Abstract
Forces are increasingly recognized as major regulators of cell structure and function, and the mechanical properties of cells, such as cell stiffness, are essential to the mechanisms by which cells sense forces, transmit them to the cell interior or to other cells, and transduce them into chemical signals that impact a spectrum of cellular responses. Here we reported a new whole-cell cell stiffness measurement technique with a subcellular spatial resolution. This technique was based on a novel cell stretching device that allowed for quantitative control and real-time measurements of mechanical stimuli and cellular biomechanical responses. Our strategy involved a microfabricated array of silicone elastomeric microposts integrated onto a stretchable elastomeric membrane. Using a computer-controlled vacuum, this micropost array membrane (mPAM) was activated to apply equibiaxial cell stretching forces to adherent cells attached on the tops of the microposts. The micropost top positions before and after mPAM stretches were recorded using fluorescence microscopy and further utilized to quantify local cell stretching forces and cell area increments. A robust computation scheme was developed and implemented for subcellular quantifications of cell stiffness using the data of local cell stretching forces and cell area increments generated from mPAM cell stretch assays. Our cell stiffness studies using the mPAM revealed strong positive correlations among cell stiffness, cellular traction force, and cell spread area, and illustrated the important functional roles of actin polymerization and myosin II-mediated cytoskeleton contractility in regulating cell stiffness. Collectively, our work reported a new approach for whole-cell cell stiffness measurements with a subcellular spatial resolution, which would likely help explain the complex biomechanical functions and force-sensing mechanisms of cells and design better materials for cell and tissue engineering and other applications in vivo.
INTRODUCTION
The mechanical properties of the cell are essential to force responses because they determine the extent of deformation of the region of the cell where the force is applied, and therefore the range of molecular deformations that can be triggered1, 2. More recently, increased interest in cell mechanics has been generated by demonstrations that the mechanical features of the extracellular environment and application of forces to cells trigger cellular responses that are essential for many aspects of cell structure and function3-10. Such studies have strongly implicated an important functional role of cell mechanics in sensing and transducing the biophysical signals in the local cellular microenvironment (i.e., external forces and matrix mechanics) into intracellular signalling events4, 5, 8, 9. In addition, cell mechanics has recently been identified as an emerging field that can potentially make significant contributions to studies of human diseases11, 12. Cell mechanics studies have not only contributed significantly to understanding of cellular mechanisms behind the pathophysiology of human diseases, such as malaria13, sickle cell anemia14, 15 and cancer2, 16-18, since diseased cells differ physically from healthy ones, but also provided important knowledge in disease diagnosis and prognosis2, 19-21.
Current experimental methods to study the mechanical behaviors of single cells can be broadly classified into two types: whole-cell mechanical loading methods such as substrate deformation22-24, micropipette aspiration25, 26, cell poking27, cell compression28-30, and optical trapping for stretching cells31, and local probing methods such as atomic force microscopy (AFM)20, 32 and magnetic twisting cytometry (MTC)33-35. The whole-cell mechanical loading methods can generate a large global cell strain, and cells are normally modelled using simple elastic or viscoelastic models with single mechanical properties characterizing the whole cell body. Thus, it is difficult to determine the spatial subcellular distribution of cell mechanical properties using the whole-cell mechanical loading methods. In the local probing methods, only a local force is applied to generate a local deformation on the cell surface. An appropriate analysis of cell local deformations provides a local picture of the elastic properties of the cell. It remains largely unclear, however, whether the local mechanical properties of the cell measured using the local probing methods can be representative of the global cell mechanical properties or is only restricted to the subcellular region that experiences the force36. The local probing methods also suffer from variability among the data from different studies, which is suspected due to the regional heterogeneity and dynamic regulation of the cytoplasm and adhesions of cells37.
As an alternative approach, attempts have been made recently to quantify cell mechanical properties such as cellular traction forces using microfabricated devices, such as those based on a regular array of elastomeric poly-(dimethylsiloxane) (PDMS) microposts38, 39 or a cantilever beam40. A more recent development of the PDMS micropost array to apply active external forces to cells in a controlled fashion is to exert either a pointwise mechanical force using PDMS microposts incorporated with magnetic nanowires41 or global equibiaxial or uniaxial stretching forces using PDMS microposts integrated on stretchable membranes42, 43. For example, we have recently developed a stretchable micropost array membrane (mPAM) system, where the PDMS micropost array is formed at the center of a stretchable PDMS membrane42. Using a computer-controlled vacuum, the mPAM can be activated to apply cell stretching forces to adherent cells attached on the tops of the microposts. Using the mPAM, we have studied the mechanotransductive system in vascular smooth muscle cells (VSMCs), or more specifically, the live-cell subcellular dynamic responses of cellular traction forces of VSMCs to sustained static equibiaxial cell stretches42. Here, we showed that the mPAM could be utilized for real-time live-cell whole-cell cell stiffness measurements with a subcellular spatial resolution. The micropost top positions of the mPAM before and after cell stretches could be recorded using live-cell fluorescence microscopy and further utilized for quantifications of local cell stretching forces and cell area increments. In this work, we further developed a robust computational approach toinversely obtain a quantitative spatial map of subcellular cell stiffness using the finite element analysis and the Levenberg-Marquardt algorithm for nonlinear optimization44. Using the mPAM cell stretch assays and our computational approach, we performed detailed studies to examine the correlations among cell stiffness, cellular traction force and cell spread area for single live VSMCs. To demonstrate the general application of the mPAM for live-cell subcellular measurements of cell stiffness, we further examined and proved the important functional roles of actin polymerization and myosin II-mediated cytoskeleton contractility in regulating cell stiffness of live single VSMCs.
