Abstract
AIM: To verify and expand the known spectrum of serine protease inhibitor Kazal type 1 (SPINK1) gene mutations in chronic pancreatitis.
METHODS: DNA extracted from 172 chronic pancreatitis patients was assayed for SPINK1 gene mutations by PCR and DNA sequencing. A control cohort of 90 unrelated healthy individuals was analysed by the same methods for presence of common populational polymorphisms, and frequency of five-loci haplotypes was calculated. Linkages of gene aberrations in single SPINK1 gene copies were analysed by long-distance PCR followed by allele-specific PCR and DNA sequencing.
RESULTS: The most frequent SPINK1 gene mutation N34S was found at a frequency of 6%. Furthermore, we detected the heterozygous intervening sequence (IVS) 3 + 2 T > C mutated gene in 2 German patients and 1 Macedonian chronic pancreatitis patient. In all three SPINK1 gene copies an additional rare base substitution was found: 5’untranslated region (UTR)-215 G > A. Polymorphism analysis revealed that all three affected genes carried the same five-loci haplotype. DNA sequencing of another chronic pancreatitis-related gene PRSS1 (cationic trypsinogen) did not reveal any mutations in these 3 patients.
CONCLUSION: We found in 3 (2%) of 172 chronic pancreatitis patients an IVS3 + 2 T > C SPINK1 gene mutation and a base substitution 5’UTR-215 G > A in the same gene copy. Most probably the 5’UTR-215 G > A represents a rare polymorphism and not a mutation as previously concluded. Haplotype analysis suggests a common origin of the IVS3 + 2 T > C mutation in these patients.
Keywords: Chronic pancreatitis, Serine protease inhibitor Kazal type 1, Gene mutations
INTRODUCTION
In approximately one-third of all patients with chronic pancreatitis, no etiological factor can be found, and these patients are classified as having idiopathic disease. Recent genetic discoveries have added much to our understanding of chronic pancreatitis[1]. The identification of a clear’ gain-of-function’ mutation, R122H, in a cationic trypsinogen gene (PRSS1) that causes hereditary pancreatitis has been of high significance[2]. Loss of trypsin inhibitor function may cause pancreatitis. Several studies have demonstrated mutations in the serine protease inhibitor Kazal type 1 (SPINK1) gene (MIM#167790), also known as pancreatic secretory trypsin inhibitor gene (PSTI). This gene is approximately 7.5 kb long, consists of 4 exons and is located on chromosome 5[3]. It encodes a polypeptide of 79 amino acids that is processed into a mature active protein consisting of 56 amino acids. Due to its molar ratio of 1:5 to trypsin, it inhibits about 20% of its activity. SPINK1 gene variations are clearly associated with chronic pancreatitis[4,5]. They are probably a disease modifying factor, possibly by lowering the threshold for development of chronic pancreatitis caused by other genetic or environmental factors[4,6,7].
The most frequent mutation of human SPINK1 gene is a heterozygous or homozygous missense mutation of codon 34 in exon 3 (c.101 A > G), and its incidence is about 80% in familial and idiopathic chronic pancreatitis[4,5]. This mutation leads to an A to G transition resulting in substitution of asparagine by serine (N34S). The high prevalence of this mutation in chronic pancreatitis led to the hypothesis that the N34S mutation could reduce the antiproteolytic activity of SPINK1 subjects[7,8].
To date, many variants of the SPINK1 gene have been reported, however other mutations than N34S are rare (for an up-to-date list of pancreatitis-associated SPINK1 mutations visit the website[9]). Concerning other SPINK1 gene sequence abnormalities, only two of them can probably be recognised as true mutations. Mutation c.2 T > C (M1T) destroys the translation initiation codon[5]. The base substitution intervening sequence (IVS)3 + 2 T > C (also named c.194 + 2 T > C) destroys the splicing of exons 3 and 4, and was described in approximately 10 cases[9]. A splicing mutation IVS2 + 1 G > A (c.87 + 1 G > A) and the frameshift mutations c.27delC and 98insA, both detected in single families, may be fatal for protein synthesis because of preliminary termination of the translation[10]. The base substitution 5’ untranslated region (UTR)-215 G > A in the untranslated 5’-region of SPINK1 has been reported in two Japanese chronic pancreatitis families up to now, but in no healthy individuals[11].
