Abstract
In this study, we provided molecular evidences that IL-6 contributed to the decreased capacity of oxidative biotransformation in human liver by suppressing the expression of CYP3A4. After human hepatocytes were treated with IL-6, DEC1 expression rapidly increased, and subsequently, the CYP3A4 expression decreased continuously. Furthermore, the repression of CYP3A4 by IL-6 occurred after the increase of DEC1 in primary human hepatocytes. In HepG2 cells, knockdown of DEC1 increased the CYP3A4 expression and its enzymatic activity. In addition, it partially abolished the decreased CYP3A4 expression as well as its enzymatic activity induced by IL-6. Consistent with this, overexpression of DEC1 markedly reduced the CYP3A4 promoter activity and the CYP3A4 expression as well as its enzymatic activity. Using sequential truncation and site directed mutagenesis of CYP3A4 proximal promoter with DEC1 construct, we showed that DEC1 specifically bound to CCCTGC sequence in the proximal promoter of CYP3A4, which was validated by EMSA and ChIP assay. These findings suggest that the repression of CYP3A4 by IL-6 is achieved through increasing the DEC1 expression in human hepatocytes, the increased DEC1 binds to the CCCTGC sequence in the promoter of CYP3A4 to form CCCTGC-DEC1 complex, and the complex downregulates the CYP3A4 expression and its enzymatic activity.
Keywords: cytochrome P450 3A4 (CYP3A4), interleukin 6 (IL-6), differentially expressed in chondrocytes 1 (DEC1), transcription regulator
1. Introduction
The liver is the richest source of drug metabolism [1], which is regulated by the expression of drug-metabolism enzymes [1,2]. Cytochrome P450 3A4 (CYP3A4) is the most important human CYP in the liver and small intestine and plays a major role in the biotransformation of many drugs. Actually, it is responsible for the oxidative metabolism of more than 60% of all pharmaceuticals [3,4] and its activity shows a wide inter-individual variability, which forms a basis for clinically significant drug interactions and toxicities [5,6]. Cytokines such as tumor necrosis factor-α (TNFα) and interleukin-6 (IL-6), which increase in inflammatory diseases and some cancers [7,8], have been shown to down-regulate the expression of a variety of drug-metabolizing enzymes including CYP3A4 [7,9,10,11]. Transactivation by nuclear receptors such as pregnane X receptor (PXR) and constitutive androstane receptor (CAR), is largely responsible for the increased expression of these genes [2]. Some studies have reported that the decrease of CYP3A4 enzyme expression is associated with the repression of CAR and PXR in mouse liver during the acute phase response [12] and in human hepatocytes mediated by IL-6 [11]. However, other mechanisms may also contribute to the down-regulation of CYP3A4 in inflammation, because CYP3A4 decreases to a larger extent compared to PXR in response to cytokines such as IL-6 [11].
Human DEC (differentially expressed in chondrocytes, DEC), mouse STRA13 (stimulated with retinoic acid 13), and rat SHARP (split and hairy related protein) constitute a new and structurally distinct class of basic helix-loop-hellix (bHLH) proteins [13,14,15]. The bHLH proteins are intimately associated with the developmental events such as cell differentiation and lineage commitment [16,17,18]. The HLH domain in the bHLH motif is responsible for dimerization, whereas the basic region mediates DNA binding [19]. Two members of DEC/STRA/SHARP proteins are identified in each mammalian species studied with a sequence identity of >90% in the bHLH region and ~40% in the total proteins, respectively [14]. They are called DEC1 and DEC2, exhibiting an overlapping tissue distribution. DEC/STRA/SHARPs have been implicated in cell differentiation [14,20], maturation of lymphocytes [21], regulation of molecular clock [22], and involvement in maintaining the homeostasis of metabolism [23]. In addition, DEC1 expression is highly elevated in response to environmental stimuli such as hypoxia and cytokines such as TNFα [24,25].
In this study, we report that DEC1 is a transcriptional repressor of CYP3A4, a major human enzyme that plays an important role in oxidative biotransformation [26]. After human hepatocytes were treated with IL-6, the DEC1 expression rapidly increased, then, the expression of CYP3A4 decreased at both mRNA and protein levels. Meanwhile, the repression of CYP3A4 by IL-6 occurred after the increase of DEC1 in primary human hepatocytes. So we hypothesize DEC1 is a transcriptional repressor of CYP3A4, and DEC1 induction is one of important mechanisms of the reduction of CYP3A4 expression by IL-6 in primary human hepatocytes.
2. Materials and methods
2.1. Chemicals and supplied
IL-6 was purchased from R & D Systems (Minneapolis, MN, USA). FlagCMV2 vector, rifampicin and Williams'E medium were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco's modified Eagle's medium, high-fidelity platinum Taq DNA polymerase, and insulin-transferrin-selenium G supplement were purchased from Invitrogen (Carlsbad, CA, USA). GeneJet™ DNA VitroTransfection Reagent (Ver II) was from SignaGen Labortories (Gaithersburg, MD, USA). Dual-luciferase reporter assay system and P450-Glo™ Luminescent cytochrome P450 3A4 Assay system were purchased from Promega (Madison, WI, USA). Fetal bovine serum was from Hyclone Laboratories (Logan, UT, USA). The antibody against glyceraldehyde-3- phosphate dehydrogenase (GAPDH) was from Abcam (Cambridge, UK). The goat anti-rabbit IgG conjugated with horseradish peroxidase from Pierce Chemical (Pierce, Rockford, IL, USA). Polyclone antibody (from rabbit) against DEC1 and CYP3A4 were kindly provided by Dr.Yan Lab (University of Rhode Island). Nitrocellulose membrane was from Bio-Rad Laboratories (Hercules, CA, USA). DEC1 shRNA plasmid and IgG were from Santa Cruz (Santa Cruz, CA, USA). Nuclear extraction kit, Chromatin Immunoprecipitation Assay Kit was from Active Motif, Inc (Carlsbad, CA, USA). [γ-32P]ATP was from FuRui Bioengineering company (Beijing, China). All other reagents were from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Culture and treatment of human primary hepatocytes and hepatoma cells
Human primary hepatocytes in 6-well plates were obtained from the Liver Tissues Procurement and Distribution System (University of Minnesota, Minneapolis, MN or CellzDirect, Pittsboro, NC, USA). The ten hepatocyte donors were all non-smokers of four males (21-65 years old) and six females (35-72 years old) with seven white and three black. Upon the arrival of the hepatocytes, the culture media were replaced with Willians'E medium containing insulin-transferrin-selenium supplement and penicillin/streptomycin [27]. After incubation at 37 °C with 5% CO2 for 24 h, the hepatocytes were treated with 10 ng/ml IL-6 for 24 h (for mRNA level) or 48 h (for protein level) [27]. Hepatoma (HepG2) cells were purchased from American Type Culture Collection (Mannassas, VA, USA), and maintained in the Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, penicillin/streptomycin, and 1 × nonessential amino acids. HepG2 cells were seeded at the density of 106 cells/well (6-well plates), 2.5 × 105 cells/well (12-well plates) and that of 8 × 104 cells/well (48-well plates )in a regular medium, and the treated cells were cultured in a 1% serum-reduced medium.
2.3. Quantitative reverse transcription-polymerase chain reaction
The total RNA was isolated by using a RNA-Bee (Tel-Test Inc., Friendswood, TX, USA) according to the manufacturer's instruction and checked by formaldehyde gel electrophoresis for quality control. The first-strand cDNA was synthesized using total RNA (1 μg) at 25 °C for 10 min, 42 °C for 50 min, and 70 °C for 10 min by using random primers and moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI, USA). The cDNAs were then diluted eight times and the quantitative PCR was conducted with TaqMan Gene Expression Assay kits (Applied Biosystems, Foster City, CA, USA). The TaqMan assay identification numbers are: CYP3A4, Hs00604506_m1; DEC1, Hs00186419_m1; GAPDH, 4352934E. A 20 μl PCR mix contained 10 μl of universal PCR master mixture, 1 μl of gene-specific TaqMan assay mixture (probe), 6 μl of diluted cDNA as template and 3 μl of water. The PCR amplification and quantification were done in an Applied Biosystems 7900 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with one cycle at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 min. The signals from each target gene were normalized based on the signal from GAPDH.
2.4. Plasmid constructs and site-directed mutagenesis
A cDNA encoding the full-length DEC1 was isolated by a cDNA-trapping method [19]. DEC1 mutant constructs were prepared by PCR with full-length DEC1 as the template as previously described [19]. CYP3A4-DP and CYP3A4P promoter reporters (−7836/−6093 to −362/+53, −362/+53) were prepared as previously described [27,28]. A set of deleted and mutated constructs targeted on proximal promoter, which were presented in Fig. 4A, were prepared by PCR using the targeting primers shown in Tab. 1 and Tab. 2. The fragments harboring these elements were amplified by PCR with primers that were extended to include appropriate endonucleases (Xhol/HindIII) to facilitate the subsequent ligation. The sequences of all of the CYP3A4 reporter gene constructs were verified by direct DNA sequencing.
