Abstract
Here we present a set of measurements using Differential Scanning Fluorimetry (DSF) as an inexpensive, high throughput screening method to investigate the folding of silk protein molecules as they abandon their first native melt conformation, dehydrate and denature into their final solid filament conformation. Our first data and analyses comparing silks from spiders, mulberry and wild silkworms as well as reconstituted ‘silk' fibroin show that DSF can provide valuable insights into details of silk denaturation processes that might be active during spinning. We conclude that this technique and technology offers a powerful and novel tool to analyse silk protein transitions in detail by allowing many changes to the silk solutions to be tested rapidly with microliter scale sample sizes. Such transition mechanisms will lead to important generic insights into the folding patterns not only of silks but also of other fibrous protein (bio)polymers.
Differential Scanning Fluorimetry is a high throughput screening method for protein solutions using interactions of proteins with low-molecular-weight ligands1,2,3. Often also called the thermofluor assay or thermal shift assay, DSF is an inexpensive, simple and quick method that probes the refolding of a protein as it experiences progressive denaturation4,5. The technique relies on using specific dyes that fluoresce as they interact with proteins in their denaturation transition between a strongly hydrated solvated state and the final aggregated solid state. State-of-the-art quantitative polymerase chain reaction (qPCR) instruments can automatically process many controlled conditions and interactions in a single run and hundreds in a day1.
During a DSF experiment, denaturation is controlled by slowly heating the protein solution from a base-line, natural (i.e. stabilised and stabilising) temperature. This heating induces changes in the water shells and the hydrogen bonds, thus leading to associated adjustments in the conformation of the protein (Figure 1). Specifically designed dyes can interact with the bonds and signal this interaction by changes in their fluorescence spectrum. Here we present a set of measurements aimed to investigate the folding of silk protein molecules as they abandon their first native melt conformation, dehydrate and denature into their final solid filament conformation. We conclude that DSF provides a powerful tool to analyse silk protein transitions in depth and with the potential of generic insights also into the structures, properties and functions of silks and other fibrous proteins.
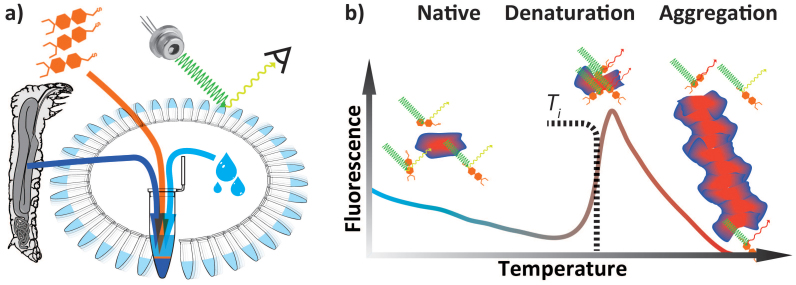
Figure 1. Differential Scanning Fluorimetry (DSF) aka Thermofluor Assay aka Thermal Shift Assay technique and signal.
(a) Schematics illustrating the sample preparation for high throughput dynamic scanning fluorimetry thermal stability assay (b) Cartoon representing the native, denaturated and aggregated states of protein alongside its typical exogenous fluorescence response.
Such information on transitions is of fundamental importance for better understanding a protein's biochemistry, its structure-function relationships and its resistance to chemical, mechanical, and thermal denaturation6. Most diagnostic methods rely on the establishment of equilibrium between the folded and unfolded forms of a protein, and on an analysis of the ratio between the various states as conditions change and the protein slowly denatures in response. For example, optical techniques such as circular dichroism (CD)7,8,9, infrared spectroscopy (IR)10,11,12 and dynamic light scattering (DLS)13 are routinely used for studies of protein thermodynamics. Since the absorption of circularly polarized light is weak and decreases upon protein precipitation, this technique typically requires concentration higher than 1 mg/ml. For infrared spectroscopy, the strong absorption of water and its temperature dependency limits the applicability of technique for thermodynamic studies. Although sensitive to increasing radius of protein gyration during aggregation, DLS can be insensitive to unfolding mechanisms not affecting the size of the protein. In addition to optical techniques, the exo-endothermic probing of differential scanning calorimetry (DSC) has become a key tool to examine a protein's denaturation and glass transition temperature14,15,16. While providing valuable information, all these techniques can generally only thermally probe one sample at a time, which imposes severe limitations on screening studies or for analytical statistics.