MATERIALS AND METHODS
Fabrication of stretchable micropost array membranes (mPAMs)
Silicon mould masters containing positive micropost array structures were first fabricated using high-resolution photolithography and deep reactive ion etching techniques8, 45. The silicon masters were silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapour (United Chemical Technology, Bristol, PA) under vacuum for 2 hr to facilitate subsequent release of moulded PDMS structures. Replica moulding of PDMS (Sylgard 184, Dow-Corning, Midland, MI) was performed by baking thoroughly mixed PDMS prepolymer (10:1 w/w ratio between PDMS base monomer and curing agent) on the silicon master at 110°C for 20 hr to obtain fully cured PDMS. After peeled off, negative PDMS micropost array substrates were silanized for the second replica moulding process. PDMS prepolymer with a 10:1 w/w PDMS base monomer to curing agent ratio was poured onto the negative PDMS mould. After covering the PDMS prepolymer with a silanized glass coverslip, the whole assembly was thermally cured at 110°C for 20 hr. The elastomeric PDMS micropost array was then generated by peeling off the thin PDMS film containing the PDMS microposts identical to the microstructures on the silicon master from the PDMS negative mould. At this step, PDMS microposts often collapsed, and we regenerated the PDMS microposts by sonication in 100% ethanol followed by dry-release with liquid CO2 using a critical point dryer (Samdri®-PVT-3D, Tousimis, Rockville, MD).
The stretchable PDMS micropost array membrane (mPAM) was fabricated by spin-coating PDMS prepolymer (10:1 w/w ratio between the PDMS base monomer and curing agent) on the lids of Petri dishes (100 mm diameter; BD Falcon, Franklin Lakes, NJ) at 500 rpm for 1 min to obtain a 100 μm thick PDMS membrane, followed by baking the Petri dish lids at 60°C for 48 hr. The thin PDMS film containing PDMS microposts was then assembled onto the central area of the 100 mm PDMS membrane using the O2 plasma-assisted PDMS-PDMS bonding process (pressure: 700 mTorr; energy: 1 kJ; Plasma Prep II, West Chester, PA) to generate the mPAM. It is important to note that prior to experiments, the mPAM should be stored at room temperature for at least 2 days to eliminate the effect of residual PDMS surface activation.
Surface functionalization of PDMS microposts
Microcontact printing was used to functionalize the tops of the PDMS microposts on the mPAM with ECM proteins to promote cell attachment8, 38. Briefly, PDMS stamps with a 30:1 w/w PDMS base monomer to curing agent ratio were utilized as printing stamps. These PDMS stamps were inked with a fibronectin solution in water (50 μg/ml; BD Biosciences, San Jose, CA) for 1 hr and brown dry with nitrogen. After treating the mPAM with ultraviolet ozone (UV-ozone cleaner; Jelight, Irvine, CA) for 7 min to oxidize the PDMS surface, fibronectin-coated PDMS stamps were placed in conformal contact with the PDMS microposts for at least 30 sec to allow a complete transfer of fibronectin from the PDMS stamps to the PDMS micropost tops. After mounting the mPAM onto the cell stretching device (CSD), the PDMS microposts were stained with 10 μg/ml 1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate (DiI, Invitrogen, Carlsad, CA) in distilled water for 60 min. To eliminate non-specific protein adsorption on uncoated PDMS surfaces of the mPAM, the blocking agent pluronics F127 NF (P2443-250G, Sigma-Aldrich, St. Louis, MO) with a 0.2% w/v concentration in water was used to treat the mPAM for 30 min. The mPAM was then rinsed thoroughly with water before seeding cells on the mPAM.
Cell culture
Primary human vascular smooth muscle cells (VSMCs; CC-2583, Lonza, Walkersvilla, MD) were cultured in the smooth muscle basal medium (SmBM-2, CC-3182, Lonza) supplemented with soluble factors (SmGM-2 SingleQuot, CC-4149, Lonza) including insulin, fetal bovine serum, gentamicin, amphotericin, human fibroblast growth factor (hFGF), and hFGF-B. Once cells became confluent, they were washed with HEPES buffered saline solution (CC-5022, Lonza) and trypsinized (CC-5012, Lonza), followed by neutralization with a trypsin neutralization solution (CC-5002, Lonza). Cells were then subcultured to a new culture plate in the growth medium at a density of 5 × 103 cells/cm2. Only early passages of VSMCs (passage 3-5) were used in experiments.
