Abstract
AIM: To explore the anti-hepatitis B virus (HBV) effects of Boehmeria nivea (B. nivea) root extract (BNE) by using the HepG2 2.2.15 cell model system.
METHODS: Hepatitis B surface antigen (HBsAg), hepatitis B virus e antigen (HBeAg), and HBV DNA were measured by using ELISA and real-time PCR, respectively. Viral DNA replication and RNA expression were determined by using Southern and Northern blot, respectively.
RESULTS: In HepG2 2.2.15 cells, HBeAg (60%, P < 0.01) and particle-associated HBV DNA (> 99%, P < 0.01) secretion into supernatant were significantly inhibited by BNE at a dose of 100 mg/L, whereas the HBsAg was not inhibited. With different doses of BNE, the reduced HBeAg was correlated with the inhibition of HBV DNA. The anti-HBV effect of BNE was not caused by its cytotoxicity to cells or inhibition of viral DNA replication and RNA expression.
CONCLUSION: BNE could effectively reduce the HBV production and its anti-HBV machinery might differ from the nucleoside analogues.
Keywords: Boehmeria nivea, Medicinal herb, Antiviral agent, Hepatitis B virus, Anti-hepatitis B virus, HepG2 2.2.15
INTRODUCTION
Hepatitis B virus (HBV) has infected more than 4 million people worldwide. About 20% of patients infected with HBV may lead to chronic hepatitis, liver cirrhosis, and hepatocarcinoma. HBV, belonging to the Hepadnaviridae family, is a non-cytopathic DNA virus with an icosahedral capsid that replicates via reverse transcription of an RNA intermediate[1]. Although the molecular biology aspects of the HBV genome have been described in detail, the mechanisms of viral packaging and transport remain to be elucidated[2]. Nevertheless, HBV-DNA-transfected cells and virus infection animal models have aided the efforts to reveal the mechanism behind the HBV replication cycle. These results led to the identification of the first antiviral agents targeting the reverse transcription process[3].
Chronic hepatitis type-B patients are clinically treated with interferon alpha (INF-α) and nucleoside analogue lamivudine (3TC), adefovir or entecavir[4,5], which are analogues of reverse-transcriptase inhibitors[6,7]. INF-α inhibits viral replication and acts as an immuno-modulator. However, its disadvantages include limited effectiveness (40% response rate)[8], low efficacy with respect to cost, and serious side effects. For 3TC, its inhibition is reversible, and continuous treatment often leads to the development of drug-resistant HBV variants (70% of patients after 4 years of treatment)[9]. Both adefovir and entecavir are used against 3TC-resistant viruses; however, resistances to these two drugs have been reported in lamivudine-resistant patients[10-12]. The use of combination therapy, such as INF-α plus 3TC, 3TC plus adefovir and 3TC plus entecavir, may yield additive or synergistic effects or reduce the emergence of resistance, though serious side effects and unsatisfactory efficacy still present problems. Undeniably, there is a demand for new and improved therapies.
The large repertoire of herbal compounds may show potential in developing new ways to combat previously considered “incurable” diseases, provided that these compounds (or often, mixtures of compounds) could satisfy current government regulations. At present, alternative or traditional medical resources are used by more than 80% of the population in developing countries and by an increasing number of people in other parts of the globe[13,14]. Complementary and alternative therapies for chronic hepatitis are also intensively explored and the results appear promising[15]. Patients with chronic liver diseases are treated with some medicinal herbs exhibiting strong anti-viral activities[16], including daphnoretin from Wikstroemia indica[17], costunolide and dehydrocostus lactone from Saussurea lappa Clarks[18], osthole from Angelica pubescens[19], and the extracts of genus Phyllanthus of the Euphobiaceae family[20]. Furthermore, genus Phyllanthus exhibited a positive effect on the clearance of serum HBsAg in clinical trials conducted on chronic HBV infections, and a synergistic effect when administered with IFN-α[20]. B. nivea has been distributed and used therapeutically in China and Taiwan for diuretic, antipyretic, and hepatoprotective purposes. Recently, it has been reported that root extracts of B. nivea exhibited hepatoprotective activities against CCl4-induced liver injuries, and anti-oxidant effects on FeCl2-ascorbate-induced lipid peroxidation in rat liver homogenate[21].
