Abstract
Aim:
Histone lysine demethylases (KDMs) control the lineage commitments of stem cells. However, the KDMs involved in the determination of the cardiomyogenic lineage are not fully defined. The aim of this study was to investigate the expression profiles of KDMs during the cardiac differentiation of mouse embryonic stem cells (mESCs).
Methods:
An in vitro cardiac differentiation system of mESCs with Brachyury (a mesodermal specific marker) and Flk-1+/Cxcr4+ (dual cell surface biomarkers) selection was used. The expression profiles of KDMs during differentiation were analyzed using Q-PCR. To understand the contributions of KDMs to cardiomyogenesis, the mESCs on differentiation d 3.5 were sorted by FACS into Brachyury+ cells and Brachyury− cells, and mESCs on d 5.5 were sorted into Flk-1+/Cxcr4+ and Flk-1−/Cxcr4− cells.
Results:
mESCs were differentiated into spontaneously beating cardiomyocytes that were visible in embryoid bodies (EBs) on d 7. On d 12–14, all EBs developed spontaneously beating cardiomyocytes. Among the 16 KDMs examined, the expression levels of Phf8, Jarid1a, Jhdm1d, Utx, and Jmjd3 were increased by nearly 2–6-fold on d 14 compared with those on d 0. Brachyury+ cells showed higher expression levels of Jmjd3, Jmjd2a and Jhdm1d than Brachyury− cells. A higher level of Jmjd3 was detected in Flk-1+/Cxcr4+ cells, whereas the level of Jmjd2c was lower in both Brachyury+ cells and Flk-1+/Cxcr4+ cells.
Conclusion:
KDMs may play important roles during cardiomyogenesis of mESCs. Our results provide a clue for further exploring the roles of KDMs in the cardiac lineage commitment of mESCs and the potential interference of cardiomyogenesis.
Keywords: embryonic stem cell, cardiac differentiation, cardiomyocyte, histone lysine demethylase, expression profile, Brachyury, Flk-1+/Cxcr4+
Introduction
Embryonic stem cells (ESCs) are capable of differentiating into three germ layers, ie, ectoderm, endoderm, and mesoderm, and their derivatives1,2. This differentiation mimics the early stages of embryonic development, which includes the cardiomyogenic lineage commi tment3. During cardiac differentiation, ESCs silence pluripotency genes and acquire the phenotype of cardiomyocytes by activating cardiac-specific genes4,5. This is achieved by intensive cross talk between the transcription factor networks and epigenetic modulators that occurs through the regulation of the chromatin structure of the packaged eukaryotic genome6,7. Therefore, understanding the regulatory mechanisms underlying cardiac lineage commitment would enable better control of ESC differentiation, which is required for therapy and drug development.
Increasing evidence suggests that post-translational modifications of histone proteins represent essential epigenetic control mechanisms in gene expression programs and cell fate decisions of ESCs, including cardiac lineage determination8,9. Methylation/demethylation is an important histone modification that is mediated by histone methyltransferases/histone demethylases, which are emerging as important players in biological processes10,11,12. H3K4 trimethylation (H3K4me3) is found in euchromatic regions and has transcriptional activity, whereas H3K27 trimethylation (H3K27me3) is associated with transcriptional repression13. In undifferentiated ESCs, a subset of developmental genes exhibit a “poised status”, which is marked by H3K27me3 and H3K4me314,15. After differentiation, most of the marks are removed15, implying that the enzymes responsible for demethylation may play a role in ESC differentiation. It has been shown that histone lysine demethylases (KDMs) play an important role in the self-renewal of ESCs16,17,18 and in cardiac differentiation19,20. However, the specific KDMs that control the differentiation of ESCs are largely unknown.
Cardiomyocyte differentiation involves the specification of pluripotent cells to mesodermal and cardiac progenitors prior to terminal differentiation5,6. In contrast to the relatively advanced knowledge of signaling pathways6,21, the epigenetic regulators, especially KDMs, that control cardiac differentiation are largely unknown. Recent studies have revealed that the Kdm6 family members Utx and Jmjd3 play important roles in the cardiovascular lineage specification of ESCs through different mechanisms. Jmjd3 reduces the H3K27me3 marks at the Brachyury promoter and facilitates the recruitment of b-catenin, which is critical for Wnt signal-induced mesoderm differentiation and cardiovascular lineage commitment19. Unlike Jmjd3, Utx controls mesoderm differentiation and Brachyury expression independent of H3K27 demethylase activity22; Utx also associates with core cardiac transcription factors and demethylates H3K27 residues in cardiac-specific genes20. However, other KDMs that are critical for the early lineage specification and subsequently cardiac differentiation of ESCs need to be further explored.
In the present study, we performed a stage-based screening of mRNA expression profiles for KDMs during the cardiomyocyte differentiation of mESCs. Our results revealed various expression patterns of KDMs during cardiac lineage formation, and we identified several KDMs as being potentially essential for mESC cardiac differentiation. Our findings provide new insight into the role of KDMs in the regulation of cardiac lineage commitment and may provide information for the further development of cell therapy23 and drug discovery24.
