Abstract
AIM: To investigate chromosome 8 numerical aberrations, C-MYC oncogene alterations and its expression in gastric cancer and to correlate these findings with histopathological characteristics of gastric tumors.
METHODS: Specimens were collected surgically from seven patients with gastric adenocarcinomas. Immunostaining for C-MYC and dual-color fluorescence in situ hybridization (FISH) for C-MYC gene and chromosome 8 centromere were performed.
RESULTS: All the cases showed chromosome 8 aneuploidy and C-MYC amplification, in both the diffuse and intestinal histopathological types of Lauren. No significant difference (P < 0.05) was observed between the level of chromosome 8 ploidy and the site, stage or histological type of the adenocarcinomas. C-MYC high amplification, like homogeneously stained regions (HSRs) and double minutes (DMs), was observed only in the intestinal-type. Structural rearrangement of C-MYC, like translocation, was observed only in the diffuse type. Regarding C-MYC gene, a significant difference (P < 0.05) was observed between the two histological types. The C-MYC protein was expressed in all the studied cases. In the intestinal-type the C-MYC immunoreactivity was localized only in the nucleus and in the diffuse type in the nucleus and cytoplasm.
CONCLUSION: Distinct patterns of alterations between intestinal and diffuse types of gastric tumors support the hypothesis that these types follow different genetic pathways.
Keywords: Chromosome 8 aneuploidy, C-MYC amplification, Immunostaining, Gastric adenocarcinoma
INTRODUCTION
Gastric cancer is the third most frequent type of cancer in the world[1]. In Northern Brazil, the State of Pará presents a high incidence of this neoplasia type and its capital, Belém, is ranked eleventh in number of gastric cancers per inhabitant among all cities in the world with cancer records[2]. Food factors may be related to the high incidence of this neoplasia in Pará, specially high consumption of salt-conserved food, reduced use of refrigerators and little consumption of fresh fruit and vegetables[3].
Gastrointestinal tract tumors are notorious for being difficult to analyze by standard cytogenetic techniques[4-8]. Fluorescence in situ hybridization (FISH) technique with specific DNA probes allows rapid detection of chromosome aberrations in tumor interphase nuclei. In FISH studies, numerical aberrations in chromosomes 1, 7, 8, 9, 17, 20, X and Y are common[9-12]. Chromosome 8 abnormalities are frequent, not only in gastric neoplasias, but also in several types of hematopoietic proliferations and solid tumors[13-15].
Among the genes found on chromosome 8, C-MYC, located at 8q24, has been the most studied. C-MYC gene is a regulator of cell cycle and plays a major role in control of cell growth, differentiation, apoptosis and neoplastic transformation[16]. C-MYC gene overexpression is a frequent alteration and has been described in several types of human cancer[17-19]. An increased C-MYC gene expression has been found in gastric neoplasias[20-23].
The aim of this study was to investigate chromosome 8 numerical aberrations, C-MYC oncogene alterations and its expression in gastric cancer samples from the State of Pará, using the FISH technique and immunohistochemistry. Possible correlations between these findings and histopathological characteristics were also evaluated.
MATERIALS AND METHODS
Cases studied
Seven samples of primary tumors submitted to surgical resection were obtained from male patients in Pará State João de Barros Barreto University Hospital (HUJBB). Patients’ ages and tumors’ anatomical sites were obtained from tumor registries (Table 1). The patients had never been submitted to chemotherapy or radiotherapy prior to surgery, nor had they any other diagnosed cancer. Genetic study of samples was approved by the Ethics Committee of HUJBB. A fraction of each sample was used for routine histopathological diagnosis according to Laurén’s classification[24].
Table 1.