The aim of this study was to verify and extend the known spectrum of SPINK1 gene mutations. We found 3 chronic pancreatitis patients carrying an IVS3 + 2 T > C SPINK1 gene mutation together with a base substitution 5’UTR-215 G > A in the same gene copy. Our finding implicates that the 5’UTR-215 G > A substitution most probably represents a rare polymorphism and not a mutation as previously concluded[11]. The same haplotypes in these genes suggest a common origin of the IVS3 + 2 T > C mutation in all 3 patients.
MATERIALS AND METHODS
Patients and controls
This study was approved by the ethics committee of the Chamber of Physicians in Hamburg, Germany. Written informed consent was obtained from all patients for use of tissue and blood samples. One hundred and seventy-two patients with chronic pancreatitis who underwent surgery in the Clinic for General, Visceral and Thoracic Surgery, University Medical Center Hamburg-Eppendorf, Germany between 1992 and 2002 were included. Pancreatic disease was confirmed by histopathological evaluation of all cases. Blood samples of 90 unrelated and healthy individuals (donors) were used as a source of control DNA and for the study of populational polymorphism.
DNA extraction and PCR
Genomic DNA from peripheral blood leukocytes and tissue specimens was extracted and purified according to the established protocols using the QIAamp blood-tissue kit (Qiagen, Hilden, Germany). All four exons of the SPINK1 gene were amplified by polymerase chain reaction (PCR) using Taq DNA-polymerase (Perkin-Elmer, Foster City, USA) and oligonucleotide primers as previously described[5]. PCR products were analysed by electrophoresis on a 2% agarose gel and purified by the QIAquick spin PCR-purification kit (Qiagen). Direct DNA sequencing of the PCR products was carried out using the BigDye terminators sequencing kit (Perkin-Elmer, Foster City, USA) and nested primers on an automatic sequencer ABI 377 (Perkin-Elmer). The presence of the 5’UTR-215 G > A substitution was also tested by performing BglI restriction endonuclease digestion and gel electrophoresis analysis. SPINK1 genes from chronic pancreatitis patients carrying the substitutions IVS3 + 2 T > C and 5’UTR-215 G > A were amplified by PCR to generate two fragments (5’-UTR-exon 3 and exon 3-3’-UTR). The long-distance PCR was performed in a 100 μL reaction mix supplemented with the TaqPlus long polymerase mixture, a high-salt buffer (Stratagene, La Jolla, USA) and the following primers: SPINK1 pr-F 5’-TTTGAGTTCATCTTACAGGTGAG and SPINK1 3-R 5’-GTTTGCTTTTCTCGGGGTGAG, SPINK1 3-F 5’-CCAATCACAGTTATTCCCCAGAG and SPINK1 4-R 5’-CCAAAGTCCCCTGACCCTGG. PCR products were analysed electrophoretically on an 0.8%-1% agarose gel and purified as described above. Allele-specific PCRs were performed in a 100 μL reaction with 1 μL of long-distance PCR products as templates and Taq DNA-polymerase. The following primers were used: Spl3-T-R 5’-AAGAAACTCAAGTTTGTACTCA or Spl3-C-R 5’-AAGAAACTCAAGTTTGTACTCG and SPINK1 pr-F, Pr-215G-F 5’-CATGTTTCAGGCCCACCTGG or Pr-215A-F 5’-CATGTTTCAGGCCCACCTGA and SPINK1 3-R for amplification of the 5’ region of the SPINK1 gene; Spl3-T-F 5’-GTGTTATGTTTTGAAAATCGGT or Spl3-C-F 5’-GTGTTATGTTTTGAAAATCGGC and SPINK1 4-R for amplification of the 3’ region of the SPINK1 gene. The amplification was performed by 10-15 PCR cycles using 0.5 units of Taq DNA-polymerase. The products of the allele-specific PCRs were analysed by gel electrophoresis on an 0.8%-1% agarose gel and purified as described above. For detection of the IVS3 + 2 T > C and 5’UTR-215 G > A substitutions and polymorphisms the amplified DNA was sequenced.