In contrast, fluorescence spectroscopy is extremely sensitive, often requiring less than parts per trillion for detection17 and can ultimately detect a single molecule18. As we shall demonstrate, differential scanning fluorimetry (DSF) is a highly sensitive analysis tool that allows low protein concentrations to be probed at remarkable resolutions of temperature transitions. Importantly, the sample throughput using DSF can be substantial in multi-well thermal cycling machines, which have been perfected in the service of DNA screening analysis. Key to such DNA analysis is its amplification via the PCR technology, which uses temperature cycling in order to allow the controlled reaction of heat-sensitive polymerase enzymes. With the rapid expansion of DNA cloning, real-time PCR machines with accurate temperature control have become ever more affordable and are now wide spread tools. The high throughput enables the concurrent study of many experimental conditions, and it has for example been used to evaluate the stabilising effect of an array of ionic liquids on a common protein4.
In addition to its features of high throughput and low costs, DSF technology has led to the development of a wide variety of specific exogenous fluorescent molecules, which makes it an extremely versatile technique19. In a typical application, temperature transitions are used to probe the interactions of a protein with a fluorescent dye as it binds with specific sites such as residues exposed during unfolding20,21. Sypro® orange is well suited as dye of choice as it was designed specifically to activate fluorescence when binding to hydrophobic patches exposed as a result of conformational changes22. Using such a specific dye, DSF elicits and identifies key temperature transitions and protein stability features23, which in turn allows the probing and analysis of molecular interactions. Other dyes designed to bind to hydrophobic regions of amyloid fibrils24 might also be of interest because silk and amyloid fibrils share important folding characteristics25. Moreover, highly specific antibodies with labels such as the green fluorescent protein (GFP) could also be used with DSF in order to target specific domains of a protein26,27.
Silk is a perfect model material for the study of protein denaturation. Firstly, Nature has evolved many thousands of silks, which between them sport many minor as well as major sequence differences28,29 in addition to displaying key differences in processing (i.e. dehydration/denaturation) parameters30,31,32. Secondly, unlike most other proteins, silk proteins have evolved to denature ‘on demand' in order to assume their 3rd functional ‘state' i.e. that of a thread. A silk's 1st state has the molecule in its long-term storage conformation. Its 2nd state occurs during the rapid structural reconfiguration during spinning. This three state transition progress is genetically pre-programmed, which makes silk protein conformation transitions interesting and valuable generic research tools. For example, there is overlap with amyloid formation33 yet in the case of silks this is not an errant ‘mis'-folding pathway but the result of design by evolution. One could consider the 1st state to be classed as active (alive) - for the protein molecules stored in the gland are labile and easily denatured if perturbed. The 2nd state could be called re-active - for each molecule refolds and reconfigures according to the environmental conditions it experiences in the controlled environment of the spinning duct. And the 3rd state could be deemed inactive (dead) - for the denatured protein molecules no longer respond actively as individuals to environmental conditions. Instead, they are now firmly locked into a polymer network, responding passively and collectively to stresses and strains imparted on the bulk material. Whether one agrees with this view or not, it is well established that silk molecules have two different states linked by a distinct, one-way transition34,35. We may assume that 400 M years of selective tweaking and tuning will have evolved a ‘spinning' process that is highly advanced. The three major independent evolutionary pathways (in insects, arachnids and crustaceans36) that have led to more or less the same proteins and processes suggest to us that silks can provide insights into protein denaturation that are likely to be fundamental and of generic relevance.
Results
Here we examine the transition from native to denatured in 3 specific silk protein complexes. We note that a silk typically consists of two distinct, different and large fibrous proteins (in the 3 cases presented here well characterised37,38) with some small molecules (mostly unknown and of unknown function39,40) in the mix. While the fibres of all three examples are inherently not very different when mechanically tested, the folding pathways, and thus the assembly mechanisms, clearly are distinct - as our DSF data demonstrate.
Convention assigns a protein a ‘melt' temperature Tm to denote the onset of molecular reconfiguration during denaturation; this is potentially confusing with the concept of Tm in the polymer literature. To avoid misunderstanding we call the point at which fluorescence changes most rapidly Ti- to denote the temperature induced instability of hydrogen interactions so fundamental to the protein ‘melt' concept41.
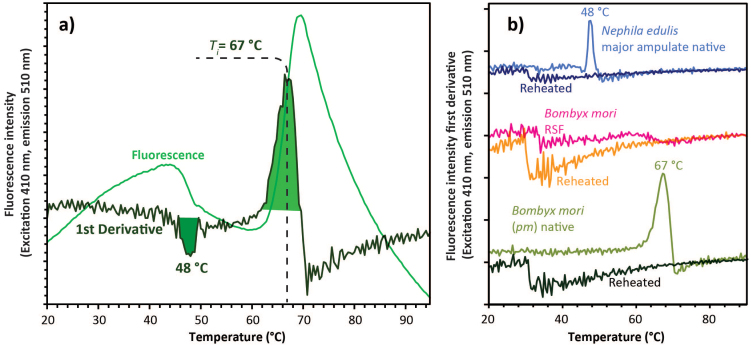
Shortly after reaching the instability temperature, Ti, the rapid protein aggregation causes dye dissociation and an associated decrease of fluorescence4. Comparable to the analysis of DSC signals42, also in DSF the first derivative peak highlights these key transitions. Outlined in Fig. 2, these transitions vary between silks. In Nephila edulis dragline spider silk, the key transition occurred around 48°C and in Bombyx mori mulberry worm silk it occurred around 67°C. Confirmation on the irreversibility of the denaturation process in our silks was provided by re-runs of the samples after 24 hours in a refrigerator (Fig. 2). RSF (Reconstituted Silk Fibroin) feedstock/dopes lack any denaturation related fluorescence peak (Fig. 2); importantly this observation strongly supports the argument (based on rheology measurements) that RSF dopes ‘denature' significantly differently from native silks31.