Number of signals by percentage of analyzed nuclei
| Case | Age (yr) | Origin | HT | UICC | Immunostaining | Probe |
Percentage of nuclei |
||||||
|
Number of signals | |||||||||||||
| 1 | 2 | 3 | 4 | 5 | ≥ 6 | HA | |||||||
| 1 | 77 | Antrum | Intestinal | T2N1M0 | Nuclear | C-MYC | 0 | 34 | 23 | 25 | 14 | 2 | 2 |
| Chrom.8 | 0 | 58 | 33 | 9 | 0 | 0 | - | ||||||
| 2 | 48 | Antrum | Intestinal | T4N0M0 | Nuclear | C-MYC | 0 | 35 | 10 | 20 | 29 | 2 | 4 |
| Chrom.8 | 0 | 70 | 21 | 6 | 3 | 0 | - | ||||||
| 3 | 74 | Antrum | Diffuse | T1N0M0 | Nuclear/Cytoplasmatic | C-MYC | 1 | 38 | 29 | 30 | 2 | 0 | 0 |
| Chrom.8 | 2 | 59 | 19 | 18 | 2 | 0 | - | ||||||
| 4 | 74 | Antrum/body | Intestinal | T4N0M0 | Nuclear | C-MYC | 0 | 40 | 12 | 28 | 17 | 2 | 1 |
| Chrom.8 | 0 | 63 | 25 | 1 | 1 | 0 | - | ||||||
| 5 | 41 | Antrum | Intestinal | T2N0M0 | Nuclear | C-MYC | 0 | 36 | 18 | 30 | 13 | 2 | 1 |
| Chrom.8 | 4 | 61 | 30 | 4 | 1 | 0 | - | ||||||
| 6 | 56 | Antrum/body | Diffuse | T2N1M0 | Nuclear/Cytoplasmatic | C-MYC | 0 | 31 | 38 | 17 | 14 | 0 | 0 |
| Chrom.8 | 3 | 60 | 32 | 5 | 1 | 0 | - | ||||||
| 7 | 55 | Antrum | Intestinal | T4N2M1 | Nuclear | C-MYC | 2 | 48 | 27 | 7 | 7 | 3 | 6 |
| Chrom.8 | 2 | 54 | 27 | 5 | 4 | 8 | |||||||
| Control | 77 | Stomach tissue | - | - | Without staining | C-MYC | 2 | 98 | 0 | 0 | 0 | 0 | 0 |
| Chrom.8 | 1 | 96 | 3 | 0 | 0 | 0 | - | ||||||
| Control | 37 | Lymphocytes | - | - | Without staining | C-MYC | 0.5 | 99.5 | 0 | 0 | 0 | 0 | 0 |
| Chrom.8 | 1 | 98.5 | 0.5 | 0 | 0 | 0 | - | ||||||
HT: Histological type; UICC: Union Internationale Contre le Cancer; HA: High amplification.
Immunostaining
For antigen retrieval, deparaffinized sections (5 μm) were pretreated by heating in a microwave oven in citrate buffer 10 mmol/L, pH 6.0 for 20 min. After cooling, sections were immersed in PBS containing 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity. Sections were then incubated in a humid chamber overnight at 4°C with primary antibody C-MYC (clone 9E10.3; dilution 1:100). After rinsing with PBS, slides were incubated with secondary antibody followed by streptavidin-biotin-peroxidase complex, both for 30 min at room temperature with a PBS wash between each step. Slides were visualised with diaminobenzidine-H2O2 and counterstained with Harry’s hematoxylin.
Fluorescence in situ hybridization
Tumor samples from all patients were processed for cytogenetic study as described previously[25]. FISH assay was performed on slides with cells fixed in methanol/acetic acid. A directly labeled dual-color probe was used for chromosome 8 alpha-satellite region (8q11) and for C-MYC gene region (8q24). Slides were washed in 2 × saline sodium citrate solution (SSC) and dehydrated in 70%, 80% and 95% ethanol, respectively. Samples were then denatured with 70% formamide/2 × SSC (pH 7.0) at 70°C for 2 min and transferred to an iced ethanol (-20°C) series at 70%, 80% and 95%. Probes were denatured at 96°C for 5 min. Then, 10 μL was applied onto the slide under a glass coverslip. In situ hybridization occurred at 37°C in a moist chamber overnight. Post-hybridization washings were done, and nuclei were counterstained with DAPI/antifade. Molecular cytogenetic analysis was carried out under an Olympus BX41 fluorescence microscope with triple DAPI/FITC/TRICT filter and an Applied Spectral Imaging image analysis. For each sample, 200 interphase nuclei were analyzed. To avoid misinterpretation due to technical error, normal lymphocyte nuclei and normal gastric tissue were used as a control.