Cell stretch arrays
VSMCs were seeded as single cells on the mPAM and were cultured for 2 days before mPAM stretch assays. During mPAM stretch assays, the PDMS micropost array was imaged using a monochrome charge-coupled device (CCD) camera (AxioCam, Carl Zeiss MicroImaging, Thornwood, NY) attached to an inverted microscope (Zeiss Axio Observer Z1, Carl Zeiss MicroImaging) with a 40× EC Plan-Neofluar objective (0.75 Ph2 D=0.71 M27, Carl Zeiss MicroImaging). The microscope was enclosed in an environmental incubator (XL S1 incubator, Carl Zeiss MicroImaging) to maintain the experimental environment at 37°C and 5% CO2. For each single VSMC, fluorescent images of the Δ9-DiI-stained PDMS microposts were acquired by focusing on a plane intersecting the micropost array tips. Cellular traction forces of VSMCs were recorded before and after activating a vacuum system (DigiVac Model 450, DigiVac, Matawan, NJ) to equibiaxially stretch the mPAM and thus the cells attached on the PDMS micropost array. Cells were considered viable if they remained attached to the PDMS microposts for the duration of the experiment and did not round up or detach. Cells were then immediately trypsinized with the radii R of the cell bodies measured right before the cells were detached from the PDMS microposts.
For drug treatment assays, after seeding VSMCs on the mPAM, different drugs or small molecule inhibitors were added to the culture medium for 1 hr before mPAM cell stretch assays. These drugs or small molecule inhibitors included blebbistatin (100 μM; EMD Chemicals, Darmstadt, Germany), Y-26632 (10 μM; Enzo Life Sciences, Farmingdale, NY), and cytochalasin D (40 μM; Invitrogen).
Quantification of subcellular traction force and cell stiffness
The microscopic images of the PDMS micropost tops captured in the experiments were processed using a customized MATLAB program (Mathworks, Novi, MI) to compute micropost displacements and cellular traction forces8, 45. In brief, a previously described localized thresholding algorithm (LT) was used to determine the centroid of each PDMS micropost. A selected region of the image was converted to black and white, where the sum of white pixels representing the expected size of the post cross-sectional area was determined with thresholding. A 2D Gaussian fit was used to model the grayscale intensity profile of the PDMS post and determined the centroid by nonlinear least squares fitting. This step determined positions of the deflected microposts by comparing the deflected positions of the microposts against a regular grid, aligned using undeflected “free” microposts as reference points. A deflection vector (r, s) was established for each “attached” PDMS micropost underneath the cell, where r and s denoted the post deflection components in the x-and y-directions, respectively. r and s were related to the horizontal traction force, fc, applied on the micropost top using the Euler-Bernoulli beam theory and fc = K (r2 + s2)1/2, where K was the effective spring constant of the PDMS micropost8.
Another custom-developed MATLAB program was used to compute subcellular cell stiffness by converting the defection vector maps of the “attached” PDMS microposts underneath the cell before and after mPAM stretches to obtain subcellular stretching forces and resulting cellular deformations. Using these subcellular stretching forces and cellular deformations, the MATLAB program would compute the optimal subcellular profiles of cell mechanical properties, i.e. elastic Young’s modulus and Poisson’s ratio, using a finite element-based model and the Levenberg-Marquardt algorithm (discussed in detail in Results and Discussion). The MATLAB program also calculated the whole-cell Young’s modulus and Poisson’s ratio.
Immunofluorescence staining
Cells were incubated for 1 hr in an ice-cold cytoskeleton buffer containing 50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 1 μg ml−1 aprotinin, 1 μg ml−1 leupeptin, and 1 μg ml−1 pepstatin. Cells were permeablized by adding 0.5% v/v Triton X-100 (Roche Applied Science, Indianapolis, IN) in the cytoskeleton buffer for 1 min. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) in PBS and incubated in 10% goat serum (Invitrogen) to eliminate non-specific binding in the following steps. Primary antibody to non-muscle myosin IIA produced in mouse (Abcam, Cambridge, MA) was added to cells, which were then stained by IgG secondary antibody (Invitrogen) with fluorescence Alexa Fluor 488 (anti-mouse). Cells were also co-stained with Alexa Fluor 555 conjugated phalloidin (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) for visualization of actin microfilaments and nuclei, respectively.
Statistics
p-values were calculated using the Student’s t-test in Excel (Microsoft, Seattle, WA).