Materials and methods
Culture and in vitro differentiation of mESCs
SCR012 mESCs and Brachyury-green fluorescent protein (Brachyury-GFP) E14 mESCs25 were cultivated and differentiated into spontaneously beating cardiomyocytes as previously described26,27. Briefly, undifferentiated mESCs were cultivated on mitomycin C-inactivated mouse feeder layers (MEFs) in the presence of leukemia inhibitory factor (LIF, 1000 U/mL; Millipore, Billerica, MA, USA). The differentiation of mESCs into cardiac cells was initiated by a hanging drop technique to form EBs28, and after 6 d in suspension, EBs were plated onto gelatin-coated 48-well tissue culture dishes for cardiac differentiation. All cultivation medium and other reagents for cell culture were from Invitrogen (Carlsbad, CA, USA) unless indicated otherwise.
Reverse transcription (RT)-PCR
Total RNA was extracted from cells and purified using the RNeasy Mini kit (QIAGEN, Valencia, CA, USA) and transcribed into cDNA using ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) and oligo(dT) primers. The PCR primers are listed in Table 1. Samples were amplified in the linear range by PCR. The PCR products were size-fractionated on 1%–1.5% agarose gels containing ethidium bromide.
Table 1. Primer sequences for RT-PCR.
| Gene | Primer sequence (5′–3′) | AT (°C) | PS (bp) |
|---|---|---|---|
| Brachyury | F-CCACCGCTGGAAATATGTG R-CAGCTATGAACTGGGTCTCG | 57 | 286 |
| Nkx2.5 | F-GCCAACAGCAACTTCGTGA R-CCGGTCCTAGTGTGGAATC | 59 | 351 |
| Tbx5 | F-GGAGCCTGATTCCAAAGACA R-TTCAGCCACAGTTCACGTTC | 59 | 153 |
| Isl1 | F-TCATCCGAGTGTGGTTTCAA R-CCTCATGTCTCTCCGGACT | 59 | 154 |
| Mef2c | F-AGATACCCACAACACACCACGCGCC R-CATTATCCTTCAGAGAGTCGCATGCGCTT | 59 | 195 |
| m28s | F-AGCAGCCGACTTAGAACTGG R-TAGGGACAGTGGGAATCTCG | 55 | 150 |
F, forward; R, reverse; AT, annealing temperature; PS, product size.
Quantitative RT-PCR (Q-PCR)
Total RNA was extracted from cells and analyzed by kinetic real-time PCR using the ABI PRISM 7900 system (Applied Biosystems, Foster City, CA, USA) with SYBR Green Realtime PCR Master Mix plus (Toyobo, Osaka, Japan) for the relative quantification of the indicated genes. The transcript of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for internal normalization. The Q-PCR primers used for the KDMs are listed in Table 2, and other primers are listed in Table 3.
Table 2. Primer sequences of KDMs for Q-PCR.
| Gene | Primer sequence (5′–3′) | AT (°C) | PS (bp) |
|---|---|---|---|
| Jarid1a/Kdm5a | F-CACAGACCCGCTGAGTTTTAT R-CTTCACAGGCAAATGGAGGTT | 59 | 107 |
| Jarid1b/Kdm5b | F-CGCTTGCTGCACCGTTATT R-TACATCAGCCTTAGAAGCCATTTTG | 60 | 72 |
| Jarid1c/Kdm5c | F-TGAACGCATTGTTTATCCCTATGA R-TCAAATGGGCGTGTGTTACAC | 59 | 75 |
| Jarid1d/Kdm5d | F-TCCATTCGCCCTTTAATATCGA R-CGTCAGATCCTGGCTTCATGT | 60 | 72 |
| Jhdm1a/Kdm2a | F-GCTGTTCCTGGTCTGCAGTATCT R-TTCCTACTGCCCACCGAAGA | 60 | 84 |
| Jhdm1b/Kdm2b | F-GATGCTGAGCGGTATCATCCG R-GAGACAGCGATCCATGAGCAG | 57 | 147 |
| Jhdm1d/Kdm7a | F-GGCAAACCAGTTAAATCTCAGGG R-AGGTTAGAAGGAGTTCGGACAT | 59 | 137 |
| Jhdm2a/Kdm3a | F-GCCTTCCAGGTTTGATGATCTG R-TGCCATCTCGCCTGGTGTA | 60 | 65 |
| Jmjd1b/Kdm3b | F-TCAATATCCACTGGGTCTGTCG R-TGCCCCTTTGCACACTTCAG | 60 | 142 |
| Jmjd2a/Kdm4a | F-CCACCGAGACCTTCTACGAAGT R-TACCACTATGTCCTCAGGATAAAGGTT | 61 | 77 |
| Jmjd2b/Kdm4b | F-AGGGACTTCAACAGATATGTGGC R-GATGTCATCATACGTCTGCCG | 60 | 114 |
| Jmjd2c/Kdm4c | F-GGCGCCATCCACTTGGT R-CGCTACCTGCTGCTTCACAA | 60 | 65 |
| Jmjd3/Kdm6b | F-CCCCCATTTCAGCTGACTAA R-CTGGACCAAGGGGTGTGTT | 59 | 199 |
| Lsd1/Kdm1a | F-TGGGACCCAAGTGTCAATTTG R-AGAGCTCACCCCTGCTAGCA | 60 | 64 |
| Phf8 | F-CGCCCAACAAATGCTAATCT R-AGAAGTTCCCTCCGAATGCT | 59 | 196 |
| Utx/Kdm6a | F-ATCCCAGCTCAGCAGAAGTT R-GGAGGAAAGAAAGCATCACG | 59 | 196 |
F, forward; R, reverse; AT, annealing temperature; PS, product size.