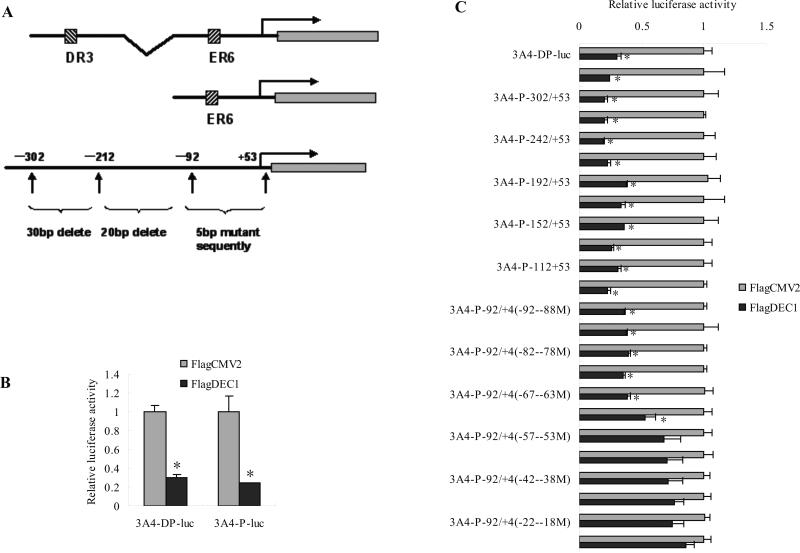
Fig. 4. Differential repression of CYP3A4-DP-Luc and deleted mutants or 5 base pairs mutants of CYP3A4-P-Luc.
(A) Diagrammatic presentation of a series of deleted mutants or 5 base pairs mutants of CYP3A4 luciferase reporter gene constructs. (B) Repressing effect of DEC1 on the CYP3A4-DP-Luc and CYP3A4-P-Luc. (C) Differential repression of CYP3A4-DP-Luc and deleted mutants or 5 base pairs mutants of CYP3A4-P-Luc. HepG2 cells were cultured in 48-well plates and transfected with CYP3A4-DP-Luc (50 ng), CYP3A4-P-Luc (50 ng), or corresponding mutated CYP3A4-P-Luc and along with 5 ng of Null-Renilla reniformis luciferase plasmid. After 48 h incubation, cells were collected and analyzed for luciferase activity. The data are expressed as fold of induction (the ratio of normalized luciferase activity from DEC1 transfection over vector transfection) (n = 4). All experiments were repeated at least three times, and data were expressed as mean ±SD. * p<0.05, statistically significant decrease by IL-6 treatment.
Table 1.
Primers used for preparation of mutated constructs of CYP3A4-P-luc reporter gene The underlined letters indicated the restriction sites.
| Primer | Sequence(5'-3') |
|---|---|
| CYP3A4-P(-302Xhols)-luc | ATACTACTCGAGAGAGAACAAGGGCAAGAGAGAG |
| CYP3A4-P(-272 Xhols)-luc | ATACTACTCGAGTAGATTTTATGCCAATGGCTC |
| CYP3A4-P(-242 Xhols)-luc | ATACTACTCGAGTTCTGATAAGAACCCAGAACC |
| CYP3A4-P(-212 Xhols)-luc | ATACTACTCGAGCCCAGTAACATTGATTGAGTT |
| CYP3A4-P(-192 Xhols)-luc | ATACTACTCGAGTGTTTATGATACCTCATAGAA |
| CYP3A4-P (-172 Xhols)-luc | ATACTACTCGAGTATATGAACTCAAAGGAGGTCA |
| CYP3A4-P(-152 Xhols)-luc | ATACTACTCGAGTAGTGAGTGGTGTGTGTGTGAT |
| CYP3A4-P(-132 Xhols)-luc | ATACTACTCGAGTTTCTTTGCCAACTTCCAAGGT |
| CYP3A4-P(-112 Xhols)-luc | ATACTACTCGAGTTGGAGAAGCCTCTTCCAACTG |
| CYP3A4-P(-92 Xhols)-luc | ATACTACTCGAGTGCAGGCAGAGCACAGGTGGCC |
| CYP3A4-P(+53HindIIIa)-luc | ATACTAAAGCTTTGTTGCTCTTTGCTGG |
“s”stands for sense and “a” stands for antisense
Table 2.
Primers used for preparation of mutated constructs of CYP3A4-P92-luc reporter gene The underlined letters indicated the restriction sites and highlight letters indicated mutant sites.
| Primer | Sequence(5’-3’) |
|---|---|
| CYP3A4(−92/−88Xhols) | ATACTACTCGAGCGTCCCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−87/−83Xhols) | ATACTACTCGAGGCAGGGTCTCCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−82/−78Xhols) | ATACTACTCGAGGCAGGCAGAGGTGTCGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−77/−73Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGCACCGCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−72/−68Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCGGACGTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−67/−63Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCATGACGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−62/−58Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGCGACGAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(+4HindIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCTAGAGAAGGAGGCAGGGCTGGAGCT |
| CYP3A4(−57/−53Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCTCGAGCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−57/−53HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCTAGAGAAGGAGGCAGGGCTGCTCGA |
| CYP3A4(−52/−48Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCGTCGGCTGCCTCCTTCTCTA |
| CYP3A4(−52/−48HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCTAGAGAAGGAGGCAGCCGACGAGCT |
| CYP3A4(−47/−43Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCGACGGTCCTTCTCTA |
| CYP3A4(−47/−43HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCTAGAGAAGGACCGTCGGCTGGAGCT |
| CYP3A4(−42/−38Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCAGGAACTCTA |
| CYP3A4(−42/−38HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCTAGAGTTCCTGGCAGGGCTGGAGCT |
| CYP3A4(−37/−33Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTGAGAT |
| CYP3A4(−37/−33HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTATATGCATCTCAAGGAGGCAGGGCTGGAGCT |
| CYP3A4(−92Xhols) | ATACTACTCGAGGCAGGCAGAGCACAGGTGGCCCTGCTACTGGCTGCAGCTCCAGCCCTGCCTCCTTCTCTA |
| CYP3A4(−32/−28HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTGTTTAATACGATCTCAAGGAGGCAGGGCTGGAGCT |
| CYP3A4(−27/−23HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTGGATTCAAATTATGCATCTCAAGGAGGCAGGGCTGGAGCT |
| CYP3A4(−22/−18HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGCTGTTCCTAAGTTTATATGCATCTCAAGGAGGCAGGGCTGGAGCT |
| CYP3A4(−17/−13HidIIIa) | ATACTAAAGCTTCAGTGATTCAGTGAGGGACAAGGATTGTTTATATGCATCTCAAGGAGGCAGGGCTGGAGCT |
2.5. Transient co-transfection experiment
HepG2 cells were plated in 48-well plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at the density of 8 × 104 cells/well. Transfection was conducted by GeneJet™ DNA VitroTransfection Reagent (Ver II). The transfection mixtures contained 50 ng of a reporter plasmid (CYP3A4-DP-luc), 50 ng of hPXR plasmid, a corresponding concentration of FlagDEC1 along with 5 ng of Null-Renilla reniformis luciferase plasmid. Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. HepG2 cells were transfected for 24 h and the transfected cells were treated with rifampicin (Rif) 10 μM for another 24 h. The cells were washed once with phosphate-buffer saline (PBS) and then lysed by 1 × passive lysis buffer (Promega, Madison, WI). The collected cells were subjected to two cycles of freeze/thaw. The reporter enzyme activities were assayed with Dual-Luciferase reporter assay system. This system contained two substrates, the firefly luminescence and Renilla luminescence, which were used to determine the activities of two luciferases sequentially. The firefly luciferase activity, which reflected the reporter activity, was evaluated by mixing an aliquot of lysates (10 μl) with Luciferase Assay Reagent II (Promega, Madison, WI). Then, the firefly luminescence was quenched and the Renilla luminescence was activated simultaneously by adding Stop & Glo reagent (Promega) to the sample tubes. The firefly luminescence signal intensity was normalized based on the intensity of Renilla luminescence signal as normalized luciferase activity, and the ratio of normalized luciferase activity from the transfected with FlagDEC1 over the transfected with vector served as relative luciferase activity.
2.6. The modulation of DEC1 expression by DEC1 shRNA and DEC1 overexpression
To define the role of DEC1 in the IL-6-mediated down-regulation of CYP3A4, the expression of DEC1 was regulated by DEC1 shRNA and overexpression. In the DEC1 shRNA experiment, HepG2 cells were plated in 6-well plates at the density of 106 and transfected with the DEC1 shRNA construct ( 800 ng/well ) or the corresponding vector for 72 h with a change of fresh medium at 36 h. The same procedure was used for overexpression experiment except for replacement DEC1 shRNA construct with the FlagDEC1 construct or the FlagCMV2 (vector) for 48 h. The transfected cells were treated with IL-6 (10 ng/ml) or the same volume of PBS for 24 h, and the expressions of CYP3A4 and DEC1 were monitored by real-time PCR and Western blot.