RESULTS AND DISCUSSION
Equibiaxial cell stretches using stretchable micropost array membranes (mPAMs)
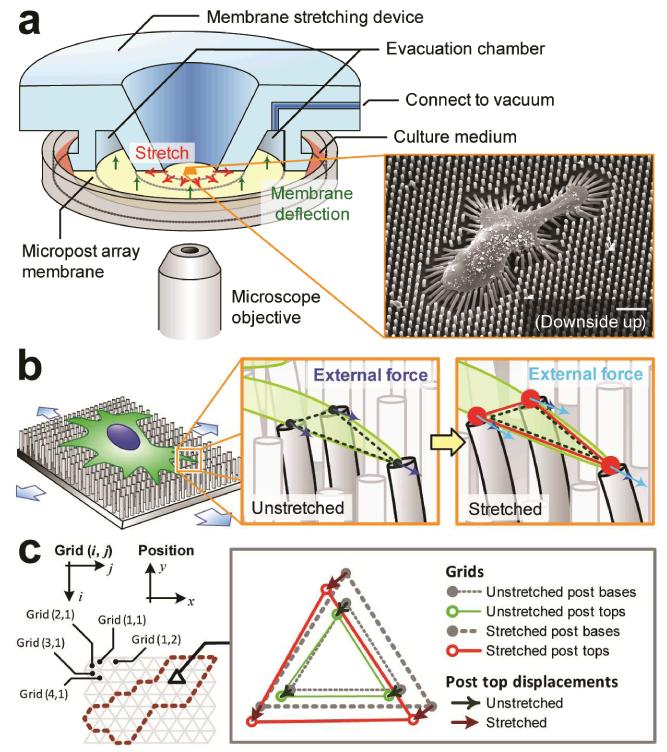
A cell stretching device (CSD) was designed to work with the stretchable micropost array membrane (mPAM) to exert equibiaxial stretches to adherent cells seeded on the PDMS micropost array formed at the center of the mPAM (Fig. 1)42. The CSD contained a circular viewing aperture surrounded by a ring-shaped evacuation chamber, and the mPAM was a 500-μm thick, 100 mm-diameter PDMS membrane with a square-shaped array of hexagonally spaced PDMS microposts at the membrane center (Fig. 1a). The PDMS micropost array was first fabricated using microfabrication and replica molding on a thin layer of PDMS before it was bonded permanently onto the 100 mm-diameter PDMS base membrane, using the O2 plasma-assisted PDMS-PDMS bonding process (see Materials and Methods for the mPAM fabrication method). During cell stretch assays, the mPAM was mounted on the CSD with the micropost array region on the mPAM aligned with the CSD viewing aperture for real-time imaging of cells seeded on the PDMS microposts (Fig. 1a). After a computer-controlled vacuum was applied to the evacuation chamber of the CSD, the periphery of the PDMS base membrane of the mPAM was drawn into the evacuation chamber and the centre region of the PDMS base membrane holding the PDMS micropost array was then stretched equibiaxially. When the mPAM was equibiaxially stretched, adherent cells attached on the tops of the PDMS microposts would also be stretched equibiaxially. Thus, the mPAM equibiaxial cell stretch platform allowed generations of whole-cell deformations over the entire cell region.
Figure 1.
Live-cell subcellular measurement of cell stiffness using a microengineered stretchable micropost array membrane (mPAM) and a cell stretching device (CSD). (a) Schematic of the CSD and its implementation for stretching the mPAM and thus single cells adhered on the tops of the PDMS microposts. Only one-half of the CSD is shown for visualization of its internal structure. A vacuum was applied to the CSD evacuation chamber to draw the periphery of the mPAM, causing the central region of the mPAM holding the PDMS micropost array and pre-seeded with adherent cells to stretch equibiaxially. Inset: Scanning electron microscopy image of a single cell seeded on the PDMS micropost array. Scale bar: 20 μm. (b) Cartoon of an adherent cell on the mPAM under a static equibiaxial stretch. Left inset shows a triangular subcellular region formed between three adjacent microposts deflected by cellular traction forces. Right inset shows the same triangular region under a static equibiaxial stretch with an increased cell area and heightened traction forces. Mechanical forces exerted on the cell by the PDMS microposts were labelled by blue and light blue arrows before and after the mPAM stretch, respectively. (c) Grid arrangement for theoretical computation of subcellular cell stiffness. Hexagonally arranged microposts formed the nodes in a regular triangular grid surface. Inset: Relative displacements of the micropost tops before and after static equibiaxial mPAM stretches used to calculate stretching forces and deformations of grid elements.
Model and calculation of subcellular cell stiffness
Using mPAM to exert equibiaxial cell stretches to adherent cells attached on the tops of the PDMS microposts would induce changes in both the positions and deflections of the PDMS microposts underneath the cells (Fig. 1b). Such position and deflection changes of the PDMS microposts could be monitored using fluorescence microscopy to quantify local stress and strain states over the entire cell body and eventually the subcellular cell stiffness.