Haplotype analysis
The frequency of polymorphic variations in the normal population was estimated from the results obtained from DNA sequencing of the appropriate amplified DNA fragments which were extracted from leukocytes of the peripheral blood from 90 healthy unrelated individuals (180 alleles). The IVS2-352 A/G polymorphism was additionally assessed by MspI restriction endonuclease digestion. The distribution of haplotypes in normal subjects for single-nucleotide polymorphic loci (SNP’s) was calculated by the expectation-maximisation-algorithms (EM-algorithms) described by Weir[12] for two-, three- and four-loci haplotypes, and according to a method proposed[13] for two- to five-loci haplotypes. The comparison of the calculated results and probabilities of both methods led to concordant data.
RESULTS
DNA sequencing revealed mutations in SPINK1 gene
To determine the frequency of SPINK1 gene aberrations, DNA was isolated from consecutive surgical specimens and blood samples of 172 patients suffering from chronic pancreatitis and analysed for SPINK1 gene mutations. The most frequent mutation found in the SPINK1 gene of chronic pancreatitis patients examined in our study was the N34S (A > G in position c.101), but with a rather low frequency of about 6% (10 of 172 patients).
In 3 (2%) of 152 patients a heterozygous IVS3 + 2 T > C splicing mutation was detected. DNA sequencing revealed the presence of another heterozygous base substitution in the same gene copy in the promoter region of exon 1. The detection of the 5’UTR-215 G > A substitution was also confirmed by BglI restriction endonuclease cleavage and subsequent gel electrophoresis analysis (Figure 1).
Figure 1.
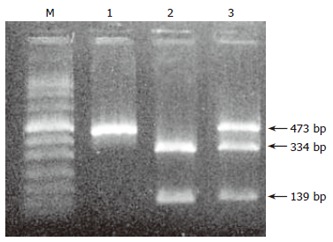
Detection of 5’UTR-215 G/A heterozygote in SPINK1 gene by BglI restriction endonuclease cleavage. 1: untreated prF-1R PCR fragment; 2: normal (-215 G/G) prF-1R PCR fragment treated with BglI endonuclease; 3: heterozygote (-215 G/A) prF-1R PCR fragment treated with BglI endonuclease [M: marker (Boehringer-Mannheim marker VIII); bp: base pairs].
Both mutations IVS3 + 2 T > C and 5’UTR-215 G > A were present in the same gene copy
Genomic DNA from IVS3 + 2 T > C and 5’UTR-215 G > A carrying patients was amplified into two long DNA fragments (each about 4 kb) by long-distance PCR using SPINK1 gene-specific primers. To examine the linkage of IVS3 + 2 T > C/5’UTR-215 G > A mutations and SPINK1 gene polymorphisms (Table 1), allele-specific PCR with a limited number of cycles was performed. Whereas the discrimination of alleles in the PCR reaction was incomplete, DNA sequencing could demonstrate the enrichment of the A peak at the position 5’-UTR-215 of the product which was synthesized with the specific primer Spl3-C-R, and, accordingly, the enrichment of the G peak of the product which was synthesized with the specific primer Spl3-T-R (Figure 2). Concordant results were found for all three patients. Based on these findings, we concluded that both single-nucleotide substitutions IVS3 + 2 T > C and 5’UTR-215 G > A were located in the same SPINK1 gene copy of the diploid genome.
Table 1.