To investigate the anti-viral mechanism of B. nivea extract (BNE), HBV-producing hepatoma HepG2 2.2.15 cells, which secrete HBsAg, HBeAg and complete Dane particles[22], were chosen for the evaluation of the anti-HBV effect of BNE. Here, we assess anti-HBV activities of BNE by measuring HBsAg, HBeAg, HBV DNA in supernatant, and replication intermediate HBV DNA and HBV RNA within the cells.
MATERIALS AND METHODS
Preparation of BNE
To prepare the B. nivea plant extract utilized in our experiments, the roots of the plants were collected and dried. One hundred gram of the dried roots was cut into pieces approximately 0.5 cm in length before boiling them in 1 L of 200 mL/L ethanol (1:10 ratio) under reflux for 3 h. The decoction was filtered through a 0.22-μm filter and lyophilized. The lyophilized powder was dissolved in normal PBS and adjusted to stock concentration (100 g/L) prior to application to the cells.
Cell lines and culture
The HepG2 2.2.15 cell line was kindly provided by Dr. Ho MS (Academia Sinica, Taipei, Taiwan, China), and Human hepatoma HepG2 cells were obtained from American Type Culture Collection (ATCC). These cells were maintained in MEM (Eagle) plus 100 mL/L fetal bovine serum (FBS) supplement with 1.5 g/L sodium bicarbonate, 0.1 mmol/L non-essential amino acids, 1.0 mmol/L sodium pyruvate, and 100 units/mL penicillin G and 100 mg/L streptomycin. A final concentration of 200 mg/L G418 was contained in the medium for the maintenance of HepG2 2.2.15 cells. Before the experiment, the cell count was adjusted to 1 × 106/mL, and cell viability to higher than 85% by trypan blue exclusion test.
Drug treatment protocols
For drug treatment, 1 × 105 cells of HepG2 2.2.15 were seeded in a 12-well plate and allowed to grow for 3 d before treatment with different concentrations of BNE. Cells were refed with drug-containing fresh medium every 3 d for up to 12 d in time-dependent experiment. In Southern and Northern blot assay, the culture condition was the same as described above (10 mg/L 3TC were also added), and in which time of drug treatments were only for 1 d.
Analysis of cytotoxicity
HepG2 2.2.15 cells were used for determining cytotoxicity of BNE. Cells were inoculated onto a 96-well plate at a density of 1 × 104 cells per 100 μL prior to drug treatment. BNE was added at concentrations of 0.1, 1, 10, and 100 mg/L and the cells were refed with drug-containing fresh medium every 3 d for up to 12 d. After drug treatment, the cytotoxicity was measured based on the reduction of MTT (Sigma, St. Louis, USA) in mitochondria[23].
Determination of HBsAg and HBeAg levels by ELISA
Conditioned medium was collected and the HBsAg and HBeAg levels were determined semi-quantitatively using ELISA assay [SURASE B-96 (TMB), General Biologicals corp., Taiwan, China] according to the manufacturer’s instructions.
Supernatant HBV DNA extraction and analysis by quantitative real-time PCR
The supernatant HBV DNA was extracted from conditioned medium as previously described[24] and stored in -20°C prior to real-time PCR analysis. The quantity of HBV DNA in culture medium was quantified with the ABI 7500 Sequence Detection System by using HBV RealQuant PCR kit (General Biologicals Corp., Taiwan, China) according to the manufacturer’s instructions. Briefly, the PCR programming was performed with an initial denaturing steps at 50°C for 2 min and 95°C for 10 min, followed by 45 amplification cycles at 95°C for 15 s and annealing/extending at 58°C for 1 min.
The 50% effective concentration (EC50), defined as the drug concentration that reduces the level of HBV DNA in the culture medium by 50%, was calculated by four parameter logistic curve equation.