In the mPAM, the hexagonal arrangement of the PDMS microposts (Fig. 1c) established a well-defined mesh composed of regularly arranged triangular grid elements. Each grid element contained three nodes representing three adjacent micropost tops and labelled with indices 1, 2 and 3 clockwise starting from an arbitrary node. Nodal displacements and stretching forces for each grid underneath the cell (enclosed by the dashed line in Fig. 1c) could be calculated from displacements of the micropost tops measured before and after mPAM stretches using fluorescence microscopy. Using one grid element shown in Fig. 1c as an example, before mPAM cell stretches, positions of the three nodes or micropost tops in one grid element, (x1, y1), (x2, y2) and (x3, y3), were measured as described in Materials and Methods, from which the PDMS micropost deflections (green arrows shown in Fig. 1c) were determined by comparing positions of the deflected micropost tops against a regular grid, aligned using undeflected “free” microposts as reference points. Similarly, micropost deflections of the grid element immediately after mPAM cell stretches were obtained (i.e. (r1, s1), (r2, s2) and (r3, s3); red arrows shown in Fig. 1c), permitting quantifications of nodal displacements or changes of micropost top positions before and after mPAM stretches, denoted by (u1x, u1y), (u2x, u2y) and (u3x, u3y) for the three nodes. Nodal displacements of the grid element were converted to local cell stretching forces, (f1x, f1y), (f2x, f2y), and (f3x, f3y), using fqx = −Kuqx and fqy = −Kuqy for q = 1, 2 and 3. Here the effective spring constant K of the PDMS micropost was given by K = 3πEPDMSD4/64L3 from the Euler-Bernoulli beam theory, with EPDMS as the Young’s modulus of PDMS and D and L as the post diameter and height, respectively.
A finite element method was applied to solve the elastic properties of each grid element in the cell region46. Here we considered each grid element elastically isotropic with respect to two mechanical parameters: (1) Eh, the product of the Young’s modulus E and the element height, and (2) ν, the Poisson’s ratio. It should be noted that the local subcellular Young’s modulus of each grid element might be determined from Eh if the grid element height could be measured using techniques such as confocal microscopy. The relation between nodal forces f = [f1xf1yf2xf2yf3xf3y]T and nodal displacements u = [u1xu1yu2xu2yu3xu3y]T of each grid is given by
| (1) |
where
| (2) |
and
| (3) |
In Eq. (3), A is the projected area of the grid element onto the x-y plane. In a matrix form, the relation between f and u in Eq. (1) can be expressed as f = s(Eh, ν)u, where s(Eh, ν) is a 6 × 6 matrix containing the unknown local cell mechanical properties, Eh and ν, in a grid element.
Since a grid element shares nodes with its neighbouring elements, we could combine relations described by Eq. (1) for all m grid elements within the cell region into a global matrix form of F = S(Eh, ν)U, where F and U are global force and displacement vectors, Eh and ν are vectors of the mechanical properties for all the grid elements, and S is the global matrix with all the unknown cell mechanical properties. We further define the total number of nodes in the cell region with m elements as n; hence both F and U are vectors of 2n components, and S is a 2n × 2n matrix.
Computation of the subcellular spatial map of cell stiffness involves calculations of Eh and ν from values of U and F measured from mPAM cell stretch assays. Because each grid element has two unknowns, Eh and v, the total number of unknowns in F = S(Eh, ν)U is 2m. However, in practice, it is often the case that m > n for a cell region. Thus, the number of unknowns, 2m, is more than the number of equations, 2n, rendering the equation F = S(Eh, ν)U unsolvable. To address this issue, we suppressed the number of unknowns, 2m, by assuming that a grid element at (i = 2n, j) and its adjacent element at (i = 2n − 1, j) in the cell region possess the same values of Eh and v, where i and j denote the row and column of the grid element as shown in Fig. 1c. Imposing such a constraint to the equation F = S(Eh, ν)U ensures that Eh and ν can be calculated mathematically when m > n.
The objective of our computation task was thus to search for optimized Eh and ν to minimize the least square deviation, , where is defined as the estimate of U computed, from . To solve for such a least-square fitting problem, we applied the iterative Levenberg-Marquardt algorithm44, a widely adopted numerical method for non-linear optimization, to obtain optimized Eh and ν. In brief, this iterative approach first imposes initial values for Eh and ν to calculate S(Eh, ν). Then it iteratively computes the approximate solution of U by and generates the new trial values of Eh and ν until the optimization condition is achieved, i.e. is minimized. The Levenberg-Marquadt method has been widely used because it can effectively calculate Eh and ν toward optimized values.
Using optimized Eh and ν, we can further calculate the whole-cell average Young’s modulus, Eave, and Poisson’s ratio, νave, of an entire single cell. Eave and νave are obtained by averaging over all grid elements, i.e. Eave = 3Σ(EhA)/(4πR3) and νave = Σ(νA)/ΣA, where R is the radius of the cell in a suspended state measured in our experiments (measurement of R is described in Materials and Methods).