Table 3. Primer sequences for Q-PCR.
| Gene | Primer sequence (5′–3′) | AT (°C) | PS (bp) |
|---|---|---|---|
| Oct-4 | F-TCAGCTTGGGCTAGAGAAGG R-TGACGGGAACAGAGGGAAAG | 59 | 254 |
| Rex-1 | F-AGCAGGATCGCCTCACTG R-GGCCACTTGTCTTTGCCG | 60 | 189 |
| α-MHC | F-GCGCATCAAGGAGCTCAC R-GGAACTTGGACAGGTTGGTG | 59 | 155 |
| Ncx1 | F-CAGCTTCCAAAACTGAAATCGA R-GTCCCTCTCATCGACTTCCAAAA | 60 | 101 |
| GAPDH | F-GTGGCAAAGTGGAGATTGTTG R-CTCCTGGAAGATGGTGATGG | 60 | 164 |
F, forward; R, reverse; AT, annealing temperature; PS, product size.
Immunocytochemical staining
EBs at d 14 were fixed with 4% paraformaldehyde for 30 min and permeabilized in 0.1% Triton X-100 (Sigma, St Louis, MO, USA) for 30 min at room temperature as described previously29. After a single wash with PBS, cells were blocked in 10% normal goat serum at room temperature for 1 h and then incubated with primary antibody against a-actinin (1:300; Sigma, St Louis, MO, USA) at 4 °C overnight, followed by the application of DyLight 549-conjugated secondary antibodies (1:1000; Jackson Lab, West Grove, PA, USA). Nuclei were stained with Hoechst 33258 dye (1:2000; Sigma, St Louis, MO, USA). A Nikon TI 2000 fluorescence microscope (Nikon, Kyoto, Japan) was used to view cells and acquire images.
Fluorescence activated cell sorting (FACS)
EBs derived from Brachyury-GFP mESCs were trypsinized, made into a single cell suspension, and sorted by FACS (FACStar Plus Flow Cytometer; BD, San Diego, CA, USA). Differentiating mESCs at d 3.5 were washed with PBS once and sorted into high GFP (Brachyury+) and low GFP (Brachyury−) populations. Differentiating mESCs at d 5.5 were washed with 4% fetal bovine serum (FBS) containing PBS once and resuspended in PBS containing an antibody to Flk-1-APC (1:200; eBioscience, San Diego, CA, USA) and Cxcr4-PE (1:200; eBioscience, San Diego, CA, USA); the mESCs were then incubated at 4 °C with gentle rotation for 45 min. The cells were then washed with PBS and sorted into Flk-1+/Cxcr4+ cells and Flk-1−/Cxcr4− cells.
Statistical analysis
The data are expressed as the mean±SEM. Statistical significance of differences was determined by Student's t-test. In all analyses, P<0.05 indicated statistical significance.
Results
Differentiation of mESCs into spontaneous beating cardiomyocytes
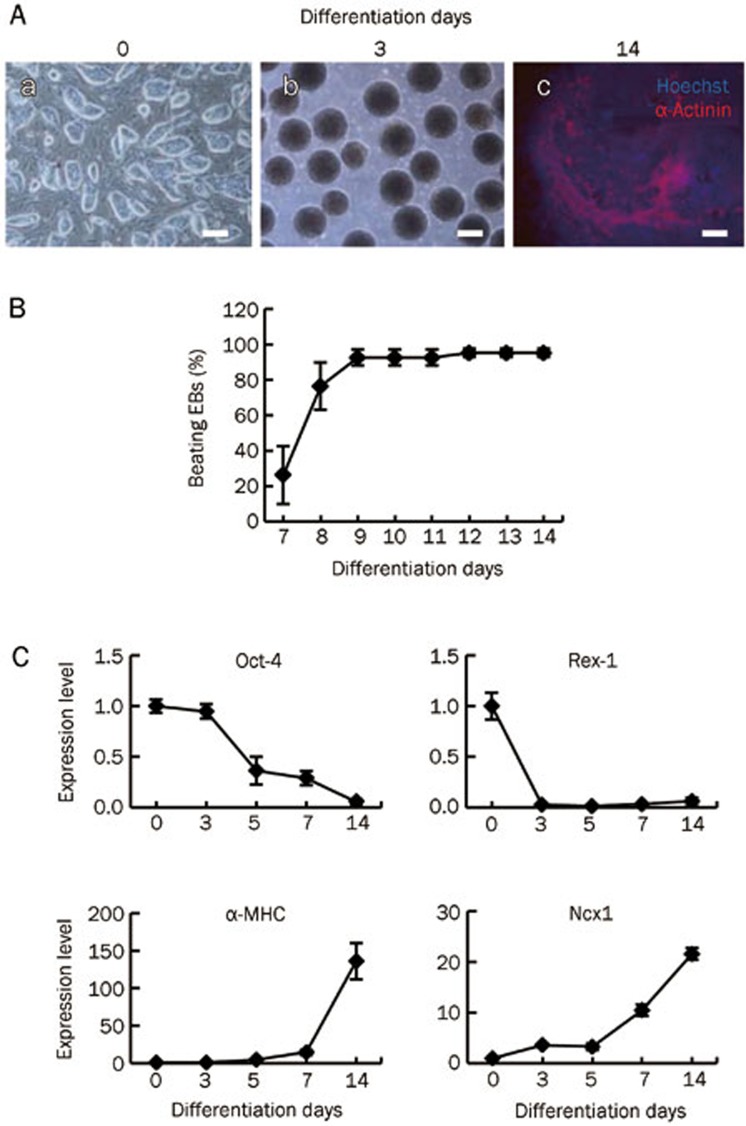