2.7. Western analysis
Human hepatocytes (8 μg) and HepG2 cell lysates (80 and 2 μg for DEC1 shRNA and DEC1 overexpression, respectively) were resolved by 7.5% SDS-polyacrylamide gel electrophoresis and electrophoretically transferred to a nitrocellulose membrane. After nonspecific binding sites were blocked with 5% nonfat milk, the blots are incubated with an antibody against CYP3A4 (1:2500), DEC1 (1:2500) and GAPDH (1:4000). The preparation of the antibody against human CYP3A4 and DEC1 was described elsewhere [19,29]. The primary antibodies were subsequently localized with goat anti-rabbit IgG conjugated with horseradish peroxidase. Horseradish peroxidase activity was detected with a chemiluminescent kit (Pierce, Rockford, IL, USA). The chemiluminescent signal was captured by KODAK Image Station 2000 (Estman Kodak, Rochester, NY, USA), and the relative intensities were quantified by KODAK Image Analysis software (Estman Kodak, Rochester, NY, USA).
2.8. Enzymatic assay
HepG2 cell lysates were prepared in 100 mM potassium phosphate buffer. The activity of CYP3A4 was determined with a P450-Glo™ kit (CYP3A4) (Promega, Madison, WI, USA) [27] according to the manufacturer's manual. Briefly, cell lysates (16.5 μg in 12.5 μl) were mixed with 12.5 μl of CYP3A4 substrate Luciferin-BE (4×). After a 10-min pre-incubation at 37°C, the NADP regeneration mixture (25 μl containing 400 mM KPO4) was added to initiate enzymatic reaction. The reaction lasted for 30 min at 37°C and was terminated by adding 50 μl of Luciferin Detection Reagent (Promega, Madison WI). After another 10 min of incubation at room temperature, the luminescent signal intensity was determined by EGΣG BERTHOLD Microplate Luminometer (PerkinElmer, Waltham, MA, USA). Several controls were performed including incubation without cell lysates or regeneration system.
2.9. Electrophoretic mobility-shift assays (EMSA)
HepG2 cells were plated in six-well plate and transfected with FlagDEC1 expression construct (800 ng/well) and FlagCMV2 (800 ng/well) for 24 h. Nuclear extracts were prepared with a nuclear extraction kit (Active Motif). Nuclear protein (10 μg) was incubated with purified 32P-labelled double-stranded oligonucleotides (5'-AGCTCCAGCCCTGCCTCCTTCTCTA -3’) in a final volume of 10 μl containing 1 × DNA-binding buffer (Pierce). For competition experiments, nuclear extracts were incubated with excess un-labelled probe (5 ×, 50 ×) or un-labelled mutants (mut1, mut2, mut3) (50 ×) and mixed with the radiolabelled probe. mut1: 5’-AGCTCCAGTTTTGCCTCCTTCTCTA-3’ (mut CCCTGC); mut2: 5’-AGCTCCAGCCCTGCCTAATTCTCTA -3’(mut +); mut3: 5’-AGCTCCAGTTTTGCCTAATTCTCTA -3’(mut CCCTGC/+). For supershift assay, an anti- DEC1 was added either before or after the nuclear extracts were incubated with the radiolabelled probe. The protein-DNA complexes were resolved in 6% polyacrylamide gel electrophoresis and visualized by autoradiography (Typhoon 8600 Variable Mode Imager )(Amersham, Silicon valley, CA). Several controls were performed including nuclear extract from the cells transfected with corresponding vector and no radiolabelled probe.
2.10. Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) assay was performed with the Chromatin Immunoprecipitation Assay Kit (Active Motif). Chromatins were prepared from the following sources: HepG2 cells were seeded at the density of 3 × 106 cells/flask (50 ml) overnight and treated with IL-6 (10 ng/ml) or the same volume of PBS for 24 h or were transfected with FlagDEC1 (3 μg) or the same amount of FlagCMV2 for 48 h. Cells were fixed with 1% formaldehyde-containing DMEM at room temperature for 10 min and then treated with Glycine Stop-Fix solution. The cells were washed with PBS, harvested, and centrifuged at 720 g for 10 min at 4 °C. Pellets were resuspended in 1 ml of lysis buffer and incubated on ice for 30 min, then processed with a Dounce homogenizer for 10 strokes and centrifuged at 2,400 g for 10 min at 4 °C. Nuclei were suspended in 0.5 ml of Shearing Buffer and sonicated with the Branson Sonifier 150 (Branson, Danbury, CT) by three strokes of 20 seconds pulse at power level 5 with a 30 seconds interval on ice to achieve optimal chromatin sizes ranging from 400 to 800 bp. The sheared chromatins were collected as the supernatant after being centrifuged at 12,000 g for 12 min. The preparation of chromatins from human liver tissues from three individual donors were obtained from Jinling Hospital (Nanjing Medical Unversity, Nanjing). The study was approved by Ethics Committee of Nanjing Medical University and was informed consent was obtained from each individual donor. The liver tissues were frozen in liquid nitrogen and pulverized into powder, followed by cross-linking with 1% formaldehyde for 15 min at room temperature and subsequent termination of cross-linking with addition of Glycine Stop-Fix solution. The soluble chromatins were prepared by following the same procedure described above. For ChIP experiments, 50 μl aliquots of sheared chromatins were used for immunoprecipitation with the addition of 25 μl of protein G magnetic bead suspension and 2 μg of DEC1 antibodies [19] or 2 μg of IgG antibodies as a negative control. The chromatins were eluted from the magnetic beads after being washed with ChIP buffers, followed by reversing the cross-link with the addition of Reverse Cross-link Buffer and being heated at 65 °C overnight. The proteins in the solution were digested by proteinase K at 37 °C for an hour, followed by the addition of proteinase K stop solution to terminate the reaction. The final volume of the eluted chromatin DNA was 100 μl. For the preparation of input DNA, 10 μl of sheared chromatins was diluted up to 100 μl to reverse the cross-link and proteinase K digestion, followed by 10-fold dilution with water.
Four sets of primers were used for PCR amplification. The primer sets the CCCTGC sequence at the position −51 bp consisted of the forward primer pCYP3A4 (−253b) 5’- CACT TGAGTTTCTG ATAAGAAC-3’ and the reverse primer pCYP3A4 (−35b) 5’- GTCTTCCTTTCAGCTCTGTGTTGCT-3’. The expected PCR product is a 218 bp fragment encompassing the sequence from nucleotides −253 to −35 in the CYP3A4 promoter. The negative control primer (No CCCTGC sequence) set consisted of the forward primer pCYP3A4 (-457b) 5’-GAGGAAAGACTCTAAGAGAA-3’ and the reverse primer pCYP3A4 (−251b) 5’- GTTCTTATCAGAAACTCAAGTG-3’ and amplified a 206 bp fragment encompassing the sequence from nucleotides −457 to −251 in the CYP3A4 promoter. PCR amplification was carried out in a final volume of 25 μl with 2 μl of eluted chromatin DNA, input DNA, or water (mock) as template and 1 unit of high-fidelity Taq DNA polymerase (Invitrogen) with the following cycling parameters: 94 °C for 3 min, 40 cycles of 94 ° C for 45 seconds, 53 °C for 45 seconds, and 68 °C for 45 seconds, followed by an extension at 68 °C for 5 min. PCR products were resolved on 2% agarose gels and visualized by Biosens sc 810 Gel Electrophoresis Image analytic system (Shanghai, China).
2.11. Other analyses
Protein concentrations were determined with BCA assay (Pierce, Rockford, IL, USA) based on the albumin standard [27]. Data are presented as mean ± SD from at least three independent experiments. Statistical analysis was performed using SAS software version 9.1, (SAS Institute, Cary, NC. USA). The significant difference between the treatments was claimed at P < 0.05 based on one-way analysis of variance followed by Duncan's multiple comparison tests.