Validation of mPAM cell stretches
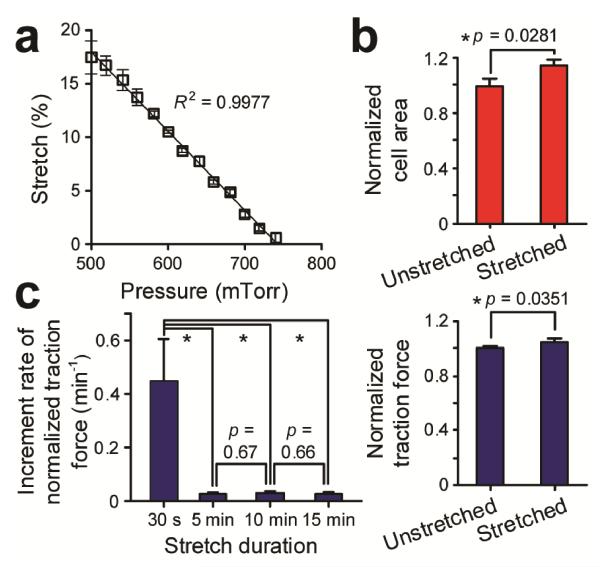
As we have recently reported42, stretch induced strain in the mPAM is mostly constrained within the PDMS base membrane and does not propagate up the PDMS micropost. Thus, stretches of the mPAM will not affect the micropost geometry and thus its spring constant42. The stretch magnitude of the mPAM can be easily determined in situ by utilizing regularly positioned PDMS microposts as fiduciary markers to quantify their increased center-to-center distance under different levels of stretches. In Fig. 2a, a detailed characterization was conducted to demonstrate that the mPAM equibiaxial stretch level from 0% - 18% could be precisely controlled with a high degree of repeatability and linearity by regulating the vacuum pressure. We further conducted experiments to quantify cell spread areas and cellular traction forces of single live VSMCs seeded on the PDMS micropost array before and immediately after the onset of mPAM stretches. Our results in Fig. 2b showed significant increases of both cell spread area and cellular traction force of single live VSMCs within 30 sec after the onset of 10% static equibiaxial mPAM stretches.
Figure 2.
(a) Equibiaxial stretch of the mPAM as a function of the level of vacuum pressure applied to the CSD evacuation chamber. Error bars represent ± standard deviations of data obtained from three independent measurements. Data trend in a was plotted using linear least square fitting (dark line), with the square of the correlation coefficient R2 indicated. (b) Instantaneous changes of cell spread area (top) and cellular traction force (bottom) of single live vascular smooth muscle cells (VSMCs) in response to 10% static equibiaxial cell stretches. Datawere recorded within 30 sec after the onset of mPAM stretches. Data were normalized to the average values of cellular traction force and cell spread area prior to mPAM stretches. Error bars represent ± standard errors of data collected from 10 individual VSMCs (n = 10). (c) Increment rate of normalized cellular traction force (min−1) after different periods of 10% static equibiaxial mPAM stretches. Asterisk indicates a statistically significant change (p < 0.05).
Previous research on cell mechanics and mechanotransduction has suggested that instantaneous biomechanical responses of mechanoresponsive cells to cell stretches (within tens of sec) are dictated by the intrinsic mechanical properties of the cells, while their long-term responses (ranging from a few min to a few hrs) in diverse cell functions are regulated by intracellular mechanotransductive signalling events and adaptive metabolic activities, e.g. cytoskeleton remodelling1, 23, 47-49. To confirm that the instantaneous response of cellular traction force of single live VSMCs to static mPAM stretches was largely due to their intrinsic mechanical properties, we quantified the increment rate of normalized cellular traction force of VSMCs as a function of durations of static mPAM stretches. The increment rate of normalized traction force of VSMCs measured within 30 sec after the onset of 10% static mPAM stretches was about 0.448 min−1, one order of magnitude greater than those occurred at later moments for sustained mPAM stretches (e.g., 0.022 min−1, 0.026 min−1, 0.024 min−1 at 5 min, 10 min and 15 min after mPAM stretches, respectively) (Fig. 2c). Thus, the instantaneous response of traction force of VSMCs should be distinct from their adaptive contractile response to long-term sustained cell stretches42 and should be related to and dictated by their intrinsic mechanical properties to a large extent.