To screen KDMs that might be involved in cardiac lineage commitment, mESCs were differentiated into cardiomyocytes by the formation of EBs (Figure 1A) as reported previously4,27. After 6 d in suspension, EBs differentiated into spontaneously beating cardiomyocytes that were visible at differentiation d 7 (Figure 1B). The percentage of beating cardiomyocytes in EBs increased gradually over time and reached 84%–100% at d 9 and 100% at d 12; thereafter, the percentage remained stable until differentiation d 14 (Figure 1B). Cardiac differentiation was confirmed by immunocytochemical staining showing the expression of the cardiac specific marker a-actinin (Figure 1A). This was consistent with the Q-PCR analysis, which showed that the expression of pluripotent marker genes POU domain class 5 transcription factor 1 (Oct-4) and zinc finger protein 42 (Rex-1) gradually decreased during mESC differentiation, while the cardiac cytoskeletal proteins myosin heavy chain (a-MHC) and sodium/calcium exchanger 1 (Ncx1) were upregulated at d 7 and had increased significantly at d 14 (Figure 1C) in patterns that correlated with the onset of spontaneous beating (Figure 1B).
Figure 1.
Differentiation of mESCs into spontaneous beating cardiomyocytes in vitro. (A) mESCs during differentiation d 0 to d 14. (a) Phase contrast image showed undifferentiated mESCs colonies on mitomycin C-inactivated MEFs. (b) Phage contrast image of d 3 EBs suspend in differentiation medium. (c) Immunostaining analysis of α-sarcomeric actinin in d 14 EBs. Bar=200 μm. (B) Differentiation profile of cardiomyocytes during EB outgrowth. EBs were plated on d 6 and checked for beating clusters during differentiation d 7 to d 14. (C) Quantitative expression analysis of pluripotency markers Oct-4 and Rex-1; cardiac specific markers α-MHC and Ncx1 during mESCs differentiation. Ct values of Q-PCR analysis were normalized against GADPH. Data are represented as mean±SEM from at least three independent experiments.
Expression profiles of KDMs during cardiac differentiation
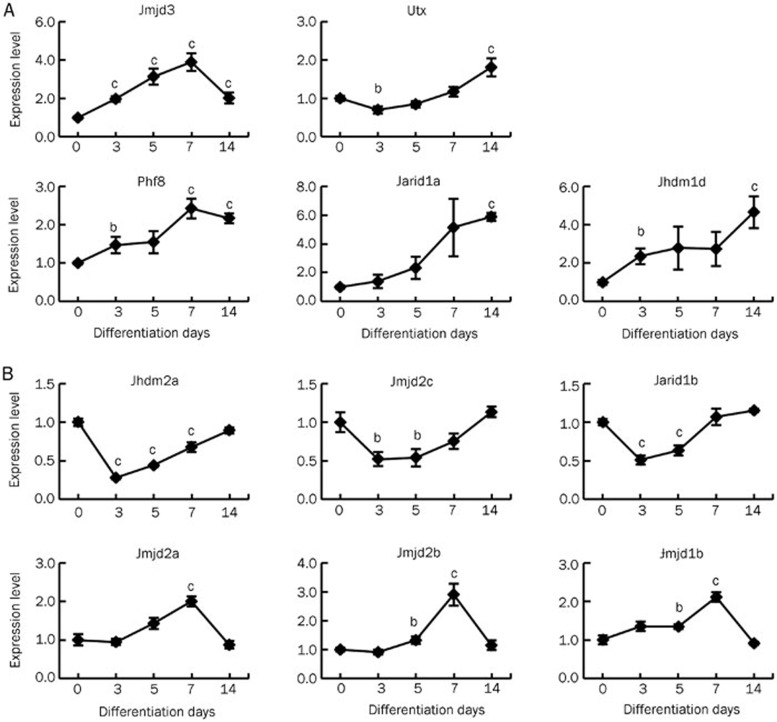
Next, we analyzed the expression profiles of KDMs that might be involved in cardiac lineage commitment by using Q-PCR to compare the expression levels in undifferentiated mESCs and differentiating mESCs at d 3, 5, 7 and 14. Different expression patterns were observed for the 16 KDMs analyzed (Table 4). Five genes (Phf8, Jarid1a, Jhdm1d, Utx, and Jmjd3) were upregulated at least 1.8-fold at differentiation d 14 compared with undifferentiated mESCs (d 0) with different patterns (Figure 2A, Table 4). The expression of Phf8, Jarid1a and Jhdm1d continued to increase during the differentiation process and was highest at differentiation d 14 (Figure 2A). Jmjd3 expression peaked at d 7 and