3. Results
3.1. IL-6 rapidly induces DEC1 expression and continuously reduces CYP3A4 expression in primary cultured human hepatocytes
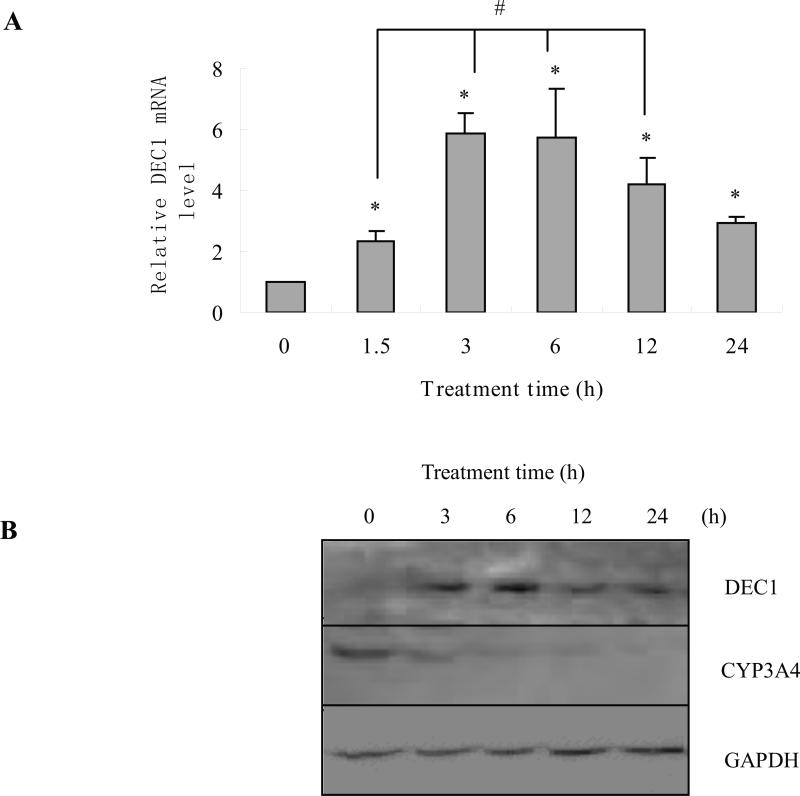
After human hepatocytes were treated with IL-6 (10 ng/ml) or the same volume of PBS for 1.5, 3, 6, 12 and 24 h, DEC1 mRNA levels were monitored by qRT-PCR. DEC1 mRNA levels significantly increased after an hour and a half treatment, and peaked at 6 h, then, the induction of DEC1 expression decreased (Fig 1A). The expression of CYP3A4 continuously decreased after 3 h treatment (data not shown). Meanwhile, the repression of CYP3A4 by IL-6 occurred after the increase of DEC1 in human hepatocytes. To further investigate whether the decrease of CYP3A4 mRNA was translated into the decrease in protein level, the expression of DEC1 and CYP3A4 at protein level for 3, 6, 12 and 24 h after being treated with IL-6 (10 ng/ml) was determined by Western blotting in cultured human hepatocytes. The data showed that DEC1 protein level increased after 3 h treatment, and peaked at 6 h, and then, the induction of DEC1 expression decreased. The expression of CYP3A4 continuously decreased after 3 h treatment (Fig. 1B) till 24 h. The changes in protein level corresponded to the changes at mRNA level.
Fig.1. Time course of DEC1 and CYP3A4 expression in primary human hepatocytes treated with IL-6.
(A) Time course of DEC1 mRNA level in human hepatocytes treated with IL-6. Human primary hepatocytes were treated with IL-6 (10 ng/ml) or the same volume of PBS for 24 h. Total RNA was isolated and subjected to qRT-PCR analysis for the level of DEC1 probe as described under Materials and Methods. The qPCR Cts were 25 for DEC1, and 20 for GAPDH. (B) Time course of DEC1 and CYP3A4 protein levels in human hepatocytes treated with IL-6. Human hepatocytes were treated with IL-6 (10 ng/ml) or the same volume of PBS for 48 h, and cell lysates were prepared and lysates (8 μg) were subjected to Western blot analyses with an antibody against DEC1, CYP3A4 or GAPDH (n = 5). All experiments were repeated at least three times, and data were expressed as mean ±SD. * p<0.05, statistically significant decrease by IL-6 treatment.
3.2. DEC1 mediates the suppression of CYP3A4 expression and its enzymatic activity induced by IL-6
There occurs a rapid increase of DEC1 expression after an hour and a half of treatment with IL-6, but the significant continuous decrease of CYP3A4 occurred after 3 h treatment, which suggests that the decrease of CYP3A4 expression occurs after the increase of DEC1 induced by IL-6 in human hepatocytes.
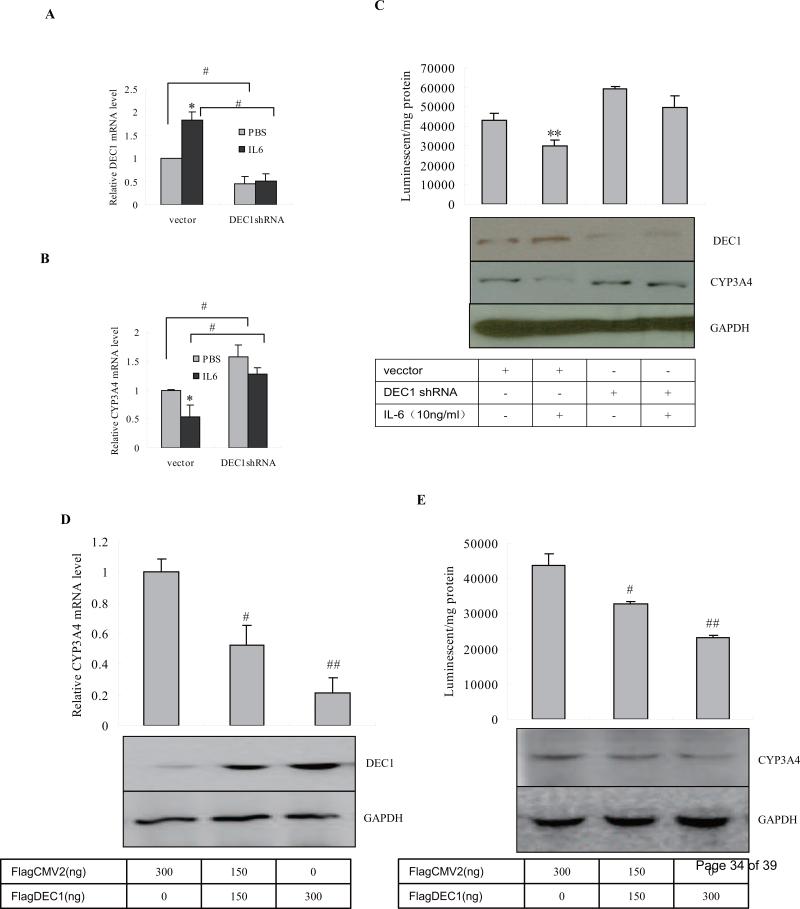
To determine the role of DEC1 in the down regulation of CYP3A4 in response to IL-6, we performed knockdown and overexpression experiments to selectively modulate the expression of DEC1. In the knockdown experiment, HepG2 cells were transfected with DEC1 shRNA construct for 72 h by using corresponding vector as a control. After being treated with IL-6 (10 ng/ml) or PBS for 24 h, the DEC1 and CYP3A4 levels in cells were analyzed by using qRT-PCR and Western blot. To test shRNA against DEC1 efficiency, the cell lysates (80 μg) were analyzed for DEC1 by Western Blot. In the results, DEC1 knockdown alone significantly increased the CYP3A4 expression both in mRNA and protein levels and enzymatic activity (Fig. 2B, 2C). And IL-6 significantly decreased CYP3A4 both in mRNA and protein levels and enzymatic activity in the cells transfected with the vector, but, not in the cells transfected with the DEC1 shRNA construct (Fig. 2B, 2C). Meanwhile, IL-6 significantly increased DEC1 expression in the cells transfected with vector, but not in the cells transfected DEC1 shRNA construct (Fig. 2A, 2C). Each experiment was confirmed to have a DEC1 shRNA knockdown effect of 60% or more on corresponding gene expression by using qRT-PCR and Western blot (Fig. 2A, 2C). These results suggest that knockdown of DEC1 partially abolishes the decrease of CYP3A4 triggered by IL-6 in hepatocytes.
Fig.2. Repression of CYP3A4 as a function of DEC1.
(A) (B) Effect of DEC1 knockdown on DEC1 and CYP3A4 mRNA level induced by IL-6. HepG2 cells in 6-well plates were transiently transfected with a mixture containing 800 ng of the DEC1 shRNA construct or the corresponding vector (per well). After 72 h incubation, the transfected cells were treated with IL-6 (10 ng/ml), or PBS for another 24 h. Total RNA was prepared and analyzed for levels of DEC1, CYP3A4 and GAPDH by qRT-PCR. The qPCR Cts were 25 for DEC1, 24 for CYP3A4 and 20 for GAPDH. (C) Effect of DEC knockdown on CYP3A4 protein and catalytic levels induced by IL-6. HepG2 cells were subjected to same procedure as above, cell lysates were analyzed for CYP3A4 (100 μg) by Western blot and its enzymatic activity as described in materials and methods. To determine the DEC1 knockdown efficiency, the cell lysates (80 μg) were analyzed for DEC1 by Western Blot. (D) (E) Effect of DEC1 overexpression on the repression of CYP3A4 and its functional activity. HepG2 cells in 6-well plates were transiently transfected with 800 ng of FlagDEC1 construct or the corresponding vector (per well). After 48 h incubation, the transfected cells, total RNA were isolated and analyzed for the expression of CYP3A4, and GAPDH by qPCR. And cell lysates (100 μg) were analyzed for CYP3A4 expression by Western bolt and its enzymatic activity. To determine the transfection efficiency, the cell lysates (2 μg) were analyzed for DEC1 by Western Blot. All experiments were repeated at least three times, and data were expressed as mean ±SD. * p< 0.05, significant difference from IL-6 treatment and PBS treatment and # p<0.05, significant difference from DEC1 shRNA- or DEC1-transfected cells and vector-transfected cells.
In the overexpression experiment, the construct encoding FlagDEC1 was used instead of the DEC1 shRNA construct in the previous experiment. HepG2 cells were seeded in 12-well plates at the density of 2.5 × 105 and transfected with FlagDEC1 150 ng or 300 ng/well for 48 h, and the transfected cells were analyzed for CYP3A4 expression and its enzymatic activity. Contrary to the DEC1 knowndown experiment, the DEC1 overexpression significantly decreased CPY3A4 expression and its enzymatic activity (Fig. 2D, 2E). Moreover, DEC1 decreased the CYP3A4 expression and its functional activity in a dose-dependent manner (Fig.2D, 2E). The data, connectively, suggest that DEC1 is a repressor of CYP3A4 and mediates the decrease of CYP3A4 expression and its functional activity triggered by IL-6 in hepatocytes.