Statistical analysis
For statistical evaluation, chi-square test was used. P < 0.05 was taken as significant.
RESULTS
All seven samples studied were histologically classified as gastric adenocarcinomas, 2 of them were diffuse type and 5 were intestinal type, according to Laurén’s classification (Table 1).
C-MYC was expressed in all cases. C-MYC immunoreactivity was localized in nucleus and cytoplasm of diffuse type samples, but only in nucleus of intestinal type (Figure 1A and 1B).
Figure 1.
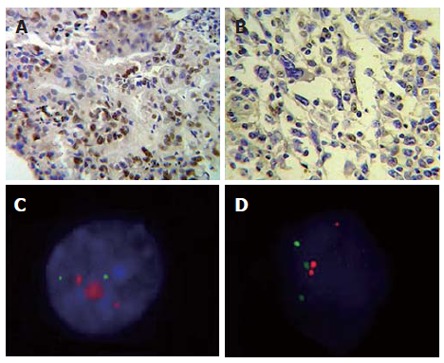
Cells submitted to immunohistochemistry and FISH techniques. A: Infiltrating gastric adenocarcinoma of intestinal type shows intense nuclear marcation for C-MYC, × 400; B: Gastric adenocarcinoma of diffuse type nuclear marcation and cytoplasmatic light marcation for C-MYC, × 400; C: Interphase nuclei presenting C-MYC high amplification (red) and chromosome 8 (green); D: Interphase nuclei presenting chromosome 8/C-MYC rearrangement.
In peripheral blood lymphocytes, two signals were observed in 98.5% of analyzed nuclei for chromosome 8 probe and 99.5% for C-MYC gene probe. Normal stomach tissue showed two signals in 96% of analyzed nuclei for chromosome 8 probe and 98% for C-MYC gene probe. All gastric adenocarcinoma cases showed numerical increase of chromosome 8 and C-MYC gene (Table 1).
Chromosome 8 trisomy was detected in all cases, varying from 19% (case 3) to 33% (case 1), and chromosome 8 tetrasomy (observed in all cases) varied from 1% (case 4) to 18% (case 3). Five signals for chromosome 8 were observed in 6 cases (85.7%) and the highest frequency was found in case 7 (4%). Six or more signals were observed in 1 case (case 7).
Presence of 6 or more signals for C-MYC gene was considered as an intermediary degree of amplification, whereas the cases which presented double minutes (DMs) and/or homogeneously stained regions (HSRs) were classified as presenting a high degree of gene amplification. Cells with more than six signals were found in 5 cases (71.42%). The frequency of cells with high amplification (Figure 1C) of this gene varied from 1% (case 4 and 5) to 6% (case 7) (Table 1). Thus, all intestinal type cases and none diffuse type presented intermediary and high amplification of C-MYC.
Rearrangements between C-MYC gene and chromosome 8 centromere were discriminated by evident separation of the two signals. In tumor samples, translocation was observed between C-MYC gene and another chromosome in case 3 (2%) and case 6 (4%), both of them of diffuse type (Figure 1D).
Analyzing the total number of signals for C-MYC gene and for chromosome 8 centromere, it was found that the number of signals for the gene was greater than the number of signals for chromosome 8 ploidy in all cases studied.
No significant difference (P < 0.05) was observed between chromosome 8 ploidy level and adenocarcinoma site, stage or histological type. Regarding C-MYC gene, a significant difference was observed between the two histological types. This difference was due to the presence of HSRs and/or DMs in Laurén’s intestinal type, where multiple (uncountable) signals were found per cell.
DISCUSSION
Gastric cancer is one of the most common neoplasias and both environmental and genetic factors contribute to its occurrence[26]. The present study used interphase dual-color FISH with direct fluorescent labeling for the chromosome 8 centromere/C-MYC gene and compared the copy number observed in 7 gastric adenocarcinoma samples with C-MYC expression by immunohistochemistry.