then declined at d 14, but it was still higher than that observed at d 0 (Figure 2A). Utx decreased slightly at an earlier differentiation stage (d 3) and was upregulated later (Figure 2A). The expression levels of six other KDM genes were similar between differentiation d 0 and d 14; however, of these genes, the expression levels of Jhdm2a, Jmjd2c, and Jarid1b decreased by more than 50% at d 3 and then gradually returned to the levels observed in the undifferentiated mESCs (Figure 2B, upper panels). In contrast, the expression levels of Jmjd2a, Jmjd2b, and Jmjd1b increased gradually and peaked at d 7, after which they declined to the levels observed in the undifferentiated ESCs (Figure 2B, lower panels). The remaining five KDM genes (Lsd1, Jhdm1a, Jhdm1b, Jarid1c, and Jarid1d) had no significant changes in their expression levels during the differentiation time examined (P>0.05, n=3–4 each. Differentiation d 14 data in Table 4, others not shown). These data suggest that the KDMs may play different roles in mESC differentiation.
Table 4. List of screened KDMs during cardiac differentiation of mESCs.
| Gene | Histone substrates | Fold changes (d 14 vs d 0) |
|---|---|---|
| Lsd1/Kdm1a | H3K4me1, H3K4me2, H3K9me1, H3K9me2 | 1.6±0.26 |
| Jhdm1a/Kdm2a | H3K36me1, H3K36me2 | 1.22±0.23 |
| Jhdm1b/Kdm2b | H3K36me1, H3K36me2, H3K4me3 | 1.13±0.05 |
| Jhdm2a/Kdm3a | H3K9me1, H3K9me2 | 0.9±0.05 |
| Jmjd1b/Kdm3b | H3K9me1, H3K9me2 | 0.95±0.16 |
| Jmjd2a/Kdm4a | H3K9me2, H3K9me3, H3K36me2, H3K36me3 | 0.88±0.05 |
| Jmjd2b/Kdm4b | H3K9me2, H3K9me3, H3K36me2, H3K36me3 | 1.17±0.25 |
| Jmjd2c/Kdm4c | H3K9me2, H3K9me3, H3K36me2, H3K36me3 | 1.20±0.24 |
| Jarid1a/Kdm5a | H3K4me2, H3K4me3 | 6.13±1.01c |
| Jarid1b/Kdm5b | H3K4me2, H3K4me3 | 1.16±0.08 |
| Jarid1c/Kdm5c | H3K4me2, H3K4me3 | 1.4±0.23 |
| Jarid1d/Kdm5d | H3K4me2, H3K4me3 | 0.60±0.29 |
| Utx/Kdm6a | H3K27me2, H3k27me3 | 1.81±0.21c |
| Jmjd3/Kdm6b | H3K27me2, H3k27me3 | 2±0.06c |
| Jhdm1d/Kdm7a | H3K9me1, H3K9me2, H3K27me1, H3K27me2 | 5.06±1.39c |
| Phf8 | H3K9me1, H3K9me2, H4K20me1 | 2.16±0.14c |
Data are represented as mean±SEM from at least three independent experiments.
cP<0.01 compared with d 0 values.
Figure 2.
Quantitative expression analysis of KDMs during cardiac differentitation of mESCs. (A) Phf8, Jarid1a, Jhdm1d, Utx, and Jmjd3. (B) Jmjd2a, Jmjd2b, Jmjd2c, Jarid1b, Jmjd1b and Jhdm2a. Ct values of Q-PCR analysis were normalized against GADPH. Data are represented as mean±SEM from at least three independent experiments. bP<0.05, cP<0.01 compared with d 0 values.