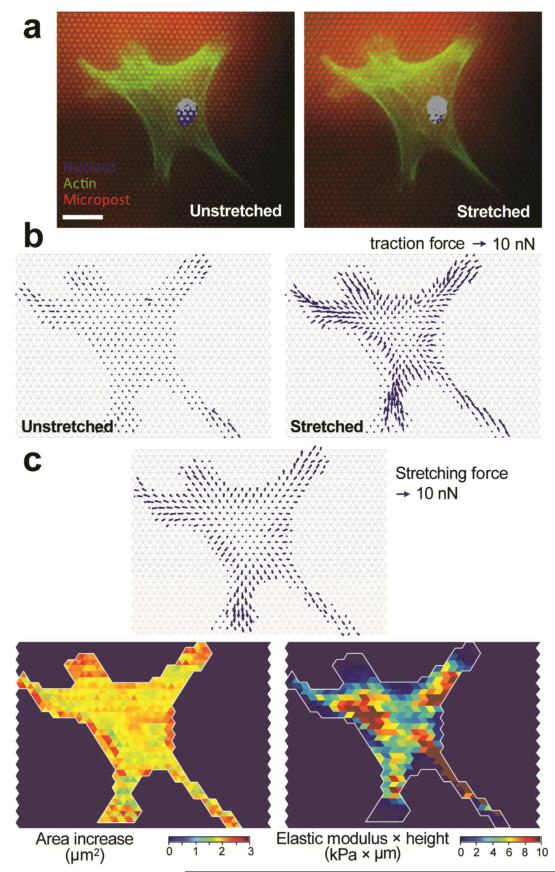
Using the computation scheme described in Model and calculation of subcellular cell stiffness, the spatial subcellular distribution of cell stiffness for single fixed VSMCs was first calculated. Figure 3a showed fluorescence images of a single fixed and stained VSMC on the PDMS micropost array before and after a 9% static equibiaxial mPAM stretch. These fluorescence images clearly demonstrated a visible increase of cell spread area due to the 9% static mPAM stretch (Fig. 3a). Using these fluorescence images, we could determine micropost top positions to quantify traction forces before and after mPAM stretches (Fig. 3b) and thus local cell stretching forces and cell area increments (Fig. 3b), from which the local subcellular cell stiffness could be calulcated using our computation scheme. Interestingly, the measured subcellular spatial profile of the product of the local cell Young’s modulus E and cell height, Eh, exhibited noticeable regional heterogeneity (Fig. 3c, lower right), presumably due to the heterogeneous microscopic spatial distribution of intracellular cytoskeletal structures and other cellular components and organelles such as nucleus.
Figure 3.
Subcellular measurement of cell stiffness for single fixed vascular smooth muscle cells (VSMCs). (a) Representative single fixed and stained VSMCs adhered on the PDMS micropost array before (left) and after (right) a 9% static equibiaxial mPAM stretch. The cell was stained for nucleus (blue) and actin microfilaments (green), and the PDMS microposts were labelled with DiI (red). Scale bar, 20 μm. (b) Spatial maps of subcellular traction forces before (left) and 30 sec after (right) a 9% static equibiaxial mPAM stretch. (c) Spatial maps of stretching force (top), cell area increment (lower left), and computed subcellular cell stiffness (lower right).
It should be noted that cell stiffness measured for fixed VSMCs in Fig. 3 should not be compared directly to cell stiffness of live VSMCs, as cross-linking reagents (such as paraformaldehyde) used as fixatives will form intermolecular bridges, normally through free amino groups, thus creating a network of linked antigens in fixed cells and increasing significantly their mechanical stiffness50. Nevertheless, our experiment in Fig. 3 for fixed VSMCs had clearly demonstrated the application of the mPAM and our computation scheme for subcellular measurements of cell stiffness.
Characteristics and correlative studies of cell stiffness of live single VSMCs
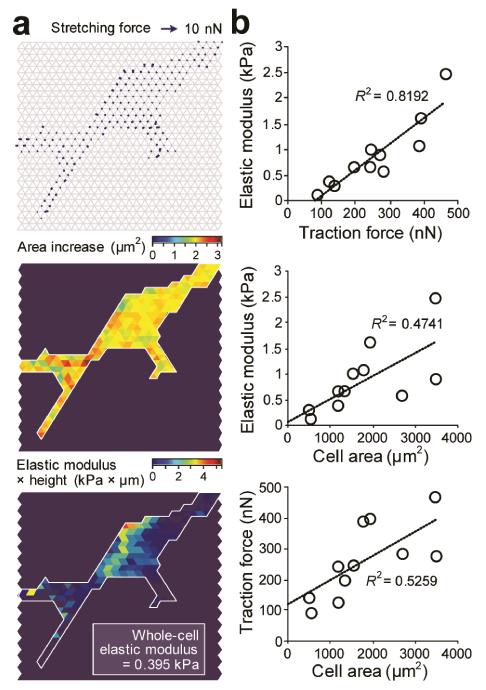
In Fig. 4, we applied the mPAM to characterize subcellular cell stiffness of live single VSMCs. Similar to our measurements in Fig. 3, local cell stretching forces (Fig. 4a, top) and cell area increments (Fig. 4a, middle) associated with static mPAM stretches were first quantified using fluorescence images of the PDMS micropost tops recorded before and immediately after mPAM stretches, from which subcellular stiffness of VSMCs was computed using our computationscheme (Fig. 4a, bottom). We further performed morphometric analysis of cell populations for live single VSMCs. Our results in Fig. 4b revealed positive correlations among the whole-cell average Young’s modulus Eave, total cellular traction force, and cell spread area for live single VSMCs, consistent with our previous study using AFM to study biomechanical properties of human mesenchymal stem cells51. Interestingly, the positive correlation between the whole-cell average Young’s modulus Eave and total cellular traction force was the strongest among the three correlations shown in Fig. 4b, indicating a potential potent role of intracellular cytoskeleton contractility in resisting deformation of cells in response to forces and thus mediating cell stiffness.