C-MYC amplification has been reported in a small percent of gastric carcinomas[21,27,28]. C-MYC expression was shown to be more frequent in diffuse than in intestinal type gastric cancer cells, and more frequent in gastric adenocarcinoma than in adenoma[29,30]. In the present study, C-MYC was expressed in all cases. C-MYC immunoreactivity was localized in the nucleus in intestinal-type and in cytoplasm and nucleus in diffuse type. More samples need to be investigated, in order to clarify if this immunostaining differences exist.
Our laboratory has previously observed the presence of chromosome 8 trisomy in all 16 cases studied by direct chromosome analysis and centromeric FISH[31] and in 60% of analyzed cells of ACP01 gastric adenocarcinoma cell line[32]. The gain of signals for the two probes analyzed in all samples in the present study corroborates with these data.
Panani et al[9] analyzed 33 gastric tumor samples by FISH, using a chromosome 8 alpha-satellite probe. Numerical aberrations in this chromosome were observed in 62.16% of studied samples, in which trisomy was detected in 43.24%, tetrasomy in 10.81% and monosomy in 8.10%. Our results confirmed that chromosome 8 trisomy is a common biological phenomenon in adenocarcinoma of stomach and can be used as a gastric mucosa malignancy marker. In our study, 100% of samples presented a gain of chromosome 8 as a clonal alteration.
Chromosome 8 numerical abnormalities, in which C-MYC is located, are suggested to be an important mechanism in C-MYC copy number increase. Xia et al[5] suggested that chromosome 8 trisomy, associated or not with other chromosomal aberrations, could occur even in less advanced stages of the disease, possibly prior to the occurrence of metastases.
Kitayama et al[12] analyzed interphase nuclei of 51 gastric cancer cases from pathology archives using 18 centromeric probes, including chromosome 8 probe, and a probe for C-MYC gene. They observed chromosome 8 numerical abnormalities in 56.9% of samples, which placed them among the most frequent alterations; in 12 cases, a C-MYC gain was observed and all of them presented chromosome 8 gain. Amplification of this oncogene was also reported in other FISH studies in gastric neoplasias[33-35].
C-MYC oncogene seems to be fundamental in the oncogenesis process. Thus, increased C-MYC allele number is directly related to the degree of tumor aggressiveness, considering that the greater the gene copy number, the higher its level of expression. This gene was amplified in all samples studied, but without numerically accompanying chromosome 8 ploidy; that is, in all cases there were more C-MYC gene alleles than this chromosome copies.
In another study conducted in our laboratory[36], 90.9% of cases presented intermediary amplification, 60% of which presented DMs and HSRs. It seems that, as our sample presented chromosome 8 gain as a clonal characteristic, C-MYC amplification is a later step, a consequence of carcinogenesis clonal expansion.
We have previously demonstrated, by comparative genomic hybridization, gains at region 8q24.1 (38.1%), which were found exclusively, in all intestinal type adenocarcinomas with systemic metastasis (M1). Results from this work also showed the highest amplification level in gastric cancer of intestinal type. C-MYC locus amplification may be an aggressiveness predictor in intestinal type gastric cancer, playing an important role in its development and progression. These results should provide useful information for developing more effective strategies in management of gastric cancer[37].
Stamouli et al[25] studied, by multicolor FISH, two primary gastric adenocarcinoma cases: a well-differentiated intestinal-type and a poorly differentiated diffuse type. The intestinal-type exhibited few structural abnormalities, in contrast to the diffuse type. In our analysis, all diffuse type cases presented at least four cells with translocation. It seems that this histological type is more susceptible to chromosomal rearrangements than intestinal type.
Correa[38] suggested that the intestinal type fits the multiple-step process. Thus, it is plausible that the intestinal type presents a greater number of DMs and/or HSRs than the diffuse type. Our findings support that these two histological types follow different genetic tumorigenesis mechanisms[39]. Moreover, it could be observed that translocations were restricted to diffuse type. This result can be explained by the fact that gene amplification is not necessarily associated or required for its overexpression. Leukemias, lymphomas and sarcomas commonly elicit specific balanced translocations which mediate proto-oncogenes activation, by their juxtaposition with promoter sequences or generating gene fusion[40].