Changes in KDM expression during mesodermal differentiation
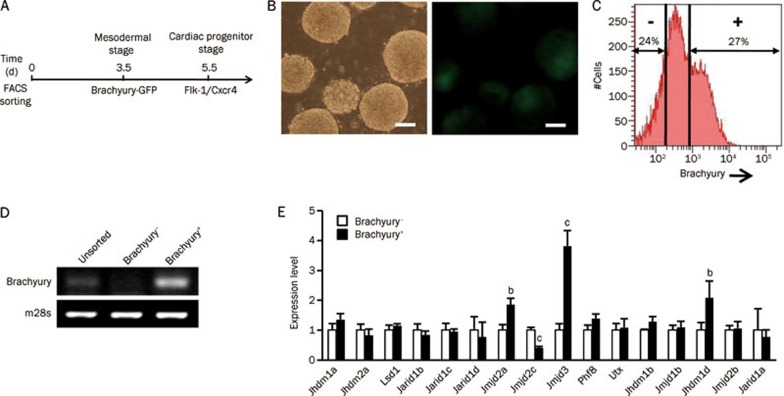
To further identify the KDMs that may be involved in the critical stage of cardiac lineage determination, we compared changes in the KDM expression levels at the mesodermal stage and the cardiac progenitor stage, two essential stages for the cardiac lineage commitment30, using stage-specific reporter systems (Figure 3A). It has been demonstrated that the transcription factor Brachyury is required for mesoderm formation, and it is widely used as a mesodermal specific marker31,32. We therefore used an ESC line with the expression of GFP driven by Brachyury25 (Figure 3B) to separate Brachyury+ and Brachyury− populations at differentiation d 3.5 by FACS (Figure 3C). RT-PCR analysis showed that the Brachyury+ subpopulation expressed a much higher level of Brachyury than did the unsorted cells and the Brachyury− subpopulation (Figure 3D). We then performed a Q-PCR analysis to evaluate the KDM expression profiles of the Brachyury+ and Brachyury− cells. The results showed that the expression levels of Jmjd2a, Jmjd3 and Jhdm1d were higher in the Brachyury+ cells, with the greatest increase occurring in Jmjd3 (Figure 3E). Another Kdm4 family member, Jmjd2c, decreased by 60% in the Brachyury+ cells compared with the Brachyury− cells (Figure 3E).
Figure 3.
Characteristics of KDM expressions in mesodermal cells. (A) Schematic diagram of analysis strategy. (B) Expression of Brachyury-GFP in d 3.5 EBs. Bar=100 μm. (C) FACS sorting of Brachyury− and Brachyrury+ subpopulations at differentiation d 3.5. (D) RT-PCR analysis of Brachyury expression in unsorted, Brachyury− and Brachyury+ cells. (E) Quantitative expression analysis of KDMs between Brachyury− and Brachyury+ subpopulations. Data are represented as mean±SEM from at least three independent experiments. bP<0.05, cP<0.01 compared with corresponding Brachyury− values.
Characteristics of KDM expression during cardiac progenitor differentiation
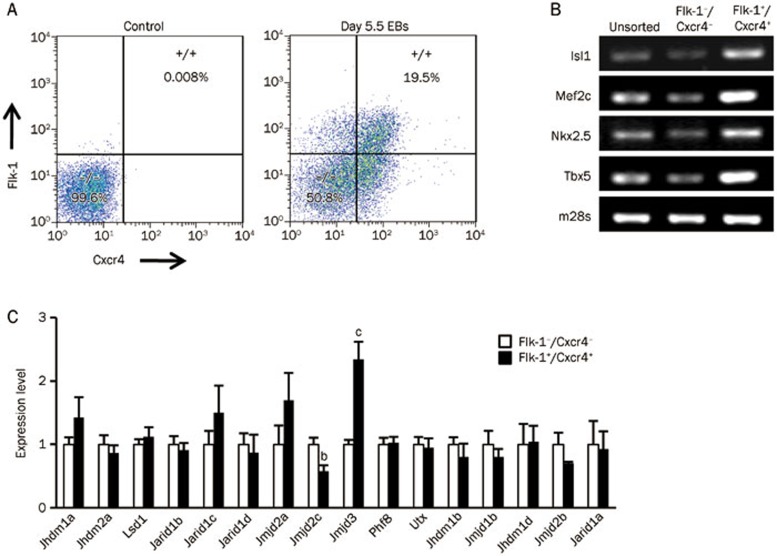
The dual cell surface biomarkers fetal liver kinase 1 (Flk-1) and chemokine (C-X-C motif) receptor 4 (Cxcr4) have been used to extract a cardiac progenitor stage-specific cytotype after 5.5 d of differentiation from pluripotent mESCs33,34. We therefore used this dual biomarker system to separate Flk-1+/Cxcr4+ cardiac progenitors from the Flk-1−/Cxcr4− cells (Figure 4A). RT-PCR analysis showed that the expression of genes that are specifically involved in cardiac development, including ISL LIM Homeobox 1 (Isl1), myocyte enhancer factor 2C (Mef2c), NK2 transcription factor related, locus 5 (Nkx2.5), and T-box transcription factor 5 (Tbx5), increased in the Flk-1+/Cxcr4+ subpopulation compared with the unsorted and Flk-1−/Cxcr4− cells (Figure 4B). We then performed a Q-PCR analysis to compare the KDM expression profiles of the Flk-1+/Cxcr4+ and Flk-1−/Cxcr4− cells. The results showed that the expression levels of Jmjd3 were significantly higher in Flk-1+/Cxcr4+ cardiac progenitors cells compared with those in Flk-1−/Cxcr4− cells. In addition, the expression levels of Jmjd2c were lower in Flk-1+/Cxcr4+ cells than they were in Flk-1−/Cxcr4− cells (Figure 4C).