Southern and Northern blot detection of HBV DNA and mRNA in HepG2.2.15 cell
HepG2 2.2.15 cells were cultured in MEM medium and treated with 0.1, 1, and 10 mg/L BNE for 1 d. Cells were lysed with 0.8 mL of 0.01 mol/L Tris-HCl (pH 8.0), 0.05 mol/L NaCl, 5 mL/L NP-40, 1 mmol/L EDTA at room temperature for 10 min as previously described[25]. For the Southern hybridizations, 20 μg of total DNA was digested with HindIII, electrophoresed on a 14 g/L agarose gel, and then transferred to nylon membrane. The probe for hybridization was synthesized from PCR amplification of a plasmid containing the full-length HBV genome (kindly provided by Dr. Ho MS, Academia Sinica, Taipei, Taiwan, China), and then the probe was labeled with digoxigenin-dUTP (DIG) by the DIG-high Prime DNA labeling and Detection Starter kitIIaccording to the manufacturer’s protocol (Roche, Basel, Switzerland). The membrane was hybridized with HBV probe at 50°C overnight. For Northern blot analysis, total RNA was isolated from BNE-treated and untreated HepG2 2.2.15 cells by using the TRIZOL kit (Invitrogen, CA, USA). A total of 20 μg of RNA was resolved in 12 g/L denatured gel and then transferred onto the nylon membrane and the membrane was hybridized with DIG-labeled HBV DNA fragment described above. For hybridization of glyceraldehyde-3-phosphate dehydrogenase (G3PDH), the full-length HBV DNA probe was removed from the membrane by washing twice at 37°C in 0.2 mol/L NaOH containing 1 mL/L sodium dodecyl sulfate (SDS) solution for 15 min and then re-hybridized with DIG-labeled probe for G3PDH.
Statistical analysis
Data were expressed as mean ± SE of three independent experiments. Statistical analysis was performed using Student’s t-test. A P value less than 0.05 was considered statistically significant.
RESULTS
Effects of BNE on secreted HBsAg, HBeAg and HBV DNA from HepG2 2.2.15 cultures
Anti-HBV activity of BNE was investigated by using HepG2 2.215 cells, which can secrete HBV particles. When HepG2 2.2.15 cells were treated with various concentrations of BNE, secretion of HBeAg, but not HBsAg, was significantly suppressed compared to vehicle controls (Figure 1A and B). The suppression of HBeAg was dose-dependent and approximately 20% of inhibition was observed in cells treated with 10 mg/L BNE. Moreover, a significant suppression of HBeAg secretion (approximately 60%) was observed in cells treated with 100 mg/L BNE (Figure 1B).
Figure 1.
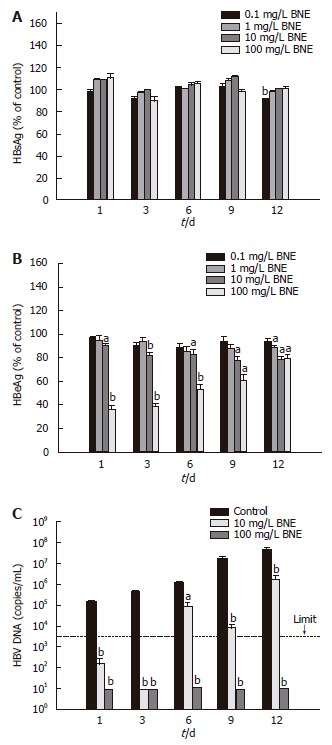
Effects of BNE on secreted HBsAg, HBeAg and HBV DNA from HepG2 2.2.15 cell cultures. A: HBsAg level; B: HBeAg level; C: Viral DNA. The dotted line presents the limitation of this kit (3000 copies/mL). Data are expressed as mean ± SE of three independent experiments. aP < 0.05; bP < 0.01 vs the corresponding controls (Student’s t-test).
We also measured the amount of viral DNA secreted in the medium (supernatant HBV DNA) by using real-time PCR. As shown in Figure 1C, supernatant HBV DNA was dramatically decreased compared to vehicle control, since the first day of treatment, in 10 and 100 mg/L BNE-treated culture medium. Although the amounts of supernatant HBV DNA increased while cells continued to grow during the experiment, approximately 95% inhibition of DNA secretion was observed in the 12-d culture treated with BNE (10 mg/L) (Figure 1C). Notably, these results showed that the production of HBsAg and HBeAg was only slightly suppressed by 10 mg/L BNE, but supernatant HBV DNA levels were dramatically suppressed by BNE at the same dose.
Cytotoxic effects of BNE on HepG2 2.2.15 cells
Suppression of HBV production by BNE might be a result of its cytotoxicity, and this possibility was examined in HepG2 2.2.15 cells by using MTT assay. No apparent cytotoxicity was detected in HepG2 2.2.15 cells up to 12 d after exposure to BNE at different concentrations (0.1, 1, 10 and 100 mg/L) (Figure 2), suggesting that the suppression of supernatant viral DNA levels by BNE was not caused by its cytotoxicity.
Figure 2.