Enhanced C-MYC protein expression contributes to almost every aspect of tumor cell biology. Although the ability of C-MYC to drive unrestricted cell proliferation and to inhibit cell differentiation had been well recognized, a recent work showed that deregulated C-MYC expression can drive cell growth and vasculogenesis, reduce cell adhesion, and promote metastasis and genomic instability. On the other hand, C-MYC loss not only inhibits cell proliferation and cell growth, but can also accelerate differentiation, increase cell adhesion and lead to an excessive response to DNA damage. Studies in animal models suggest that C-MYC may be a target for human cancer treatment, but it is still unknown whether such drugs will be useful[41].
The alterations found in this study have been described in the literature, even though their frequency was higher in our sample. Considering that external factors, such as eating habits and other environmental agents, have a direct influence on the development of this neoplasia, many genetic alterations may be regional characteristics of a given population.
Based on our findings we could also affirm that gastric adenocarcinomas of differing histopathological features are associated with distinct patterns of genetic alterations, suggesting that they evolve through different genetic pathways.
Footnotes
Supported by Financiadora de Estudos e Projetos (FINEP CT-INFRA/FADESP), No. 0927-03 and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) No. 2003/06540-5; DQC had a master fellowship, No. 151127/2002-6, granted by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
S- Editor Liu Y L- Editor Zhu LH E- Editor Bi L
References
- 1.Instituto Nacional do Câncer (INCA) Estimativa 2005: incidência de câncer no Brasil. Rio de Janeiro: INCA; 2004. p. 33. [Google Scholar]
- 2.Cancer Databases and Other Resourcess. International Agency for Research on Cancer (IARC) page. Available from URL: http: //www.iarc.fr. Accessed January 28; 2005. [Google Scholar]
- 3.Pereira LP, Waisberg J, André EA, Zanoto A, Mendes Júnior JP, Soares HP. Detection of Helicobacter pylori in gastric cancer. Arq Gastroenterol. 2001;38:240–246. doi: 10.1590/s0004-28032001000400006. [DOI] [PubMed] [Google Scholar]
- 4.Ferti-Passantonopoulou AD, Panani AD, Vlachos JD, Raptis SA. Common cytogenetic findings in gastric cancer. Cancer Genet Cytogenet. 1987;24:63–73. doi: 10.1016/0165-4608(87)90083-5. [DOI] [PubMed] [Google Scholar]
- 5.Xia JC, Lu S, Geng JS, Fu SB, Li P, Liu QZ. Direct chromosome analysis of ten primary gastric cancers. Cancer Genet Cytogenet. 1998;102:88–90. doi: 10.1016/s0165-4608(97)00293-8. [DOI] [PubMed] [Google Scholar]
- 6.Ochi H, Douglass HO Jr, Sandberg AA. Cytogenetic studies in primary gastric cancer. Cancer Genet Cytogenet. 1986;22:295–307. doi: 10.1016/0165-4608(86)90022-1. [DOI] [PubMed] [Google Scholar]
- 7.Gomyo Y, Osaki M, Kaibara N, Ito H. Numerical aberration and point mutation of p53 gene in human gastric intestinal metaplasia and well-differentiated adenocarcinoma: analysis by fluorescence in situ hybridization (FISH) and PCR-SSCP. Int J Cancer. 1996;66:594–599. doi: 10.1002/(SICI)1097-0215(19960529)66:5<594::AID-IJC2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Kitayama Y, Igarashi H, Sugimura H. Different vulnerability among chromosomes to numerical instability in gastric carcinogenesis: stage-dependent analysis by FISH with the use of microwave irradiation. Clin Cancer Res. 2000;6:3139–3146. [PubMed] [Google Scholar]
- 9.Panani AD, Ferti AD, Avgerinos A, Raptis SA. Numerical aberrations of chromosome 8 in gastric cancer detected by fluorescence in situ hybridization. Anticancer Res. 2004;24:155–159. [PubMed] [Google Scholar]