3.3. DEC1 represses the transactivation of CYP3A4-DP promoter
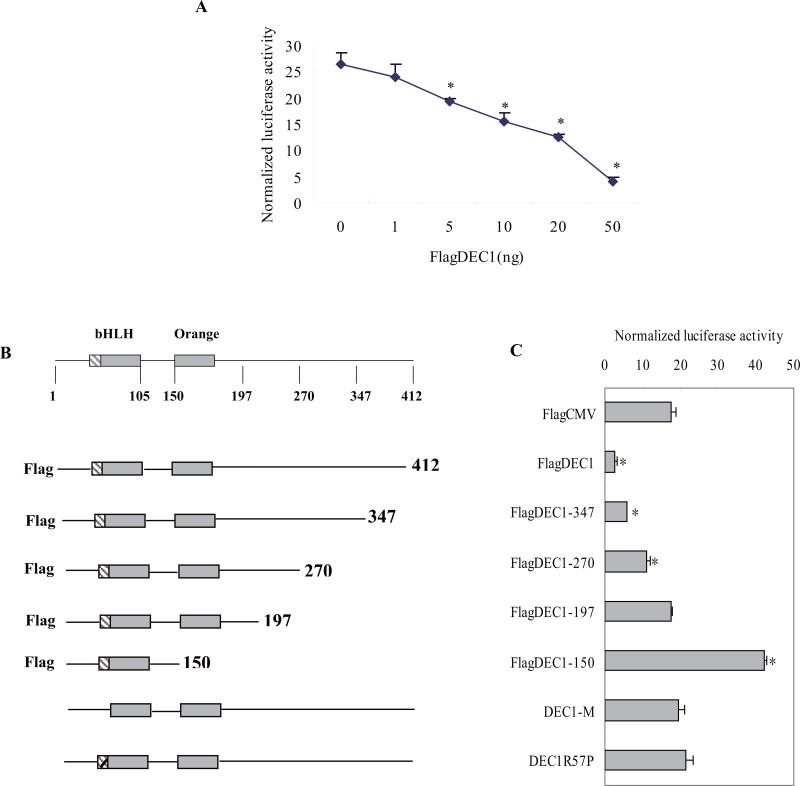
Our previous data showed that the IL-6-mediated repression of CYP3A4 occurs at the transcriptional level in hepatocytes [11]. In the present study, we found that DEC1 is required for the IL-6 induced repression of CYP3A4 and the increased DEC1 is likely to lead to the CYP3A4 decrease in hepatocytes. Next, we investigated whether DEC1, as a transcriptional factor, repressed the CYP3A4 promoter. HepG2 cells were transfected with mixtures containing 50 ng of a reporter plasmid (CYP3A4-DP-luc), 50 ng of hPXR and a corresponding concentration of FlagDEC1 along with 5 ng of Null-Renilla reniformis luciferase plasmid by using GeneJet™ DNA VitroTransfection Reagent (Ver II). Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. HepG2 cells were transfected for 24 h and collected for luciferase assay. As shown in Fig. 3A, DEC1 repressed the CYP3A4-DP-luciferase in a dose-dependent manner. The repression of DEC1 on CYP3A4 reporter provides direct evidence that this transcription factor is a regulator of the CYP3A4 promoter, presumably by interacting with a certain region. To test this possibility, we examined DEC1 mutants including those without DNA binding domain (DEC1-M), substitution mutant (DEC1R58P), and C-terminal deletion mutants (DEC11-347, DEC11-270, DEC11-197, DEC11-150) (Fig.3B) for their repressive ability towards this reporter (CYP3A4-DP-luc). HepG2 cells were plated in 48-well plates and transfected with mixtures containing 50 ng of a reporter plasmid (CYP3A4-DP-luc), 50 ng of hPXR plasmid, 50 ng of FlagDEC1 or FlagCMV2 along with 5 ng of Null-Renilla reniformis luciferase plasmid for 12 h and all the transfected cells were treated with 10 μM rifampicin (Rif) for another 24 h. Then the cells were collected for luciferase assay. As shown in Fig. 3C, the wild type DEC1 repressed the CYP3A4 reporter Rif-induced transactivation, whereas DEC1R58P and DEC1-M showed no effects towards the CYP3A4-DP-Luc. The C-terminal deletion mutants such as FlagDEC11-347, FlagDEC11-270, on the other hand, showed that the repression of DEC1 decreased as deletion increased (Fig. 3B, 3C). Furthermore, FlagDEC11-197 showed no repression of CYP3A4 reporter, whereas FlagDEC11-150, which has no ligand binding domain but DNA binding domain, markedly transactivated the CYP3A4 reporter by two fold (Fig. 3C).
Fig.3. Requirement of DNA binding for DEC1 to repress CYP3A4 promoter.
(A) DEC1 repressed CYP3A4 promoter in a dose-dependent manner. HepG2 cells were cultured in 48-well plates and transfected with CYP3A4-DP-Luc (50 ng), hPXR (50 ng), corresponding concentration of DEC1 ( 1, 5, 10, 20, 50 ng ) and 5 ng of Null-Renilla reniformis luciferase plasmid. Vector plasmid was used to equalize the mount of plasmid DNA for each transfection. After 24 h incubation, the transfected cells were treated with Rif (10 μM) for another 24 h, and then, the cells were collected and analyzed for luciferase activity. (B) Diagrammatic presentation of a series of wild type and various mutated DEC1 constructs. (C) Differential effect of wild type and various mutated DEC1 on CYP3A4 promoter. HepG2 cells were cultured in 48-well plates and transfected with CYP3A4-DP-Luc (50 ng), hPXR (50 ng), wild type DEC1 or corresponding mutated DEC1 (50 ng) along with 5 ng of Null-Renilla reniformis luciferase plasmid for 24 h, and the transfected cells were treated with Rif (10 μM) for another 24 h, and then, the cells were collected and analyzed for luciferase activity. The data were expressed as normalized luciferase activity (based on Null-Renilla reniformis luminescence signal) (n = 3). All experiments were repeated at least three times, and data were expressed as mean ±SD. # p< 0.05, significantly difference from DEC1-transfected cells and vector-transfected cells.
3.4. DEC1 is specifically recruited to the CCCTGC sequence in the CYP3A4 proximal promoter
DEC1 represses the CYP3A4 promoter and the DNA binding domain is required for DEC1 to regulate the CYP3A4 promoter, which suggests that DEC1 interact with certain sequence in the CYP3A4 promoter. Next, we located the sequence of the CYP3A4 promoter interacting with DEC1 by using sequential truncation and site directed mutagenesis of CYP3A4 promoter. As shown in Fig. 4B, DEC1 repressed both CYP3A4-DP-luc and CYP3A4-P-luc, which suggests that the DEC1 response element(s) is (are) located in CYP3A4-P-luc. So we prepared a series of 30 base pairs deletion mutants from −302 to −212 of CYP3A4-P-luc, 20 base pairs deletion mutants from −212 to −92 of CYP3A4-P-Luc and every 5 base pairs mutants from −92 to +4 of CYP3A4-P-luc. The diagrammatic presentation of reporters was shown in Fig. 4A. As shown in Fig. 4C, DEC1 repressed all the CYP3A4 proximal promoter deletion mutants and 5 base pairs mutants. However, the reduction of every 5 base pairs mutants from −52 to +4 of CYP3A4-P decreased, which suggested that DEC1 probably bound to this region (Fig. 4C).