Figure 4.
Characteristics of KDMs in Flk-1+/Cxcr4+ cardiac progenitor cells. (A) FACS sorting of Flk-1−/Cxcr4− and Flk-1+/Cxcr4+ subpopulations at d 5.5. (B) RT-PCR analysis of cardiac marker expression in unsorted, Flk-1−/Cxcr4− and Flk-1+/Cxcr4+ cells. (C) Quantitative expression analysis of KDMs between Flk-1−/Cxcr4− and Flk-1+/Cxcr4+ subpopulations. Data are represented as mean±SEM from at least three independent experiments. bP<0.05, cP<0.01 compared with corresponding Flk-1−/Cxcr4− values.
Discussion
KDMs are emerging as important players in both the maintenance of ESCs stemness and lineage commitment11. In the present study, we present a global gene expression profile of KDMs during the cardiac differentiation process of mESCs. We then analyzed the specific expression of KDMs at the mesodermal and cardiac progenitor stages, two critical stages for cardiac lineage commitment. These findings provide new insight into the role of KDMs in cardiac lineage commitment.
We found that the Jmjd3 expression levels increased significantly during cardiac differentiation, suggesting that Jmjd3 may play an important role in this process. This result is consistent with the recent finding that Jmjd3 functions in controlling mesodermal and cardiovascular differentiation19. Jmjd3 also has demethylase-independent functions: Miller et al35 reported that Jmjd3 mediates a functional interaction between the lineage-defining T-box transcription factor family member and its target genes. T-box transcription factor family members, such as Brachyury and Tbx5, are critical regulators of cardiac differentiation36. Consistent with these findings, our study showed that Jmjd3 has much higher expression levels in Brachyrury+ cells than it does in Brachyury− cells (Figure 3E). In addition, we found that the expression of Jmjd3 was upregulated in the Flk-1+/Cxcr4+ cardiac progenitor cells (Figure 4C) with a higher expression level of Tbx5 (Figure 4B), suggesting that Jmjd3 might control cardiac progenitor commitment via other T-box transcription factor family members. This possibility needs to be further investigated.
The expression levels of Utx, another Kdm6 family member, are also upregulated during cardiac differentiation, suggesting its potential role in cardiac differentiation. This is supported by the observation that Utx acts as a critical switch to activate the expression of cardiac-specific genes20. However, unlike Jmjd3, Utx is not found to be enriched in either Brachyury+ or Flk-1+/Cxcr4+ cells (Figures 3E and 4C), and its expression levels do not change significantly before d 7 (Figure 2A). These findings suggest that Utx and Jmjd3 might regulate cardiac differentiation at different stages. This possible mechanism of regulation needs to be further investigated.
Phf8, Jhdm1d, and Jarid1a have been shown to be involved in differentiation, but little is known regarding their roles in cardiac differentiation. Phf8 preferentially acts on H3K9me2 and H3K9me1 and associates with transcriptional activation and retinoic acid signaling pathways in neuron differentiation37. Jhdm1d controls brain development by activating follistatin gene transcription38. Jarid1a controls the osteogenic differentiation of human adipose-derived stromal cells39. Interestingly, we found that all of these genes are constitutively upregulated during cardiac differentiation (Figure 2A, Table 4). This expression pattern is similar to the expression patterns of genes related to the cardiac lineage decision, such as Flk-1, Nkx2.5, Mef2c, and cardiac specific marker Ncx1, as found in our previous40,41 and present data (Figure 1C), suggesting that these KDMs may be involved in lineage determination.
Jmjd2a and Jmjd2c are histone H3K9 and H3K36 demethylases that belong to the same subclass; our results show that they have opposite expression patterns during cardiac differentiation (Figure 2B). Jmjd2a promotes the transcriptional activation of the Myog gene and thus contributes to skeletal muscle differentiation42. Its expression is upregulated in human hypertrophic cardiomyopathy patients and is found to promote cardiac hypertrophy in pathological conditions43. Our data reveal that the expression of Jmjd2a is upregulated in Brychyury+ cells, suggesting that Jmjd2a might participate in skeletal muscle and cardiomyocyte differentiation as early as the mesodermal stage. Jmjd2c is preferentially expressed in undifferentiated ESCs44 and regulates self-renewal in ESCs along with Jmjd1a45. We found in this study that the expression levels of Jmjd2c are decreased at the early days of differentiation upon the withdrawal of LIF and feeder layers, which is consistent with its role in self-renewal. Furthermore, we found that the expression levels of Jmjd2c are downregulated in both Brachyury+ and Flk-1+/Cxcr4+ cells, which is the opposite of Jmjd3. These results imply that Jmjd2c might exert a function distinct from that of Jmjd3 in cardiac differentiation. This possibility needs to be explored further.