Figure 2. Temperature induced exogenous fluorescence response of Sypro® to the heating of native and denatured-reconstituted silk dope feedstocks.
(a) Representative fluorescence for Bombyx mori native silk dopes along first derivative response. (b) Comparative signals for spider, silkworm and reconstituted ‘silk' (RSF) dope shown in the first derivative responses for Nephila edulis spider major ampullate silk, Bombyx mulberry silkworm native silk and a dope reconstituted from spun silk fibres (RSF). Responses of the materials were measured in a first run immediately shadowed by a second run (reheated), both following the same temperature ramps of 0.4°C/min increasing from 20 to 90°C using excitation of 470 nm and emission of 510 nm.
We assert that DSF is a powerful tool to assign silk and silk-derived bio-protein conversions and for this we need to demonstrate that the fluorescence signal does indeed occur only during a true transition. After all, irreversible denaturation, as outlined in the introduction, is supposed to be the key feature in the 3-stage active-to-inactive silk protein extrusion process of spinning a fibre from an aqueous dope. Our data (Fig. 2 and SM2) clearly demonstrate the irreversibility of the fluorescence signal on both spider and worm silks. Upon heating the sample a second time, the transitions observed at 48 and 67°C resp. completely disappear. The only remarkable feature that emerged during all repeating runs (i.e. reheating a dope a 2nd time) was a small drop i.e. decrease in fluorescence around 31°C, which we provisionally assign to a softening of the presumably gelled samples. Importantly, the DSF signal curve of the reconstituted silk feedstock (RSF) completely lacks any Ti peak at 67°C. Moreover, this material displayed rather similar responses to both the initial and the reheating runs with the small reheating drop at 31°C (Fig. 3), which suggests that the reconstituted silk prepared lacks the denaturation process of native dopes, be they from spider or worm silks. DSF thus corroborates previous rheology and scattering reports31,43,44 and supports the theory that reconstituted silk lacks the molecular assembly behaviour characteristic of native silk dopes.
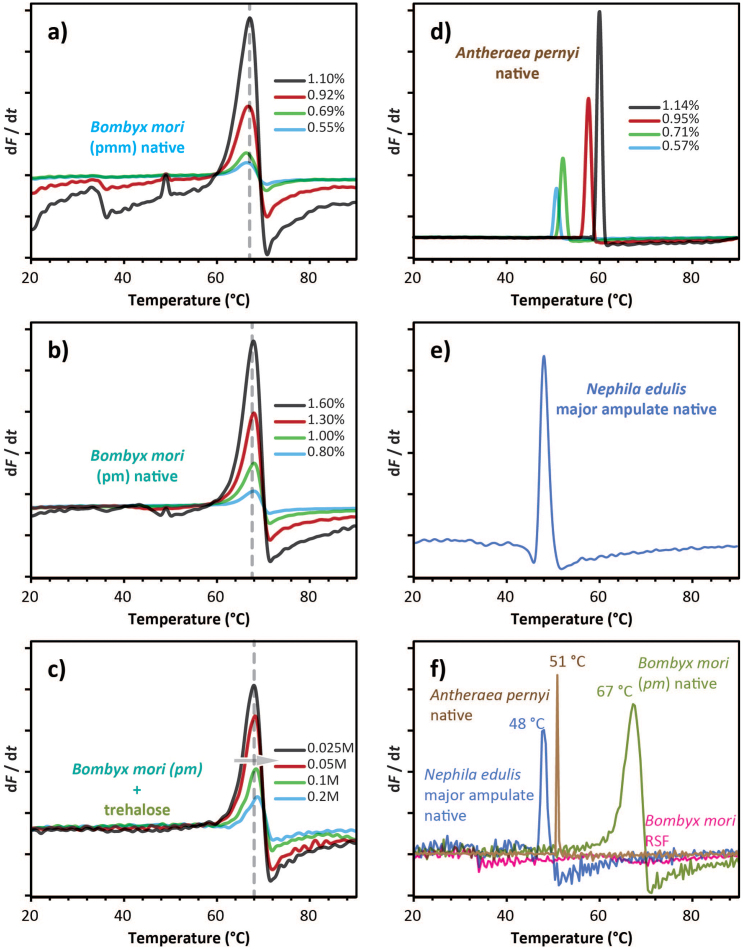
Figure 3. DSF fluorescence signals of various silks feedstock/dopes and dope concentrations.