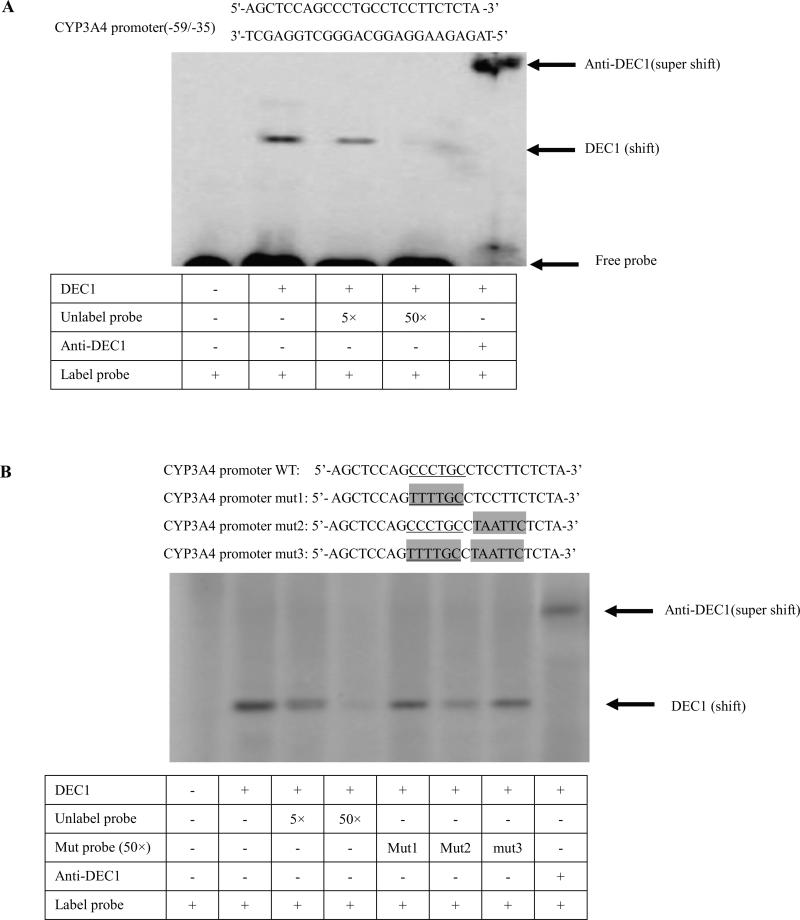
To validate the direct binding of the DEC1 to CYP3A4 proximal promoter, EMSA experiments were carried out in vitro with DEC1 and the three double-stranded oligonucleotides from −79 to +4 of CYP3A4 promoter. The data showed that the double-stranded oligonucleotides from −59 to −35 of the CYP3A4 promoter bound to DEC1 (Fig.5A), excessively unlabeled probe (5×, 50×) partially or completely competed with the binding band, and shifted band was completely supershifted by an anti-DEC1 antibody (Fig.5A). In order to test whether the probe (−59 to −35) in the CYP3A4 promoter is a specific sequence to bind to DEC1, three mutated probes (−59 to −35) were determined to compete with the binding band of DEC1, as well as wild type probe. As shown in Fig. 5B, excessive unlabeled wild type probe (50 ×) competed with the binding band, but excessive mutated probes containing CCCTGC sequence (mut1 and mut3) (50 ×) did not compete totally, while mut2 (50 ×) partially did (Fig.5B). The data suggest that DEC1 bound directly to CCCTGC sequence in the CYP3A4 proximal promoter.
Fig. 5. DEC1 binding to the CCCTGC sequence in the proximal promoter of CYP3A4 by EMSA.
(A) DEC1 binding to region (−59/−35) of CYP3A4 proximal promoter. (B) DEC1 binding to the CCCTGC sequence of CYP3A4 proximal promoter. HepG2 cells were plated in six-well plates and transfected with Flag-DEC1 or Flag CMV2 800 ng/well overnight. Nuclear extracts were prepared with a nuclear extraction kit (Active Motif). Nuclear proteins (10 μg) were incubated with 32P-labelled oligonucleotides (5'-AGCTCCAGCCCTGCCTCCTTCTCTA -3’) in a final volume of 10 μl containing 1 × DNA-binding buffer. For competition experiment, nuclear extracts were incubated with excess unlabelled wild type probe (5×, 50×) or each unlabelled mutated probe (mut1, 2, 3) (50×) and then mixed with the radioactive-belled probe. For supershift assays, an anti-DEC1 antibody was added after the nuclear extracts were incubated with the radioactive-labelled probe. The protein-DNA complexes were resolved on 6% polyacrylamide gel and visualized by Typhoon 8600 Variable Mode Imager. Three independent experiments were performed.
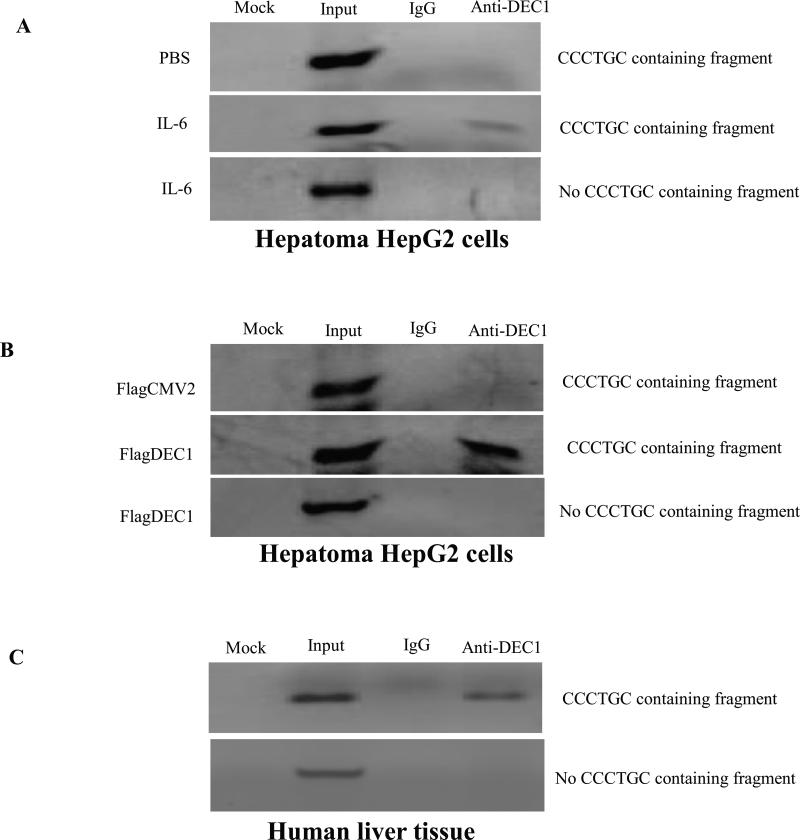
After demonstrating the specific binding of DEC1 to the CCCTGC sequence by EMSA, we next performed the ChIP experiments to determine whether DEC1 is specifically recruited to the CCCTGC sequence in the CYP3A4 promoter in vivo. Chromatins were prepared from HepG2 cells treated with IL-6 or transfected with DEC1 expressed plasmid and immunoprecipitated with anti-DEC1 antibodies. A primer set, pCYP3A4 (-253b) and pCYP3A4 (-35b), flanking the CCCTGC sequence, was used to detect the presence of CCCTGC -containing chromatin DNA. As shown in Fig. 6, a CCCTGC -containing PCR fragment was readily detected by using precipitated chromatins with antibodies against DEC1, whereas no PCR products were amplified in a sample precipitated with IgG antibodies, which indicates the specificity of the assay. To further validate this finding, a second set of primers, pCYP3A4 (−457b) and pCYP3A4 (−251b), locating 450 bp upstream of the CCCTGC sequence, was used as a negative control to amplify a 206 bp fragment. As shown in Fig. 6B, a PCR product (no CCCTGC-containing fragment) with the expected size was readily detected with input chromatin DNA. However, no PCR products were detected with chromatin DNA immunoprecipitated with IgG or DEC1 antibodies, which confirmed the specificity of the ChIP assay.
Fig.6. DEC1 being specifically recruited to the CCCTGC sequence in the CYP3A4 promoter by ChIP.
(A) HepG2 cells treated with IL-6. (B) HepG2 cells transfected with DEC1 expression construct. The chromatins with optimized size ranging from 400 to 800 bp were prepared as described in Materials and Methods. After immunoprecipitation with antibodies against DEC1 or IgG as a negative control, one set of PCR amplification was performed to specifically detect the presence of CCCTGC sequence-containing chromatin DNA. A negative control primer (no CCCTGC sequence containing fragment) set was included to amplify a 206 bp fragment 450 bp upstream of the CCCTGC sequence. PCR amplification was carried out in a final volume of 25 μl with 2 μl of eluted chromatin DNA, input DNA, or water (mock) as template with 40 cycles. PCR products were resolved on a 1.5% agarose gel. Three independent experiments were performed. (C) Chromatins were prepared from normal human liver tissues and immunoprecipitated with antibodies against DEC1 or IgG. The presence of CCCTGC sequence-containing chromatin DNA was detected by amplification using a set of primers flanking CCCTGC sequence. PCR amplification using the no CCCTGC sequence primers set was also performed as a control (n = 3).
To confirm the finding from HepG2 cells, we next performed ChIP assays using chromatins prepared from human liver. Consistent with the data from HepG2 cells, a CCCTGC-containing PCR fragment was robustly detected. While no signal was detected using negative control primer set or chromatins immunoprecipitated with IgG antibodies (Fig. 6C). Taken together, these data clearly demonstrated that DEC1 was specifically recruited to the CCCTGC sequence.
4. Discussion
It has been reported that drug biotransformation is impaired in patients with liver conditions such as hepatitis and cirrhosis [30,31]. In those conditions, as the production of various proinflammatory cytokines (e.g., IL-6) markedly increased [32,33], the metabolisms of many drugs decreased [34,35,36]. Our studies have shown that IL-6 suppresses the CYP3A4 expression at both mRNA and protein levels, and suppresses the CYP3A4 activity as well in human hepatocytes.