Haplotype frequencies at five positions of the SPINK1 gene from 90 individuals (180 chromosomes) as estimated by EM-algorithm
| Haplotype | IVS2-352 A/G | IVS3-1643 G/C | IVS3-1568 C/A | IVS3-476 T/G | IVS3-321 C/T | nhplt1 | Haplotype frequency2 |
| 1 | A | G | C | T | C | 61 | 0.338889 |
| 2 | G | G | C | T | C | 35 | 0.194444 |
| 3 | A | G | C | T | T | 4 | 0.022222 |
| 4 | A | G | A | T | C | 2 | 0.011111 |
| 5 | G | G | C | T | T | 6 | 0.033333 |
| 6 | G | G | C | G | C | 2 | 0.011111 |
| 7 | G | G | A | T | C | 7 | 0.038889 |
| 8 | A | G | A | T | T | 1 | 0.0055556 |
| 9 | A | C | A | T | C | 2 | 0.011111 |
| 10 | G | G | A | T | T | 28 | 0.155556 |
| 11 | G | G | A | G | C | 1 | 0.0055556 |
| 12 | G | C | A | T | C | 4 | 0.022222 |
| 13 | G | G | A | G | T | 1 | 0.0055556 |
| 14 | G | C | A | T | T | 25 | 0.138889 |
| 15 | G | C | A | G | T | 1 | 0.0055556 |
| Total | 70/110 | 148/32 | 108/72 | 175/5 | 114/66 | 180 | 1.0 |
Estimated number of haplotypes.
The remaining 17 possible haplotypes could not be found in any individual.
Figure 2.
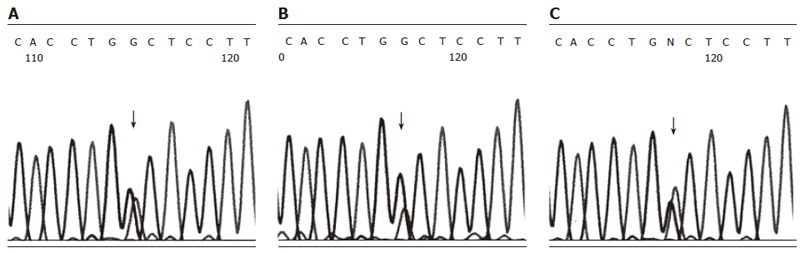
Linkage between 5’UTR-215 G > A and IVS 3 + 2 T > C mutations in SPINK1 gene. A: sequencing of -215 G/A heterozygote; B: sequencing of IVS 3 + 2 T (normal allele) enriched by allele-specific PCR; C: sequencing of IVS 3 + 2 C (mutant allele) enriched by allele-specific PCR.
Family history of IVS3 + 2C/5’UTR-215A gene carriers was disclosed
The investigation of the relatives revealed that one patient inherited both gene aberrations of the same SPINK1 gene copy from his mother. Interestingly however, the patient did not suffer from chronic pancreatitis but from celiac disease. These results might be a hint for a role of SPINK1 gene mutations in celiac disease. Unfortunately, the family history of the other 2 patients was not available.
Haplotype analysis suggested a common origin of IVS3 + 2C/5’UTR-215A mutations
The haplotypes, composed of five common SNP markers, were evaluated in a population of 90 healthy individuals (blood donors) and calculated by using an EM-algorithm[12,13]. The observed frequencies of 15 out of 32 possible haplotypes are presented in Table 1. The remaining 17 haplotypes could not be found in any individual. Long-distance PCR followed by allele-specific PCR and subsequent DNA sequencing of the IVS3 + 2C/5’UTR-215A-mutant genes showed that all three genes carried the most frequent haplotype 1. In contrast, the haplotype of the other IVS3 + 2C/5’UTR-215A-free SPINK1 gene copy of the heterozygous gene set of all three patients appeared to be random. One of them represented the same most frequent haplotype 1, one belonged to haplotype 2 and the third to the relatively rare haplotype 6. In spite of the low number of mutant samples, these findings suggested a common origin of the IVS3 + 2C/5’UTR-215A mutation in these patients. The conclusion of a common origin is supported by the assumption that 5’UTR-215A is a rare polymorphism and not a mutation as previously proposed[11].
No mutations of PRSS1 gene could be detected in IVS3 + 2C/5’UTR-215A carriers
DNA sequencing did not reveal any mutation in the other chronic pancreatitis-related gene PRSS1 in these three patients.
