via green fluorescent protein based flow cytometry and cell sorting. Microb Biotechnol 1, 68–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S., Salahshour S. & Nygren P. A. Monitored whole gene in vitro evolution of an anti-hRaf-1 affibody molecule towards increased binding affinity. Nat Biotechnol 29, 534–542 (2012). [DOI] [PubMed] [Google Scholar]
- Mustafi N., Grunberger A., Kohlheyer D., Bott M. & Frunzke J. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng 14, 449–457 (2012). [DOI] [PubMed] [Google Scholar]
- Chu B. C. & Vogel H. J. A structural and functional analysis of type III periplasmic and substrate binding proteins: their role in bacterial siderophore and heme transport. Biol Chem 392, 39–52 (2011). [DOI] [PubMed] [Google Scholar]
- Binnie R. A., Zhang H., Mowbray S. & Hermodson M. A. Functional mapping of the surface of Escherichia coli ribose-binding protein: mutations that affect chemotaxis and transport. Protein Sci 1, 1642–1651 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev 57, 862–952 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner J. W. et al. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol 176, 1157–1163 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M. Signaling by transmembrane proteins shifts gears. Cell 126, 829–831 (2006). [DOI] [PubMed] [Google Scholar]
- Srividhya K. V. & Krishnaswamy S. A simulation model of Escherichia coli osmoregulatory switch using E-CELL system. BMC Microbiol 4, 44 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer M. A. & Hellinga H. W. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr Opin Struct Biol 14, 495–504 (2004). [DOI] [PubMed] [Google Scholar]
- Breijo E. G. et al. TNT detection using a voltammetric electronic tongue based on neural networks. Sensors and Actuators A: Physical 192, 1–8 (2013). [Google Scholar]
- Alper H., Fischer C., Nevoigt E. & Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102, 12678–12683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer M. A., Looger L. L. & Hellinga H. W. Computational design of a Zn2+ receptor that controls bacterial gene expression. Proc Natl Acad Sci U S A 100, 11255–11260 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier B., Stumpp C., Wiesner S. & Hocker B. Computational design of ligand binding is not a solved problem. Proc Natl Acad Sci U S A 106, 18491–18496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes M. S. et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS One 6, e16292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service R. F. Protein designers go small. Science 341, 1052 (2013). [DOI] [PubMed] [Google Scholar]
- Boas F. E. & Harbury P. B. Design of protein-ligand binding based on the molecular- mechanics energy model. J Mol Biol 380, 415–424 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoitei M. L. et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334, 373–376 (2011). [DOI] [PubMed] [Google Scholar]
- Kiss G., Çelebi-Ölçüm N., Moretti R., Baker D. & Houk K. N. Computational enzyme design. Angew Chem Int Ed Engl 52, 5700–5725 (2013). [DOI] [PubMed] [Google Scholar]
- Tinberg C. E. et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature 501, 212–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. Molecular cloning: a laboratory manual. third edn, (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- Rosenberg A. H. et al. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56, 125–135 (1987). [DOI] [PubMed] [Google Scholar]
- Studier F. W. & Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189, 113–120 (1986). [DOI] [PubMed] [Google Scholar]
- Zaslaver A. et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3, 623–628 (2006). [DOI] [PubMed] [Google Scholar]
- Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2, 2006 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information