(a, b) For the ‘domesticated' silk of Bombyx mori the signal decreases in strength as a function of silk dope concentration for two distinct major parts of the gland (pmm for middle section and pm for posterior section, details in SM). (c) For the ‘wild' silk of Antherea pernii the picture is rather different with the signal shifting not only changing in strength but also shifting along the temperature ramp as a function of dope concentration. (d) The signals for dope from the Major Ampullate gland of the Nephila edulis spider resemble those for Antherea dopes at high concentration. (e) Trehalose affects the signal of Bombyx dope (shown by the arrow) dependent on the concentration of the additive. (f) Overlay of the exogenous fluorescence intensity first derivative for native dope of Bombyx, Nephila and Antheraea as well as for reconstituted silk dope made using Bombyx silk fibres.
In comparison to the Bombyx dope, native dope from Nephila edulis major ampulate gland showed a weaker Ti, at the much lower temperature of 48°C. This feature could be explained by either (i) a smaller fraction of the protein undergoing denaturation or (ii) the spider silks having a much greater thermal instability45. Or indeed (iii) the spider silk dopes might have been at much lower concentrations, which was difficult to assess in this study due to a combination of small gland sizes and the variability in concentrations (C. Dicko personal communication). Fortunately we were able to source wild silk worms Antheraea pernyi that have silks with proteins more alike to spider silks than Bombyx silks despite a much closer taxonomic relationship with the latter46. So, to evaluate the effect of protein conformation and concentration on the Ti, we compared native silk dopes of Bombyx mori and Antheraea pernyi, which had been diluted in different ratios to provide dopes of specific concentrations. Importantly, the Antheraea wild silk dopes behaved much more like the spider silks dopes, although here we could also examine the effect of concentration.
Firstly, the overall signal pattern of Antheraea dope resembled the pattern of Nephila dragline dope (see SM3). Secondly, Fig. 3a shows that the concentration affected strongly the signal integrated intensity for all native silk dope measured. Hence, it stresses the need for determining the sample concentration in order to compare intensity values. Figure 3b shows that the temperature trigger for both parts of the Bombyx mori silk gland is only weakly influenced by the protein. In contrast, the Ti of A. pernyi dope is much more dependent on the concentration whilst being at significantly lower temperatures.
Last but not least, we examined the effect of compounds known to stabilise molecules under hydration stress. After all, the study aimed to examine dehydration, and we hypothesised that a cryo/denaturation protectant such as trehalose47 should have a measureable effect. Indeed, this turned out to be the case by decreasing the transition integrated intensity (Fig. 3c) and significantly shifting the folding temperature (Fig. 3d). This experiment demonstrates that trehalose somehow shields the bonds and thus manages to protect the folding conformation. As importantly for the concept, it demonstrates that DSF can indeed be used also to test compounds that might affect folding parameters in one way or another.
Discussion
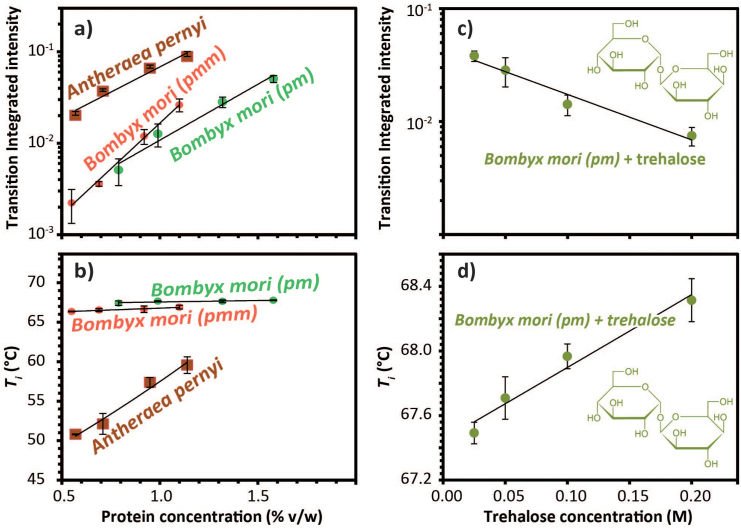
Our data demonstrate that DSF is a very powerful technique for detailed observations of clearly defined denaturation transitions in silks (Fig. 4). Not only were the quantitatively measured transitions highly specific for each of the silks tested, they also showed significant concentration dependences, which in themselves reveal underlying molecular folding differences between these silks. The signature of mulberry silk was significantly different from that of wild silk, which in turn was rather close to that of spider silk. The universal cryoprotectant trehalose shifted the folding signature slightly but significantly, thus demonstrating its shielding action. Last but not least, the technology clearly showed that reconstituted silks showed a folding pattern comparable to that of its denatured progenitor, which has important implications for the argument that RSF is not a silk sensu stricto and should neither be treated - nor called - as such48.