It is well-known that inflammation decreases the drug metabolism enzymes including CYP3A4, but the mechanism(s) of such action is (are) still controversial. Several studies proposed possible mechanisms which were limited to the increases of signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinases (MAPKs), or nuclear factor-κB (NF-κB), which directly impaired the action of nuclear receptors involved in the regulation of CYP3A4 [37]. For example, if the actions of PXR and constitutive androstane receptor (CAR), which are responsible for the induction of CYP3A4 by xenobiotics [35], are impaired, the expression of their target genes such as CYP3A4 will be influenced. Gu et al [38] found that lipopolysaccharide (LPS) and proinflammatory cytokines (TNFα) increased the level of NF-κB that forms heterodimers with retinoid X receptor-α (RXR-α). The formation of NF-κB - RXR-α heterodimer consumes the RXR-α to disrupt the combination of PXR and RXR-α and therefore represses the CYP3A4 transcription. Pregnane X receptor (PXR) was abundantly expressed in liver and intestine, which are predominately responsible for the entry and metabolism of chemicals [5,39]. Pregnane X receptor has been recognized as a key regulator that mediates the induction of many chemical elimination genes including CYP3A4 [39,40].
Some studies have reported that the decrease of the CYP3A4 enzyme expression is associated with the repression of CAR and PXR in mouse liver during the acute phase response [12] and in human hepatocytes [11]. However, other mechanisms may also contribute to the downregulation of CYP3A4 in inflammation because the much bigger decrease of CYP3A4 was compared to the decrease of PXR in response to cytokines such as IL-6 [11].
Differentiated embryo-chondrocyte expressed gene 1 (DEC1), an important transcription factor that has a basic helix-loop-helix domain and is ubiquitously expressed in both human embryonic and adult tissues, has a pivotal function in various biological phenomena, including neurogenesis, neuroregulation, chondrogenesis, cell growth, oncogenesis [19,41], immune balance and circadian rhythm. It has been paid an increasing attention for its role in maintaining the homeostasis of metabolism and energy [23,42].
Recently, it has been reported that DEC1 repressed the transactivation of RXR heterodimers, such as liver X receptor (LXR), farnesoid X receptor (FXR), vitamin D receptor (VDR), and retinoic acid receptor (RAR) [43]. One study showed that the hepatic DEC1 mediates the ligand-dependent LXR signal to regulate the expression of genes involved in the hepatic clock system and metabolism [44].
In the present study, we found that IL-6 rapidly induced the DEC1 expression and continuously reduced the CYP3A4 expression in primary cultured human hepatocytes. Meanwhile, the repression of CYP3A4 by IL-6 occurred after the increase of DEC1 in human hepatocytes (Fig. 1A, 1B). So we hypothesize that the decrease of CYP3A4 expression in primary human hepatocytes by IL-6 is due to the rapid increase of the DEC1 expression. In order to investigate the role of DEC1 in the downregulation of CYP3A4 in response to IL-6, experiments of both knockdown and overexpression of DEC1 gene were conducted to selectively modulate the expression of DEC1.
Our results also indicated that DEC1 knockdown alone significantly increased the CYP3A4 expression and enzymatic activity (Fig. 2B, 2C). Furthermore, IL-6 significantly decreased CYP3A4 expression and enzymatic activity in the cells transfected with the vector, but, not in the cells transfected with the DEC1 shRNA construct (Fig. 2B, 2C). IL-6 significantly increased DEC1 expression in the cells transfected with vector, but not in the cells transfected DEC1 shRNA construct (Fig. 2A, 2C). The DEC1 knockdown efficiency was at least more than 60% in each experiment (Fig. 2A, 2C). The results indicate that the DEC1 knockdown can partially abolish the decrease of CYP3A4 expression and its enzymatic activity induced by IL-6 in hepatocytes.
Contrary to the DEC1 knockdown experiment, the overexpression of DEC1 significantly decreased CPY3A4 expression and its enzymatic activity (Fig. 2D, 2E). Moreover, DEC1 decreased the CYP3A4 expression and its enzymatic activity in a dose-dependent manner (Fig. 2D, 2E). The data suggest that DEC1 directly down regulates the expression of CYP3A4 in hepatocytes. The decrease of CYP3A4 mRNA suggests two possibilities: (1) DEC1 suppresses the transcription and/or (2) DEC1 increases the degradation of mRNA.
We have presented evidences to support the first possibility. First, the CYP3A4 promoter reporters were repressed by transfection with DEC1 in a dose-dependent manner in HepG2 cells (Fig. 3A and 4B). Furthermore, the DNA-binding domain is required for DEC1 to repress the CYP3A4 promoter (Fig. 3C) activated by Rif. On the other hand, the repression of DEC1 decreased as the C-terminal deletion increased, such as FlagDEC11-347, FlagDEC11-270 (Fig. 3B, 3C). Moreover, FlagDEC11-197 presented no repression of CYP3A4 reporter, whereas FlagDEC11-150, which has no ligand binding domain but has DNA binding domain, markedly transactivated the CYP3A4 reporter activity by two fold (Fig.3C) activated by Rif. FlagDEC11-150, with only DNA binding domain but without ligand binding domain, combined with target genes but had no effect and then prevent from the action of endogenous DEC1. FlagDEC11-150 possibly acted as an antagonist of DEC1 (Fig.3C). More importantly, the repression of the promoter mediated by DEC1 was comparable with the extent of the decreased mRNA (Fig. 2D, 3A and 4B), which provided direct evidence to confirm that the transcriptional repression is responsible for the decrease of the CYP3A4 expression. Second, we have located a genomic sequence in CYP3A4 gene to response to DEC1. We studied the CYP3A4 gene promoter transcriptional repression via reporter assay. DEC1 repressed all the CYP3A4 proximal promoter deletion mutants and 5 base pairs mutants. However, the reduction of every 5 base pairs mutants from −52 to +4 of CYP3A4-P decreased, which suggests that DEC1 binds to this region (Fig. 4C). To examine the direct binding of DEC1 to CYP3A4 proximal promoter, EMSA experiments were carried out. We found that the double-stranded oligonucleotides from −59 to −35 of the CYP3A4 promoter bound to DEC1 (Fig.5A), the excessively unlabeled probe (5×, 50×) partially or completely competed with the radioactive-labelled probe (weak band or no band), and the shifted band was completely supershifted by an anti-DEC1 antibody (Fig. 5A). The data suggest that DEC1 binds to the region from −59 to −35 in the CYP3A4 promoter.
Many studies have demonstrated that DEC1 negatively regulated target genes through binding E-box (CACGTG) element in their promoters [19,45,46,47]. While other studies reported that DEC1 regulated target genes such as survivin and mPer1 through binding to the Sp1 site in promoter [48,49]. In the region (−59 to −35) of the CYP3A4 promoter, there is no E-box but a similar Sp1 sequence (CCCTGC). Next, three mutated probes (Fig.5B) were tested to compete with wild type probe. As a result, the excessive mutated CCCTGC probes (mut1 and mut3) did not compete with the radioactive-labelled probe totally (presented bands), while the excessive mutated 6 base pairs next to CCCTGC probe (mut2) partially did that (presented weak band) (Fig. 5B). The data suggest that DEC1 binds to the CCCTGC sequence to form DNA-protein binding complex, which downregulates the CYP3A4 promoter.
Alternatively, Sp1 interacts with other proteins and the resultant complexes collectively determine DNA binding selectivity. In support of this possibility mSHARP, a DNA related mouse protein has been shown to interact with Sp1 [50]. In this study, we have demonstrated that anti-DEC1 antibody completely disrupts the shifted bands with wild type probe including CCCTGC sequence (Fig.5A, B), which suggests that DEC1 is part of the CCCTGC-DEC1 complex and this complex represses the promoter of CYP3A4. This finding is also strongly confirmed by ChIP assays (Fig. 6A, 6B, 6C).
In summary, our work points out several important conclusions. First, DEC1 is induced rapidly by IL-6. Potentially, DEC1 is an inducible protein during acute-phase inflammation. Second, the repression of CYP3A4 by IL-6 is achieved through increasing theDEC1 expression in human hepatocytes, and the increased DEC1 binds to the CCCTGC sequence in the CYP3A4 promoter to form CCCTGC-DEC1 complex which downregulates the activity of the CYP3A4 promoter. Third, DEC1 transcriptionally downregulates the expression of CYP3A4 in response to IL-6, which proves, for the first time, that DEC1 has been implicated in metabolizing xenobiotics including the vast majority of clinically used drugs, environmental procarcinogens and toxins.
Acknowledgement
This work is supported by the Natural Science Foundation of China (No. 30772616, 81173128), the National Key Basic Research Program of China (No.2009CB521906), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the National Institutes of Health of USA (USA, F05AT003019).