- 10.Han K, Oh EJ, Kim YS, Kim YG, Lee KY, Kang CS, Kim BK, Kim WI, Shim SI, Kim SM. Chromosomal numerical aberrations in gastric carcinoma: analysis of eighteen cases using in situ hybridization. Cancer Genet Cytogenet. 1996;92:122–129. doi: 10.1016/s0165-4608(96)00165-3. [DOI] [PubMed] [Google Scholar]
- 11.Beuzen F, Dubois S, Fléjou JF. Chromosomal numerical aberrations are frequent in oesophageal and gastric adenocarcinomas: a study using in-situ hybridization. Histopathology. 2000;37:241–249. doi: 10.1046/j.1365-2559.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- 12.Kitayama Y, Igarashi H, Watanabe F, Maruyama Y, Kanamori M, Sugimura H. Nonrandom chromosomal numerical abnormality predicting prognosis of gastric cancer: a retrospective study of 51 cases using pathology archives. Lab Invest. 2003;83:1311–1320. doi: 10.1097/01.lab.0000087622.80751.c5. [DOI] [PubMed] [Google Scholar]
- 13.Bofin AM, Ytterhus B, Fjøsne HE, Hagmar BM. Abnormal chromosome 8 copy number in cytological smears from breast carcinomas detected by means of fluorescence in situ hybridization (FISH) Cytopathology. 2003;14:5–11. doi: 10.1046/j.1365-2303.2003.01132.x. [DOI] [PubMed] [Google Scholar]
- 14.Paulsson K, Fioretos T, Strömbeck B, Mauritzson N, Tanke HJ, Johansson B. Trisomy 8 as the sole chromosomal aberration in myelocytic malignancies: a multicolor and locus-specific fluorescence in situ hybridization study. Cancer Genet Cytogenet. 2003;140:66–69. doi: 10.1016/s0165-4608(02)00628-3. [DOI] [PubMed] [Google Scholar]
- 15.Mark HF, Feldman D, Samy M, Sun C, Das S, Mark S, Lathrop J. Assessment of chromosome 8 copy number in cervical cancer by fluorescent in situ hybridization. Exp Mol Pathol. 1999;66:157–162. doi: 10.1006/exmp.1999.2256. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 18.Escot C, Theillet C, Lidereau R, Spyratos F, Champeme MH, Gest J, Callahan R. Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci USA. 1986;83:4834–4838. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZR, Liu W, Smith ST, Parrish RS, Young SR. c-myc and chromosome 8 centromere studies of ovarian cancer by interphase FISH. Exp Mol Pathol. 1999;66:140–148. doi: 10.1006/exmp.1999.2259. [DOI] [PubMed] [Google Scholar]
- 20.Han S, Kim HY, Park K, Cho HJ, Lee MS, Kim HJ, Kim YD. c-Myc expression is related with cell proliferation and associated with poor clinical outcome in human gastric cancer. J Korean Med Sci. 1999;14:526–530. doi: 10.3346/jkms.1999.14.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii H, Gobé G, Kawakubo Y, Sato Y, Ebihara Y. Interrelationship between Epstein-Barr virus infection in gastric carcinomas and the expression of apoptosis-associated proteins. Histopathology. 2001;38:111–119. doi: 10.1046/j.1365-2559.2001.01037.x. [DOI] [PubMed] [Google Scholar]
- 22.Xu AG, Li SG, Liu JH, Gan AH. Function of apoptosis and expression of the proteins Bcl-2, p53 and C-myc in the development of gastric cancer. World J Gastroenterol. 2001;7:403–406. doi: 10.3748/wjg.v7.i3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang GX, Gu YH, Zhao ZQ, Xu SF, Zhang HJ, Wang HD, Hao B. Coordinate increase of telomerase activity and c-Myc expression in Helicobacter pylori-associated gastric diseases. World J Gastroenterol. 2004;10:1759–1762. doi: 10.3748/wjg.v10.i12.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 25.Stamouli MI, Ferti AD, Panani AD, Raftakis J, Consoli C, Raptis SA, Young BD. Application of multiplex fluorescence in situ hybridization in the cytogenetic analysis of primary gastric carcinoma. Cancer Genet Cytogenet. 2002;135:23–27. doi: 10.1016/s0165-4608(01)00635-5. [DOI] [PubMed] [Google Scholar]