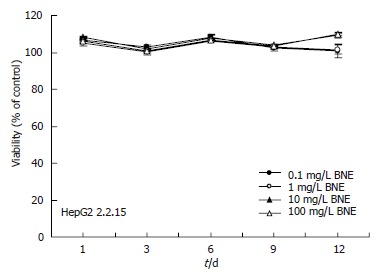
Cytotoxic effects of BNE on HepG2 2.2.15 cells. Data are expressed as mean ± SE of three independent experiments (MTT assay).
Determination of the effective concentration of BNE anti-HBV activity
The effective concentration of BNE to suppress 50% of secreted HBV DNA (EC50) was determined with various concentrations of BNE on d 1. The suppression of secreted HBV DNA was shown in a dose-dependent manner with BNE treatment (70% at 0.1 mg/L and 100% at ≥ 10 mg/L) (Figure 3); accordingly, the EC50 was 0.0462 mg/L. Here, the concentration of BNE (10 mg/L) used for the study of anti-HBV activity was about 200-fold of EC50.
Figure 3.
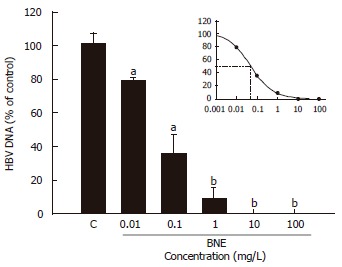
Determination of the effective concentration of BNE anti-HBV activity. Cells were treated for 24 h. Data are expressed as mean ± SE of three independent experiments. aP < 0.05, bP < 0.01 vs the corresponding controls (Student’s t-test).
Effects of BNE on intracellular HBV DNA replication and transcription in HepG2 2.2.15 cells
To address the mechanism of inhibition by BNE in HepG2 2.2.15 cells, we analyzed the viral mRNA after 24 h exposure to BNE by Northern blot using full-length HBV genome as a probe (Figure 4A) and intracellular relaxed circular (RC) and single-stranded (SS) forms of HBV DNA by Southern blot in parallel (Figure 4B). The levels of viral mRNA were not affected in either BNE-treated or 3TC-treated cells (Figure 4A). Moreover, Southern blot results indicated that BNE did not suppress intracellular RC and SS forms of HBV DNA. In Contrast, an apparent inhibition of intracellular RC and SS forms of HBV DNA was observed in 3TC-treated cells, which was significantly different from the BNE group (Figure 4B). These results suggested that BNE did not apparently decrease viral DNA replication and viral mRNA expression in HepG2 2.2.15 cells, and that the anti-HBV mechanism of BNE seemed to be different from that of 3TC.
Figure 4.
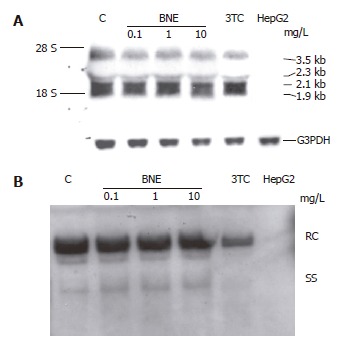
Effects of BNE on intracellular HBV DNA replication and transcription in HepG2 2.2.15 cells. Cells were treated for 1 d. A: Northern analysis; B: Southern analysis.
DISCUSSION
In this study, we first demonstrated that BNE had anti-HBV activity of inhibiting the supernatant HBV DNA levels in a dose-dependent manner in HepG2 2.2.15 cells without blocking HBsAg secretion. This inhibition was caused neither by the toxicity of BNE to HepG2 2.2.15 cells, nor by blocking HBV gene expression and replication. The significant inhibition of supernatant HBV DNA levels was observed at a concentration greater than 10 mg/L of BNE in HepG2 2.2.15 cells. In addition, BNE could also dose-dependently inhibit the secretion of HBeAg. Although BNE had higher inhibitory ability on HBeAg secretion at 100 mg/L than that at 10 mg/L, it still could efficiently inhibit secreted HBV DNA at the latter concentration. This result might be due to the fact that BNE comprise multiple compounds, and the effective concentration of those compounds to inhibit secreted HBV DNA was lower than that of the compounds required for inhibition of HBeAg.