Abbreviations
- DEC1
differentially expressed in chondrocytes 1
- IL-6
interleukin-6
- CYP450
cytochrome P450
- PXR
pregnane X receptor
- hPXR
human pregnane X receptor
- Rif
rifampicin
- TNF-α
tumor necrosis factor α
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PCR
polymerase chain reaction
- DMEM
Dulbecco's modified Eagle's medium
- PBS
phosphate-buffered saline
- bHLH
basic helix-loop-hellix
- HLH
helix-loop-hellix
- SHARP
split and hairy related protein
- STRA13
stimulated with retinoic acid 13
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
REFERENCE
- 1.Parkinson A. Biotransformation of xenobiotics, in The Casarett and Doull's Toxicology: The Basic Science of Poisons. McGraw-Hill; New York: 2001. pp. 139–62. [Google Scholar]
- 2.Poso A, Honkakoski P. Ligand recognition by drug-activated nuclear receptors PXR and CAR: structural, site-directed mutagenesis and molecular modeling studies. Mini Rev Med Chem. 2006;6:937–47. doi: 10.2174/138955706777935008. [DOI] [PubMed] [Google Scholar]
- 3.Cooper BW, Cho TM, Thompson PM, Wallace AD. Phthalate induction of CYP3A4 is dependent on glucocorticoid regulation of PXR expression. Toxicol Sci. 2008;103:268–77. doi: 10.1093/toxsci/kfn047. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shou M, Hayashi M, Pan Y, Xu Y, Morrissey K, Xu L, et al. Modeling, prediction, and in vitro in vivo correlation of CYP3A4 induction. Drug Metab Dispos. 2008;36:2355–70. doi: 10.1124/dmd.108.020602. [DOI] [PubMed] [Google Scholar]
- 7.Charles KA, Rivory LP, Brown SL, Liddle C, Clarke SJ, Robertson GR. Transcriptional repression of hepatic cytochrome P450 3A4 gene in the presence of cancer. Clin Cancer Res. 2006;12:7492–7. doi: 10.1158/1078-0432.CCR-06-0023. [DOI] [PubMed] [Google Scholar]
- 8.Neuhaus J, Jacobs DR, Jr., Baker JV, Calmy A, Duprez D, La Rosa A, et al. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J Infect Dis. 2010;201:1788–95. doi: 10.1086/652749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frye RF, Schneider VM, Frye CS, Feldman AM. Plasma levels of TNF-alpha and IL-6 are inversely related to cytochrome P450-dependent drug metabolism in patients with congestive heart failure. J Card Fail. 2002;8:315–9. doi: 10.1054/jcaf.2002.127773. [DOI] [PubMed] [Google Scholar]
- 10.Prandota J. Important role of proinflammatory cytokines/other endogenous substances in drug-induced hepatotoxicity: depression of drug metabolism during infections/inflammation states, and genetic polymorphisms of drug-metabolizing enzymes/cytokines may markedly contribute to this pathology. Am J Ther. 2005;12:254–61. [PubMed] [Google Scholar]
- 11.Yang J, Hao C, Yang D, Shi D, Song X, Luan X, et al. Pregnane X receptor is required for interleukin-6-mediated down-regulation of cytochrome P450 3A4 in human hepatocytes. Toxicol Lett. 2010;197:219–26. doi: 10.1016/j.toxlet.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beigneux AP, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Reduction in cytochrome P-450 enzyme expression is associated with repression of CAR (constitutive androstane receptor) and PXR (pregnane X receptor) in mouse liver during the acute phase response. Biochem Biophys Res Commun. 2002;293:145–9. doi: 10.1016/S0006-291X(02)00196-1. [DOI] [PubMed] [Google Scholar]
- 13.Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052–65. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto K, Shen M, Noshiro M, Matsubara K, Shingu S, Honda K, et al. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem Biophys Res Commun. 2001;280:164–71. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- 15.Rossner MJ, Dorr J, Gass P, Schwab MH. Nave KA SHARPs: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci. 1997;10:460–75. [PubMed] [Google Scholar]
- 16.Cronmiller C, Schedl P, Cline TW. Molecular characterization of daughterless, a Drosophila sex determination gene with multiple roles in development. Genes Dev. 1988;2:1666–76. doi: 10.1101/gad.2.12a.1666. [DOI] [PubMed] [Google Scholar]
- 17.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–40. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, Nurieva R, et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol. 2009;10:1260–6. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Xie M, Song X, Gragen S, Sachdeva K, Wan Y, et al. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J Biol Chem. 2003;278:16899–907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 20.Shen M, Yoshida E, Yan W, Kawamoto T, Suardita K, Koyano Y, et al. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem. 2002;277:50112–20. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol. 2001;2:1040–7. doi: 10.1038/ni721. [DOI] [PubMed] [Google Scholar]
- 22.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–4. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 23.Iizuka K, Horikawa Y. Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem Biophys Res Commun. 2008;374:95–100. doi: 10.1016/j.bbrc.2008.06.101. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova AV, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J Mol Biol. 2004;340:641–53. doi: 10.1016/j.jmb.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277:47014–21. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 26.Bu HZ. A literature review of enzyme kinetic parameters for CYP3A4-mediated metabolic reactions of 113 drugs in human liver microsomes: structure-kinetics relationship assessment. Curr Drug Metab. 2006;7:231–49. doi: 10.2174/138920006776359329. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Yan B. Photochemotherapeutic agent 8-methoxypsoralen induces cytochrome P450 3A4 and carboxylesterase HCE2: evidence on an involvement of the pregnane X receptor. Toxicol Sci. 2007;95:13–22. doi: 10.1093/toxsci/kfl120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Li Y, Liu J, Mukundan M, Yan B. Simultaneous substitution of phenylalanine-305 and aspartate-318 of rat pregnane X receptor with the corresponding human residues abolishes the ability to transactivate the CYP3A23 promoter. J Pharmacol Exp Ther. 2005;312:571–82. doi: 10.1124/jpet.104.074971. [DOI] [PubMed] [Google Scholar]
- 29.Lindley C, Hamilton G, McCune JS, Faucette S, Shord SS, Hawke RL, et al. The effect of cyclophosphamide with and without dexamethasone on cytochrome P450 3A4 and 2B6 in human hepatocytes. Drug Metab Dispos. 2002;30:814–22. doi: 10.1124/dmd.30.7.814. [DOI] [PubMed] [Google Scholar]
- 30.Qu ZQ, Li XD, Liu HL, He P, Zhang X, Wu MC. Impaired clearance of phenacetin in hepatic cirrhosis and fibrosis. Int J Clin Pharmacol Ther. 2007;45:55–62. doi: 10.5414/cpp45055. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya M, Echizen H, Kubo S, Tamura N, Suzuki K, Ushiama H, et al. Reduced urinary 6beta-hydroxycortisol to cortisol ratios in patients with liver cirrhosis. Hepatol Res. 2003;26:28–33. doi: 10.1016/s1386-6346(03)00005-6. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson AS, Gretzer C, Wallerstedt S. Elevation of cytokines in peritoneal fluid and blood in patients with liver cirrhosis. Hepatogastroenterology. 2004;51:505–9. [PubMed] [Google Scholar]
- 33.Zhang W, Yue B, Wang GQ, Lu SL. Serum and ascites levels of macrophage migration inhibitory factor, TNF-alpha and IL-6 in patients with chronic virus hepatitis B and hepatitis cirrhosis. Hepatobiliary Pancreat Dis Int. 2002;1:577–80. [PubMed] [Google Scholar]
- 34.Aitken AE, Morgan ET. Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab Dispos. 2007;35:1687–93. doi: 10.1124/dmd.107.015511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jover R, Bort R, Gomez-Lechon MJ, Castell JV. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB J. 2002;16:1799–801. doi: 10.1096/fj.02-0195fje. [DOI] [PubMed] [Google Scholar]
- 36.Thorn M, Finnstrom N, Lundgren S, Rane A, Loof L. Expression of cytochrome P450 and MDR1 in patients with proctitis. Ups J Med Sci. 2007;112:303–12. doi: 10.3109/2000-1967-203. [DOI] [PubMed] [Google Scholar]
- 37.Robertson GR, Liddle C, Clarke SJ. Inflammation and altered drug clearance in cancer: transcriptional repression of a human CYP3A4 transgene in tumor-bearing mice. Clin Pharmacol Ther. 2008;83:894–7. doi: 10.1038/clpt.2008.55. [DOI] [PubMed] [Google Scholar]
- 38.Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, et al. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem. 2006;281:17882–9. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- 39.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 40.Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res. 2002;43:359–64. [PubMed] [Google Scholar]
- 41.Zheng Y, Jia Y, Wang Y, Wang M, Li B, Shi X, et al. The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in gastric cancer. OMICS. 2009;13:301–6. doi: 10.1089/omi.2009.0014. [DOI] [PubMed] [Google Scholar]
- 42.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–70. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 43.Cho Y, Noshiro M, Choi M, Morita K, Kawamoto T, Fujimoto K, et al. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol Pharmacol. 2009;76:1360–9. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
- 44.Noshiro M, Usui E, Kawamoto T, Sato F, Nakashima A, Ueshima T, et al. Liver X receptors (LXRalpha and LXRbeta) are potent regulators for hepatic Dec1 expression. Genes Cells. 2009;14:29–40. doi: 10.1111/j.1365-2443.2008.01247.x. [DOI] [PubMed] [Google Scholar]
- 45.Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J. 2004;382:43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawamoto T, Noshiro M, Sato F, Maemura K, Takeda N, Nagai R, et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem Biophys Res Commun. 2004;313:117–24. doi: 10.1016/j.bbrc.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 47.Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol. 2008;28:4080–92. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Song X, Ma Y, Liu J, Yang D, Yan B. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem J. 2004;382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Xie M, Yang J, Yang D, Deng R, Wan Y, et al. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene. 2006;25:3296–306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azmi S, Sun H, Ozog A, Taneja R. mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression. J Biol Chem. 2003;278:20098–109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]