Figure 4. Analysis of DSF Data shown in Figure 3.
(a) Integrated transition intensity as a function of the protein concentration for Antheraea pernyi and Bombyx mori posterior (pm) and middle (pmm). (b) Ti as a function of the protein concentration for Antheraea and Bombyx posterior (pm) and middle (pmm) sections. (c) Integrated transition intensity as a function of trehalose concentration for B. mori posterior section (pm). (d) Ti as a function of the trehalose concentration for B. mori posterior section (pm).
DSF also now allows us to formulate specific and testable hypotheses. For example, the Ti of folding transitions was relatively independent of dope concentrations in silks of Bombyx mori, which may suggest that 6000 yrs of farming for spinning consistency may have bred out any ancestral responses to dehydration stresses. Wild silkworms like Antherea pernii and spiders like Nephila edulis, on the other hand still live in natural habitats where desiccation is a real threat49 and their silks have built-in mechanisms to shield against this. However this may be, the technique discussed here will allow us to draw possible and testable conclusions that relate not only to molecular mechanisms but also to evolutionary drivers behind them by using clade comparisons50.
Our first experiments with DSF reveals the power of this technique to quantitatively probe the underlying dynamics of a protein and protein mix with the view of identifying patterns and perhaps even assigning activity patterns. Hypothesising on such assignments is not the purpose of this article introducing a tool novel to silk studies. Hence here we simply propose that DSF provides a powerful tool to screen for specific denaturation transitions. We assert that operating this tool will have significant implications for the ‘prospecting' not only of silks but also of other natural bio-polymer proteins as well as for iterative testing procedures of synthetic or semi-synthetic ‘designer' proteins.
Methods
Preparation of the silk samples is explained in detail in the supplementary methods SM1. In essence, the samples were either obtained (i) by dissection of the gland and by subsequent washing out of the silk-dope in Ringer's solution or (ii) by following traditional recipes to make reconstituted silk dopes RSF. After silk dope preparation, various amount of Sypro® Orange Merocyanine dye (Sigma Aldrich) were added to the silk solution19 with the exception of control runs. Sypro® Orange has an absorption maxima at 470 nm and an emission maxima at 569 nm50. This dye was designed to have a weak intrinsic fluorescence but a strong florescence upon binding to protein (~500 fold). To homogenise the dyed solutions, samples were then protected from light and placed onto an orbital shaker at 60 rpm in a cold room at 5°C for 9 to 16 hours. Sample concentrations (w/v) were determined by drying 200 μL of sample on aluminium foil at 60°C overnight and measuring the residual dry weight.
For the testing of the interactions between the dope and the dye, we deployed a Rotor-Gene Q (Qiagen, Venlo, Netherlands) originally developed for qPCR reactions. This instrument offers highly controlled, steady temperature ramps as well as high florescence sensitivity. The fluorescence signal was measured for 36 samples simultaneously using 5 independent channels (green, yellow, orange, red and crimson) respectively exciting at 470, 530, 585, 625 and 680 nm and emitting at 510, 557, 610, 660 and 715 nm. The green channel (excitation at 470 nm, emission at 510 nm) was not the most sensitive (which was yellow) but green offered the best dynamic range for probing the fluorescence of Sypro® orange interacting with silk protein over the concentration range studied (see Supplementary Methods). Fluorescence was measured continuously at 1 Hz (i.e. one data point per minute per sample) while temperature was gradually raised at 0.4°C/min with the instrument spinning samples at 400 rpm, which ensures excellent mixing without causing phase separation. Gain was optimized at the beginning of each sample run and the resulting gain value was used to compare between runs; moreover, gain optimization provided the desired range of starting fluorescence at the start temperature for all channels. The standard deviations for each sample set were calculated from 4 repeats. We used the centre of gravity of the first derivative peak of the measurements in order to assure best accuracy for each calculation of instability transition temperature (Ti). All results for each sample set were obtained within a few hours of each other.