DISCUSSION
The most frequent variation of the human SPINK1 gene is the heterozygous or homozygous c.101 A > G (N34S) transition representing about 80% of all SPINK1 aberrations found in familial and idiopathic chronic pancreatitis[4,5]. In our study we detected the N34S with a frequency of 6% in chronic pancreatitis patients. In accordance with previous reports, this substitution is also the most frequent mutation of the SPINK1 gene. Other mutations of this gene are rare, but may be of high interest concerning the role of SPINK1 in development of hereditary chronic pancreatitis. More than 10 proposed SPINK1 gene mutations have been described until now[9]. However, to avoid incorrect interpretation, functional analysis has to be done to investigate the influence of the suggested mutations on the activity of the SPINK1 gene. Particularly, examination of the artificially produced N34S mutant protein demonstrated no change in its trypsin-binding ability differing from the wild-type protein[14]. On the other hand, an inactivating effect could be postulated a priori for other SPINK1 gene mutations, like IVS3 + 2 T > C, that destroys the splicing site of exons 3 and 4 and consequently leads to inactivation of the gene. We found three of these IVS3 + 2 T > C SPINK1 mutations in two German and a Macedonian patient with chronic pancreatitis. Another base substitution, 5’UTR-215 G > A, was also detected in all three mutant gene copies and for the first time in Caucasians. Up to now this substitution has only been detected as a homozygous base substitution in two chronic pancreatitis patients from Japan. Supported by the fact that this abnormality has never been found in healthy controls it has been considered to be an inactivating SPINK1 gene mutation[11]. Furthermore, 1 of the 2 Japanese patients had a family history of chronic pancreatitis. In our study, we only detected the 5’UTR-215 G > A substitution in direct linkage with the known SPINK1 gene inactivating mutation IVS3 + 2 T > C. The probability of the occurrence of 2 true mutations in the same gene copy seems to be rather seldom. Therefore, our conclusion is that the associated base substitution 5’UTR-215 G > A in the same SPINK1 gene copy is a rare polymorphism rather than a mutation. This assumption is supported by the fact that this base substitution is located outside the known coding and controlling sequences of the SPINK1 gene[11].
All 3 SPINK1 gene copies carrying these 5’UTR-215A/IVS3 + 2C aberrations had an identical haplotype of common polymorphism, namely haplotype 1. Although only 3 cases were discovered in our study, this finding hints for a single origin of the IVS3 + 2C mutation in these 3 patients. This can also substantiate that the 5’UTR-215 G > A base substitution is rather rare.
Family data were available for one patient with 5’UTR-215A/IVS3 + 2C mutant SPINK1 gene. This mutated gene was inherited from his mother who did not suffer from chronic pancreatitis, but from celiac disease. In some families the celiac disease was mapped close to the SPINK1 locus on chromosome 5 (5q31)[15,16]. Our results might be the first data suggesting a role of SPINK1 gene mutations in celiac disease.
The data from Kume et al[17] are compatible with our findings described in the present study. They showed that the 5’UTR-215 G > A and IVS3 + 2 T > C substitutions were in complete linkage in 9 of 116 pancreatitis patients. The published frequency of 8% in this report is higher than our incidence of 2% and suggests that the linkage of both substitutions may rather play a role in Japanese patients[17]. It would be interesting to compare the haplotypes of these mutant genes in both populations, Mongoloids and Caucasians, to evaluate the origin of these mutations.
Taken together, we found 3 chronic pancreatitis patients carrying an IVS3 + 2 T > C SPINK1 gene mutation and the rare base substitution 5’UTR-215 G > A in the same gene copy. Most probably, the 5’UTR-215 G > A substitution represents a rare polymorphism and not a mutation as previously concluded[11]. Future studies with a larger patient sample and functional analysis of the SPINK1 gene mutations are required to closer evaluate our findings and increase our understanding of their clinical manifestations.
ACKNOWLEDGMENTS
We thank Petra Merkert for excellent technical assistance.