Currently, 3TC and other nucleoside analogues have been shown to inhibit HBV replication both in vitro and in vivo[24,26,27]. A previous study had reported that 3TC, a viral polymerase inhibitor, reduced episomal DNA (RC and SS form), whereas HBV-specific RNAs were not affected in HepG2 2.2.15 cells[24]. In this study, we could not find any apparent reduction of HBV-specific RNAs or intracellular SS and RC DNA in response to BNE, and that these data revealed that the anti-HBV mechanism of BNE might be different from 3TC that targeted the viral polymerase. Since the HBV mRNAs were transcribed from the integrated DNA, it was not unexpected that HBV-specific transcripts were not affected by BNE treatment. The finding that supernatant HBV DNA rather than HBsAg and HBeAg was dramatically inhibited by 10 mg/L BNE after 24-h treatment might come from the possibility that exported virions have outer protein coats or HBsAg without packaging DNA. Though the mechanism of anti-viral effects by BNE remains unclear, we deduced that BNE might block coating and secretion of HBV containing nucleocapsids or destabilize HBV DNA containing nucleocapsids.
Whether the HBV-inhibiting effects of BNE could be contributed to a single component, or multiple components, is currently unknown. In order to explore the active compound for anti-HBV activity, we have attempted to analyze the chemical composition of BNE by HPLC. Very little, if any, nucleotide analogues were detected (data not shown). Fractionation experiments of BNE are currently being executed in our laboratory. Recently, Herteroaryldihydropyrimidines (HAP)[28], Bis-ANS[29], and alkylated imino sugars[30,31] have been found to block the viral production by interference with either nucleocapsid assembly or nucleocapsid maturation, and these compounds might have the potential to be developed as non-viral polymerase targeting antiviral drugs.
In conclusion, the BNE, together with other medicinal herbs that exploit different action modes to inhibit HBV, could be administered in combination with other polymerase inhibitors or cytokines, providing possibly a novel HBV treatment strategy to the current therapies.
ACKNOWLEDGMENTS
We gratefully thank to Dr. Ho MS for providing the materials, HepG2 2.2.15 cells and full-length HBV plasmid, and also thank the Dr. Ho MS and Dr. Yuan TT for giving comments on this manuscript.
Footnotes
Supported by DCB-094EN203 and NSC 93-2311-B-007-002
S- Editor Pan BR L- Editor Kumar M E- Editor Bai SH
References
- 1.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 2.Knipe DM, Howley PM. Fundamental virology. 4th ed. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 3.Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 4.Lai CL, Rosmawati M, Lao J, Van Vlierberghe H, Anderson FH, Thomas N, Dehertogh D. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology. 2002;123:1831–1838. doi: 10.1053/gast.2002.37058. [DOI] [PubMed] [Google Scholar]
- 5.Chang TT, Gish RG, Hadziyannis SJ, Cianciara J, Rizzetto M, Schiff ER, Pastore G, Bacon BR, Poynard T, Joshi S, et al. A dose-ranging study of the efficacy and tolerability of entecavir in Lamivudine-refractory chronic hepatitis B patients. Gastroenterology. 2005;129:1198–1209. doi: 10.1053/j.gastro.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 6.Buster EH, Janssen HL. Antiviral treatment for chronic hepatitis B virus infection--immune modulation or viral suppression. Neth J Med. 2006;64:175–185. [PubMed] [Google Scholar]
- 7.Thomas H, Foster G, Platis D. Mechanisms of action of interferon and nucleoside analogues. J Hepatol. 2003;39 Suppl 1:S93–S98. doi: 10.1016/s0168-8278(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 8.Niederau C, Heintges T, Lange S, Goldmann G, Niederau CM, Mohr L, Häussinger D. Long-term follow-up of HBeAg-positive patients treated with interferon alfa for chronic hepatitis B. N Engl J Med. 1996;334:1422–1427. doi: 10.1056/NEJM199605303342202. [DOI] [PubMed] [Google Scholar]
- 9.Fischer KP, Gutfreund KS, Tyrrell DL. Lamivudine resistance in hepatitis B: mechanisms and clinical implications. Drug Resist Updat. 2001;4:118–128. doi: 10.1054/drup.2001.0190. [DOI] [PubMed] [Google Scholar]
- 10.Angus P, Vaughan R, Xiong S, Yang H, Delaney W, Gibbs C, Brosgart C, Colledge D, Edwards R, Ayres A, et al. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003;125:292–297. doi: 10.1016/s0016-5085(03)00939-9. [DOI] [PubMed] [Google Scholar]