Sypro Orange ® was developed and indeed has become is the dye of choice for detecting most protein re-folding studies19,51. It also provided by far the best results also for our silks when compared to a number of other candidate dyes such as Nile Red, Congo Red and Rhodamine B, all of which are known or suspected to indicate molecular folding of silk proteins. Sypro Orange ® is part of the merocyamine dye group, which change colour with changing polarity of the solvent52. Such solvatochromism occurs when the hydrophobic dye binds to our protein's hydrophobic domains53. In addition, the sulfonate function (-SO3-) is likely to bind with buried charged residues such as arginine54. Figure 2 shows the evolution of green fluorescence (i.e. measured by the green channel) in response to increasing temperature. Denaturation starts around 60°C when water molecules progressively leave the protein backbone; this allows the dihedral angles around the amide bounds to rotate freely, which in turn leads to the adoption of progressively more stable conformations55. During this process the evolving configurations allow dye molecules to bind to hydrophobic patches as they expose themselves on the proteins surface4. Such binding delocalises the dye's electron, which substantially enhances fluorescence, in the case of Sypro® orange by more than 500 fold19 making this dye's fluorescence a superb indicator of temperature induced protein denaturation process and product.
Author Contributions
F.V. devised the technology, designed the experiments and wrote the manuscript. N.H. did many of the measurements. D.P., C.H. and M.B.A. discussed the concept and findings with F.V. as well co-analysing the experiments and reviewing the various drafts. M.B.A. finalised all figures. All authors reviewed the manuscript.
Supplementary Material
Differential Scanning Fluorimetry provides high throughput data on silk protein transitions.
Acknowledgments
We thank the United States Air Force Office of Scientific Research (FA9550-09-1-0111), the European Research Council (SP2-GA-2008-233409), Magdalen College Oxford, the UK EPSRC (EP/K005693/1; EP/G068224/1) and the Canadian NSERC (PGS 3D/6799-379132-2009) for funding, and our editor at NSR for much appreciated comments and guidance.
References
- Garner H. R., Armstrong B. & Lininger D. M. HIGH-THROUGHPUT PCR. Biotechniques 14, 112–115 (1993). [PubMed] [Google Scholar]
- Pantoliano M. W. et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6, 429–440, 10.1089/108705701753364922 (2001). [DOI] [PubMed] [Google Scholar]
- Matulis D., Kranz J. K., Salemme F. R. & Todd M. J. Thermodynamic stability of carbonic anhydrase: Measurements of binding affinity and stoichiometry using ThermoFluor. Biochem. 44, 5258–5266, 10.1021/bi048135v (2005). [DOI] [PubMed] [Google Scholar]
- Rodrigues J. V., Prosinecki V., Marrucho I., Rebelo L. P. N. & Gomes C. M. Protein stability in an ionic liquid milieu: on the use of differential scanning fluorimetry. Phys. Chem. Chem. Phys. 13, 13614–13616, 10.1039/c1cp21187k (2011). [DOI] [PubMed] [Google Scholar]
- Fan Y. C. et al. Fluorescence Spectroscopic Analysis of the Interaction of Papain with Ionic Liquids. Appl. Biochem. Biotechnol. 168, 592–603, 10.1007/s12010-012-9801-x (2012). [DOI] [PubMed] [Google Scholar]
- Wright N. T. & Humphrey J. D. Denaturation of collagen via heating: An irreversible rate process. Annu. Rev. Biomed. Eng. 4, 109–128, 10.1146/annurev.bioeng.4.101001.131546 (2002). [DOI] [PubMed] [Google Scholar]
- Arakawa T. & Tsumoto K. The effects of arginine on refolding of aggregated proteins: not facilitate refolding, but suppress aggregation. Biochem. Biophys. Res. Commun. 304, 148–152, 10.1016/s0006-291x(03)00578-3 (2003). [DOI] [PubMed] [Google Scholar]
- Safar J., Roller P. P., Gajdusek D. C. & Gibbs C. J. Thermal-stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 2, 2206–2216 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N. J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 1, 2527–2535, 10.1038/nprot.2006.204 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeller L., Rubens P. & Heremans K. Pressure effect on the temperature-induced unfolding and tendency to aggregate of myoglobin. Biochem. 38, 3816–3820, 10.1021/bi981693n (1999). [DOI] [PubMed] [Google Scholar]
- Williams S. et al. Fast events in protein folding: Helix melting and formation in a small peptide. Biochem. 35, 691–697, 10.1021/bi952217p (1996). [DOI] [PubMed] [Google Scholar]