Footnotes
Supported by research grants from the ´Werner Otto Stiftung e.V.´
S- Editor Wang J L- Editor Zhu LH E- Editor Bi L
References
- 1.Truninger K, Ammann RW, Blum HE, Witt H. Genetic aspects of chronic pancreatitis: insights into aetiopathogenesis and clinical implications. Swiss Med Wkly. 2001;131:565–574. [PubMed] [Google Scholar]
- 2.Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK Jr, Amann ST, Toskes PP, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 3.Horii A, Kobayashi T, Tomita N, Yamamoto T, Fukushige S, Murotsu T, Ogawa M, Mori T, Matsubara K. Primary structure of human pancreatic secretory trypsin inhibitor (PSTI) gene. Biochem Biophys Res Commun. 1987;149:635–641. doi: 10.1016/0006-291x(87)90415-3. [DOI] [PubMed] [Google Scholar]
- 4.Pfützer RH, Barmada MM, Brunskill AP, Finch R, Hart PS, Neoptolemos J, Furey WF, Whitcomb DC. SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000;119:615–623. doi: 10.1053/gast.2000.18017. [DOI] [PubMed] [Google Scholar]
- 5.Witt H, Luck W, Hennies HC, Classen M, Kage A, Lass U, Landt O, Becker M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 6.Witt H. Gene mutations in children with chronic pancreatitis. Pancreatology. 2001;1:432–438. doi: 10.1159/000055843. [DOI] [PubMed] [Google Scholar]
- 7.Witt H, Luck W, Becker M, Böhmig M, Kage A, Truninger K, Ammann RW, O'Reilly D, Kingsnorth A, Schulz HU, et al. Mutation in the SPINK1 trypsin inhibitor gene, alcohol use, and chronic pancreatitis. JAMA. 2001;285:2716–2717. doi: 10.1001/jama.285.21.2716-a. [DOI] [PubMed] [Google Scholar]
- 8.Rossi L, Pfützer RH, Parvin S, Ali L, Sattar S, Kahn AK, Gyr N, Whitcomb DC. SPINK1/PSTI mutations are associated with tropical pancreatitis in Bangladesh. A preliminary report. Pancreatology. 2001;1:242–245. doi: 10.1159/000055818. [DOI] [PubMed] [Google Scholar]
- 9. Available from: http: //www.uni-leipzig.de/pancreasmutation.
- 10.Le Maréchal C, Chen JM, Le Gall C, Plessis G, Chipponi J, Chuzhanova NA, Raguénès O, Férec C. Two novel severe mutations in the pancreatic secretory trypsin inhibitor gene (SPINK1) cause familial and/or hereditary pancreatitis. Hum Mutat. 2004;23:205. doi: 10.1002/humu.9212. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko K, Nagasaki Y, Furukawa T, Mizutamari H, Sato A, Masamune A, Shimosegawa T, Horii A. Analysis of the human pancreatic secretory trypsin inhibitor (PSTI) gene mutations in Japanese patients with chronic pancreatitis. J Hum Genet. 2001;46:293–297. doi: 10.1007/s100380170082. [DOI] [PubMed] [Google Scholar]
- 12.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic data. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 13.Sergeev AS, Arapova RK. [The use of the expectation-maximization (EM) algorithm for maximum likelihood estimation of gametic frequencies of multilocus polymorphic codominant systems based on sampled population data] Genetika. 2002;38:407–418. [PubMed] [Google Scholar]
- 14.Kuwata K, Hirota M, Shimizu H, Nakae M, Nishihara S, Takimoto A, Mitsushima K, Kikuchi N, Endo K, Inoue M, et al. Functional analysis of recombinant pancreatic secretory trypsin inhibitor protein with amino-acid substitution. J Gastroenterol. 2002;37:928–934. doi: 10.1007/s005350200156. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Juo SH, Holopainen P, Terwilliger J, Tong X, Grunn A, Brito M, Green P, Mustalahti K, Mäki M, et al. Genomewide linkage analysis of celiac disease in Finnish families. Am J Hum Genet. 2002;70:51–59. doi: 10.1086/338453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel RS, Johlin FC Jr, Murray JA. Celiac disease and recurrent pancreatitis. Gastrointest Endosc. 1999;50:823–827. doi: 10.1016/s0016-5107(99)70166-5. [DOI] [PubMed] [Google Scholar]
- 17.Kume K, Masamune A, Mizutamari H, Kaneko K, Kikuta K, Satoh M, Satoh K, Kimura K, Suzuki N, Nagasaki Y, et al. Mutations in the serine protease inhibitor Kazal Type 1 (SPINK1) gene in Japanese patients with pancreatitis. Pancreatology. 2005;5:354–360. doi: 10.1159/000086535. [DOI] [PubMed] [Google Scholar]