- 26.Ponz de Leon M. Genetic predisposition and environmental factors in gastric carcinoma. Recent Results Cancer Res. 1994;136:179–202. doi: 10.1007/978-3-642-85076-9_14. [DOI] [PubMed] [Google Scholar]
- 27.Peng H, Diss T, Isaacson PG, Pan L. c-myc gene abnormalities in mucosa-associated lymphoid tissue (MALT) lymphomas. J Pathol. 1997;181:381–386. doi: 10.1002/(SICI)1096-9896(199704)181:4<381::AID-PATH787>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 28.O'Leary JJ, Landers RJ, Crowley M, Healy I, Kealy WF, Hogan J, Doyle CT. Alterations in exon 1 of c-myc and expression of p62c-myc in cervical squamous cell carcinoma. J Clin Pathol. 1997;50:896–903. doi: 10.1136/jcp.50.11.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajdú J, Kozma L, Kiss I, Szentkereszty Z, Szakáll S, Ember I. Is the presence of distant metastasis associated with c-myc amplification in gastric cancer. Acta Chir Hung. 1997;36:119–121. [PubMed] [Google Scholar]
- 30.Amadori D, Maltoni M, Volpi A, Nanni O, Scarpi E, Renault B, Pellegata NS, Gaudio M, Magni E, Ranzani GN. Gene amplification and proliferative kinetics in relation to prognosis of patients with gastric carcinoma. Cancer. 1997;79:226–232. doi: 10.1002/(sici)1097-0142(19970115)79:2<226::aid-cncr5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 31.Assumpção PP, Ishak G, Chen ES, Takeno SS, Leal MF, Guimarães AC, Calcagno DQ, Khayat AS, Demachki S, Smith Mde A, et al. Numerical aberrations of chromosome 8 detected by conventional cytogenetics and fluorescence in situ hybridization in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genet Cytogenet. 2006;169:45–49. doi: 10.1016/j.cancergencyto.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Lima EM, Rissino JD, Harada ML, Assumpção PP, Demachki S, Guimarães AC, Casartelli C, Smith MA, Burbano RR. Conventional cytogenetic characterization of a new cell line, ACP01, established from a primary human gastric tumor. Braz J Med Biol Res. 2004;37:1831–1838. doi: 10.1590/s0100-879x2004001200008. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y, Shintaku K, Ishii Y, Asai S, Ishikawa K, Fujii M. Analysis of MYC and chromosome 8 copy number changes in gastrointestinal cancers by dual-color fluorescence in situ hybridization. Cancer Genet Cytogenet. 1998;107:61–64. doi: 10.1016/s0165-4608(98)00089-2. [DOI] [PubMed] [Google Scholar]
- 34.Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- 35.Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence In situ hybridization studies. Cancer Genet Cytogenet. 2000;117:97–103. doi: 10.1016/s0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- 36.Calcagno DQ, Leal MF, Taken SS, Assumpção PP, Demachki S, Smith Mde A, Burbano RR. Aneuploidy of chromosome 8 and C-MYC amplification in individuals from northern Brazil with gastric adenocarcinoma. Anticancer Res. 2005;25:4069–4074. [PubMed] [Google Scholar]
- 37.Burbano RR, Assumpção PP, Leal MF, Calcagno DQ, Guimarães AC, Khayat AS, Takeno SS, Chen ES, De Arruda Cardoso Smith M. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006;26:2909–2914. [PubMed] [Google Scholar]
- 38.Correa P. Bacterial infections as a cause of cancer. J Natl Cancer Inst. 2003;95:E3. doi: 10.1093/jnci/95.7.e3. [DOI] [PubMed] [Google Scholar]
- 39.Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ., 157: 327-349, 20044 Mitelman Database of Chromosome Aberrations in Cancer. National Center for Biotechnology Information (NCI) page. Available from URL: http: //cgap.nci.nih.gov/Chromosomes/Mitelman. Accessed January 28; 2005. [Google Scholar]
- 40.Popescu NC, Zimonjic DB. Chromosome-mediated alterations of the MYC gene in human cancer. J Cell Mol Med. 2002;6:151–159. doi: 10.1111/j.1582-4934.2002.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]