- 11.Fung SK, Chae HB, Fontana RJ, Conjeevaram H, Marrero J, Oberhelman K, Hussain M, Lok AS. Virologic response and resistance to adefovir in patients with chronic hepatitis B. J Hepatol. 2006;44:283–290. doi: 10.1016/j.jhep.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P, et al. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–3507. doi: 10.1128/AAC.48.9.3498-3507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gesler WM. Therapeutic landscapes: medical issues in light of the new cultural geography. Soc Sci Med. 1992;34:735–746. doi: 10.1016/0277-9536(92)90360-3. [DOI] [PubMed] [Google Scholar]
- 14.Bent S, Ko R. Commonly used herbal medicines in the United States: a review. Am J Med. 2004;116:478–485. doi: 10.1016/j.amjmed.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 15.Coon JT, Ernst E. Complementary and alternative therapies in the treatment of chronic hepatitis C: a systematic review. J Hepatol. 2004;40:491–500. doi: 10.1016/j.jhep.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Strader DB, Bacon BR, Lindsay KL, La Brecque DR, Morgan T, Wright EC, Allen J, Khokar MF, Hoofnagle JH, Seeff LB. Use of complementary and alternative medicine in patients with liver disease. Am J Gastroenterol. 2002;97:2391–2397. doi: 10.1111/j.1572-0241.2002.05993.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen HC, Chou CK, Kuo YH, Yeh SF. Identification of a protein kinase C (PKC) activator, daphnoretin, that suppresses hepatitis B virus gene expression in human hepatoma cells. Biochem Pharmacol. 1996;52:1025–1032. doi: 10.1016/0006-2952(96)00420-0. [DOI] [PubMed] [Google Scholar]
- 18.Chen HC, Chou CK, Lee SD, Wang JC, Yeh SF. Active compounds from Saussurea lappa Clarks that suppress hepatitis B virus surface antigen gene expression in human hepatoma cells. Antiviral Res. 1995;27:99–109. doi: 10.1016/0166-3542(94)00083-k. [DOI] [PubMed] [Google Scholar]
- 19.Huang RL, Chen CC, Huang YL, Hsieh DJ, Hu CP, Chen CF, Chang C. Osthole increases glycosylation of hepatitis B surface antigen and suppresses the secretion of hepatitis B virus in vitro. Hepatology. 1996;24:508–515. doi: 10.1002/hep.510240307. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lin H, McIntosh H. Genus Phyllanthus for chronic hepatitis B virus infection: a systematic review. J Viral Hepat. 2001;8:358–366. doi: 10.1046/j.1365-2893.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin CC, Yen MH, Lo TS, Lin JM. Evaluation of the hepatoprotective and antioxidant activity of Boehmeria nivea var. nivea and B. nivea var. tenacissima. J Ethnopharmacol. 1998;60:9–17. doi: 10.1016/s0378-8741(97)00122-0. [DOI] [PubMed] [Google Scholar]
- 22.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of radiosensitivity. Cancer Res. 1987;47:943–946. [PubMed] [Google Scholar]
- 24.Doong SL, Tsai CH, Schinazi RF, Liotta DC, Cheng YC. Inhibition of the replication of hepatitis B virus in vitro by 2',3'-dideoxy-3'-thiacytidine and related analogues. Proc Natl Acad Sci USA. 1991;88:8495–8499. doi: 10.1073/pnas.88.19.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, Hazboun T, Block T. Limited proteolysis induces woodchuck hepatitis virus infectivity for human HepG2 cells. Virus Res. 2001;73:27–40. doi: 10.1016/s0168-1702(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 26.Dienstag JL, Perrillo RP, Schiff ER, Bartholomew M, Vicary C, Rubin M. A preliminary trial of lamivudine for chronic hepatitis B infection. N Engl J Med. 1995;333:1657–1661. doi: 10.1056/NEJM199512213332501. [DOI] [PubMed] [Google Scholar]
- 27.Dixon JS, Boehme RE. Lamivudine for the treatment of chronic hepatitis B. Acta Gastroenterol Belg. 2000;63:348–356. [PubMed] [Google Scholar]
- 28.Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, et al. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res. 2002;54:69–78. doi: 10.1016/s0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnick A, Ceres P, Singh S, Johnson JM. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J Virol. 2002;76:4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Tran T, Simsek E, Block TM. The alkylated imino sugar, n-(n-Nonyl)-deoxygalactonojirimycin, reduces the amount of hepatitis B virus nucleocapsid in tissue culture. J Virol. 2003;77:11933–11940. doi: 10.1128/JVI.77.22.11933-11940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deres K, Schröder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Krämer T, Niewöhner U, Pleiss U, Stoltefuss J, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]