- Yan Y. B., Zhang J., He H. W. & Zhou H. M. Oligomerization and aggregation of bovine pancreatic ribonuclease A: Characteristic events observed by FTIR spectroscopy. Biophys. J. 90, 2525–2533, 10.1529/biophysj.105.071530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbmann U. et al. Dynamic light scattering as a relative tool for assessing the molecular integrity and stability of monoclonal antibodies. Biotechol. Gen. Eng. Rev. 24, 117–128 (2007). [DOI] [PubMed] [Google Scholar]
- Bruylants G., Wouters J. & Michaux C. Differential scanning calorimetry in life science: Thermodynamics, stability, molecular recognition and application in drug design. Curr. Med. Chem. 12, 2011–2020, 10.2174/0929867054546564 (2005). [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Intermediate states in protein folding. J. Mol. Biol. 258, 707–725, 10.1006/jmbi.1996.0280 (1996). [DOI] [PubMed] [Google Scholar]
- Ahrer K., Buchacher A., Iberer G. & Jungbauer A. Thermodynamic stability and formation of aggregates of human immunoglobulin G characterised by differential scanning calorimetry and dynamic light scattering. J. Biochem. Biophys. Methods 66, 73–86, 10.1016/j.jbbm.2005.12.003 (2006). [DOI] [PubMed] [Google Scholar]
- Richardson J. H. & Ando M. E. Sub-part-per-trillion detection of polycyclic aromatic-hydrocarbons by laser-induced molecular fluorescence. Anal. Chem. 49, 955–959, 10.1021/ac50015a021 (1977). [DOI] [PubMed] [Google Scholar]
- Garcia-Parajo M. F., Segers-Nolten G. M. J., Veerman J.-A., Greve J. & van Hulst N. F. Real-time light-driven dynamics of the fluorescence emission in single green fluorescent protein molecules. Proc. Natl. Acad. Sci. 97, 7237–7242, 10.1073/pnas.97.13.7237 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland R. P., Jones L. J., Singer V. L. & Steinberg T. H. Molecular Probes, Inc., European patent, EP 0774122 A1, Merocyanine dye protein stains. 1997 May 21.
- Vaiana A. C. et al. Fluorescence quenching of dyes by tryptophan: Interactions at atomic detail from combination of experiment and computer simulation. J. Am. Chem. Soc. 125, 14564–14572, 10.1021/ja036082j (2003). [DOI] [PubMed] [Google Scholar]
- Sonoda Y. et al. Benchmarking Membrane Protein Detergent Stability for Improving Throughput of High-Resolution X-ray Structures. Structure 19, 17–25, 10.1016/j.str.2010.12.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen F. H., Berglund H. & Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221, 10.1038/nprot.2007.321 (2007). [DOI] [PubMed] [Google Scholar]
- Vedadi M. et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. 103, 15835–15840, 10.1073/pnas.0605224103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson K. P. R. Small organic probes as amyloid specific ligands - Past and recent molecular scaffolds. FEBS Lett. 583, 2593–2599, 10.1016/j.febslet.2009.04.016 (2009). [DOI] [PubMed] [Google Scholar]
- Dicko C., Porter D. & Vollrath F. The relevance of silks to amyloids. in Functional Amyloid Aggregation. Rigacci S., & and Monica Bucciantini M. (eds) 51–70 (Research Signpost/Transworld Reasearch Network 2010) ISBN: 978-81-308-0425-5. [Google Scholar]
- Moreau M. J. J. et al. Rapid determination of protein stability and ligand binding by differential scanning fluorimetry of GFP-tagged proteins. Rsc Adv 2, 11892–11900, Doi 10.1039/C2ra22368f (2012). [Google Scholar]
- Moreau M. J. J. & Schaeffer P. M. Dissecting the salt dependence of the Tus-Ter protein-DNA complexes by high-throughput differential scanning fluorimetry of a GFP-tagged Tus. Mol. Biosys. 9, 3146–3154, Doi 10.1039/C3mb70426b (2013). [DOI] [PubMed] [Google Scholar]
- Hayashi C. Y. & Lewis R. V. Molecular architecture and evolution of a modular spider silk protein gene. Science 287, 1477–1479 (2000). [DOI] [PubMed] [Google Scholar]
- Sutherland T. D. et al. An independently evolved Dipteran silk with features common to Lepidopteran silks. Insect Biochem. Mol. Biol. 37, 1036–1043, 10.1016/j.ibmb.2007.05.016 (2007). [DOI] [PubMed] [Google Scholar]
- Vollrath F. & Knight D. P. Liquid crystalline spinning of spider silk. Nature 410, 541–548 (2001). [DOI] [PubMed] [Google Scholar]
- Holland C., Terry A. E., Porter D. & Vollrath F. Natural and unnatural silks. Polymer 48, 3388–3392 (2007). [Google Scholar]