There are some limitations in the present study. First, although we identified the KDMs that changed during cardiac differentiation, the functional relevance of these changes needs to be further demonstrated in overexpression or knockdown experiments and subsequent mechanistic studies. Second, although the Flk-1/Cxcr4 co-expressing population is enriched for cardiac progenitor cells compared with the unsorted and Flk-1−/Cxcr4− cells (Figure 4B), it is not a pure population. The reporter ESC lines driven by cardiac specific markers, such as Nkx2.5, could be more specific for the comparison of KDM expression in cardiac progenitor cells and non-cardiac progenitor cells.
In summary, we presented KDM expression profiles during cardiac differentiation and identified KDMs with altered expression levels at mesodermal and cardiac progenitor stages. Our results suggest that KDMs might play important roles at different stages of cardiomyocyte commitment. The establishment of the KDM profile provides new insight into how the post-translational modification of histone proteins by demethylation controls ESC specific germ layer commitment and subsequent lineage differentiation.
Author contribution
Yan TANG, Huang-tian YANG, and Charlie Degui CHEN designed the research; Yan TANG, Zhong-yan CHEN, Ya-zhen HONG, and Qiang WU performed the experiments; Yan TANG and Huang-tian YANG analyzed the data; Han-qing LIN designed part of the KDM primers; and Yan TANG and Huang-tian YANG wrote the paper.
Acknowledgments
This study was supported by grants from the Strategic Priority Research Program of CAS (XDA01020204), the National Basic Research Program of China (2011CB965300 and 2014CB965100), and the National Natural Science Foundation of China (31030050). We thank Dr Gordon KELLER for kindly providing the Brachyury-GFP E14 mESCs.
References
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–9. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–55. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–44. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–80. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–23. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–54. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kim J, Yuan X, Braun T. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ Res. 2011;109:1067–81. doi: 10.1161/CIRCRESAHA.111.243709. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–71. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Wen L, Chen Y, Zeng LL, Zhao F, Li R, Liu Y, et al. Triptolide induces cell-cycle arrest and apoptosis of human multiple myeloma cells in vitro via altering expression of histone demethylase LSD1 and JMJD2B. Acta Pharmacol Sin. 2012;33:109–19. doi: 10.1038/aps.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–68. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–9. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–84. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–84. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, et al. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–62. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–20. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109:15324–9. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Yi F, Pan HZ, Duan SL, Ding ZC, Yuan GH, et al. Progress and prospects in stem cell therapy. Acta Pharmacol Sin. 2013;34:741–6. doi: 10.1038/aps.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. Progress and prospects in stem cell therapy and drug discovery. Acta Pharmacol Sin. 2013;34:717–8. doi: 10.1038/aps.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–27. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Yang HT, Tweedie D, Wang S, Guia A, Vinogradova T, Bogdanov K, et al. The ryanodine receptor modulates the spontaneous beating rate of cardiomyocytes during development. Proc Natl Acad Sci U S A. 2002;99:9225–30. doi: 10.1073/pnas.142651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JD, Yu HM, Wang R, Liang J, Yang HT. Developmental regulation of intracellular calcium transients during cardiomyocyte differentiation of mouse embryonic stem cells. Acta Pharmacol Sin. 2006;27:901–10. doi: 10.1111/j.1745-7254.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods Mol Biol. 2002;185:127–56. doi: 10.1385/1-59259-241-4:127. [DOI] [PubMed] [Google Scholar]
- Cao N, Liu Z, Chen Z, Wang J, Chen T, Zhao X, et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–36. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–9. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–32. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Faustino RS, Chiriac A, Crespo-Diaz R, Behfar A, Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–73. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Chiriac A, Nelson TJ, Faustino RS, Behfar A, Terzic A. Cardiogenic induction of pluripotent stem cells streamlined through a conserved SDF-1/VEGF/BMP2 integrated network. PLoS One. 2010;5:e9943. doi: 10.1371/journal.pone.0009943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Suzuki A. Regulation of organogenesis and stem cell properties by T-box transcription factors. Cell Mol Life Sci. 2013;70:3929–45. doi: 10.1007/s00018-013-1305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–18. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–7. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Shi L, Zhou Y, Liu Y, Ma GE, Jiang Y, et al. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells. 2011;29:1112–25. doi: 10.1002/stem.663. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang YJ, Tang Y, Cao N, Wang J, Yang HT. Type 3 inositol 1,4,5-trisphosphate receptor negatively regulates apoptosis during mouse embryonic stem cell differentiation. Cell Death Differ. 2010;17:1141–54. doi: 10.1038/cdd.2009.209. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao N, Yuan M, Cui H, Tang Y, Qin L, et al. MicroRNA-125b/Lin28 pathway contributes to the mesendodermal fate decision of embryonic stem cells. Stem Cells Dev. 2012;21:1524–37. doi: 10.1089/scd.2011.0350. [DOI] [PubMed] [Google Scholar]
- Verrier L, Escaffit F, Chailleux C, Trouche D, Vandromme M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011;7:e1001390. doi: 10.1371/journal.pgen.1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest. 2011;121:2447–56. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells. Int J Mol Med. 2007;20:269–73. [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]