- Holland C., Terry A. E., Porter D. & Vollrath F. Comparing the rheology of native spider and silkworm spinning dope. Nat. Mater. 5, 870–874, 10.1038/nmat1762 (2006). [DOI] [PubMed] [Google Scholar]
- Dicko C., Kenney J. M. & Vollrath F. Beta-Silks: Enhancing and controlling aggregation. Adv. Protein Chem. 73, 17–53 (2006). [DOI] [PubMed] [Google Scholar]
- Lucas F. & Rudall K. Extracellular fibrous proteins: The silks. Compr. Biochem. 26 Part B, 475–558 (1968). [Google Scholar]
- Knight D. P., Knight M. M. & Vollrath F. Beta transition and stress-induced phase separation in the spinning of spider dragline silk. Int. J. Biol. Macromol. 27, 205–210 (2000). [DOI] [PubMed] [Google Scholar]
- Craig C. Evolution of arthropod silks. Annu. Rev. Entomol. 42, 231–267 (1997). [DOI] [PubMed] [Google Scholar]
- Tamura T., Inoue H. & Suzuki Y. The fibroin genes of Antheraea yamamai and Bombyx mori are different in their core regions but reveal a striking sequence similarity in their 5′ ends and 5′flanking regions. Mol. Gen. Genet. 207, 189–195 (1987). [Google Scholar]
- Jackson C. & O'Brian J. P. Molecular weight distribution of Nephila clavipes dragline silk. Macromol. 28, 5975–5977 (1995). [Google Scholar]
- Zhang P. et al. Proteome analysis of silk gland proteins from the silkworm, Bombyx mori. Proteomics 6, 2586–2599 (2006). [DOI] [PubMed] [Google Scholar]
- Weiskopf A., Senecal K., Vouros P., Kaplan D. & Mello C. M. The carbohydrate composition of spider silk: Nephila edulis dragline. Glycobiol. 6, 1703 (1996). [Google Scholar]
- Porter D. & Vollrath F. Water mediated proton hopping empowers proteins. Softmatter 9, 643–646 10.1039/C2SM27155A (2013). [Google Scholar]
- Zhang H., Takenaka M. & Isobe S. DSC and electrophoretic studies on soymilk protein denaturation. J. of Therm. Anal. Calorimet. 75, 719–726, 10.1023/b,jtan.0000027168.18317.78 (2004). [Google Scholar]
- Boulet-Audet M., Terry A. E., Vollrath F. & Holland C. Silk protein aggregation kinetics revealed by Rheo-IR. Acta Biomater. 10, 776–784, 10.1016/j.actbio.2013.10.032 (2013). [DOI] [PubMed] [Google Scholar]
- Greving I., Dicko C., Terry A., Callow P. & Vollrath F. Small angle neutron scattering of native and reconstituted silk fibroin. Soft Matter 6, 4389–4395, 10.1039/c0sm00108b (2010). [Google Scholar]
- Dicko C., Porter D., Bond J., Kenney J. M. & Vollratht F. Structural disorder in silk proteins reveals the emergence of elastomericity. Biomacromol. 9, 216–221, 10.1021/bm701069y (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yang H., Shao H. & Hu X. Antheraea pernyi silk fiber: a potential resource for artificially biospinning spider dragline silk. J. Biomed. Biotechnol. 2010, 683962, 10.1155/2010/683962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N. K. & Roy I. Effect of trehalose on protein structure. Protein Sci. 18, 24–36, 10.1002/pro.3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath F. & Porter D. Silks as ancient models for modern polymers. Polymer 50, 5623–5632, 10.1016/j.polymer.2009.09.068 (2009). [Google Scholar]
- Dicko C., Knight D., Kenney J. M. & Vollrath F. Conformational polymorphism, stability and aggregation in spider dragline silks proteins. Int. J. Biol. Macromol. 36, 215–224, 10.1016/j.ijbiomac.2005.06.004 (2005). [DOI] [PubMed] [Google Scholar]
- Harvey P. H. & Pagel M. D. The comparative method in evolutionary biology. Oxford University Press, Oxford (1991). [Google Scholar]
- Steinberg T. H., Jones L. J., Haugland R. P. & Singer V. L. SYPRO Orange and SYPRO Red protein gel stains: One-step fluorescent staining of denaturing gels for detection of nanogram levels of protein. Anal. Biochem. 239, 223–237, 10.1006/abio.1996.0319 (1996). [DOI] [PubMed] [Google Scholar]
- Reichardt C. R. C. Solvents and solvent effects in organic chemistry. (VCH, Weinheim, 1988). [Google Scholar]
- Toutchkine A., Kraynov V. & Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J. Am. Chem. Soc. 125, 4132–4145, 10.1021/ja0290882 (2003). [DOI] [PubMed] [Google Scholar]
- Ory J. J. & Banaszak L. J. Studies of the ligand binding reaction of adipocyte lipid binding protein using the fluorescent probe 1,8-anilinonaphthalene-8-sulfonate. Biophys. J. 77, 1107-1116 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. & Vollrath F. Water mobility, denaturation and the glass transition in proteins. Biochim. Biophys. Acta 1824, 785–791 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential Scanning Fluorimetry provides high throughput data on silk protein transitions.