Figure 4.
Live-cell subcellular measurement of cell stiffness for vascular smooth muscle cells (VSMCs) and its correlation with cellular traction force and cell spread area. (a) Cell stiffness measurement for a representative live single VSMC. Spatial maps were shown for stretching force (top), cell area increment (middle), and computed subcellular cell stiffness (bottom). (b) Whole-cell elastic modulus plotted against cellular traction force (top) or initial cell spread area before mPAM stretches (middle). The bottom figure plotted cellular traction force against cell spread area before mPAM stretches.
Roles of intracellular cytoskeleton components on cell stiffness of live single VSMCs
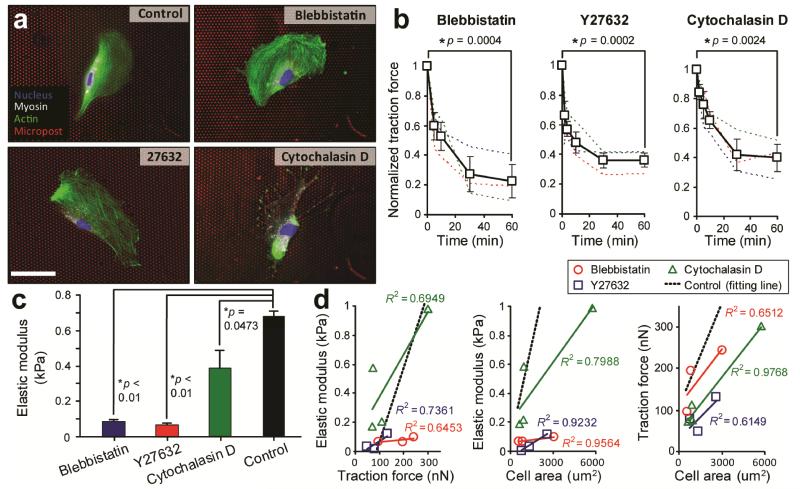
The cytoskeleton consisting of multiple molecular components such as actin microfilaments and myosin II molecules is the primary intracellular structure that determines the cell shape and mechanical stiffness2. We treated VSMCs with small-molecule inhibitors or drugs to target specific intracellular cytoskeleton components and relevant signalling pathways to investigate the functional roles of actin polymerization and myosin II-mediated cytoskeleton contractility in regulating mechanical stiffness of live single VSMCs. Here, for drug-treated VSMCs, blebbistatin (myosin II ATPase inhibitor; 100 μM for 1 hr), Y27632 (Rho-associated kinase (ROCK) inhibitor; 10 μM for 1 hr), and cytochalastin D (inhibitor of actin polymerizatrion; 40 μM for 1 hr) were applied to block myosin II and ROCK activities and actin polymerization, respectively. Immunofluorescence images of drug-treated VSMCs shown in Fig. 5a confirmed that both blebbistatin and Y27632 treatments could decrease expression levels of myosin II without affecting the overall organization of actin microfilaments, while cytochalastin D treatment caused a complete depolymerization and fragmentation of actin microfilaments in VSMCs. Cellular traction forces of drug-treated VSMCs, measured using the PDMS microposts as force sensors, decreased rapidly within the first 20 min after the onset of drug treatments and appeared to reach a static equilibrium state at the end of the 1-hr drug treatments (Fig. 5b). More specifically, after 1 hr treatments with blebbistatin, Y27632, and cytochalastin D, cellular traction forces of live single VSMCs decreased by 77.45%, 63.72%, and 59.99%, respectively.
Figure 5.
Effects of drug treatments to inhibit myosin II and ROCK activities and actin polymerization on cell stiffness of live single vascular smooth muscle cells (VSMCs). (a) Representative immunofluorescence images showing untreated (control) and drug-treated single VSMCs plated on the PDMS micropost array. For drug-treated VSMCs, blebbistatin, Y27632, and cytochalastin D were applied to block myosin II and ROCK activities and actin polymerization, respectively. Cells were stained for nucleus (blue), myosin IIA (white), and actin microfilaments (green). The PDMS microposts were labelled with DiI (red). Scale bar, 20 μm. (b) Temporal evolutions of cellular traction force (normalized to the initial value prior to drug treatments) for individual VSMCs (thin dashed lines) and population means (heavy lines with symbols) under different drug treatment conditions as indicated. Error bars represent ± standard errors of data collected from 3-5 individual VSMCs. (c) Whole-cell elastic modulus of VSMCs that were either untreated (control) or treated with different drugs for 1 hr as indicated. Each p-value calculated using the Student’s t-test was included to indicate statistical significance. (d) Correlations between whole-cell elastic modulus, cellular traction force, and cell spread area for untreated (control) and drug-treated VSMCs as indicated. Data trends in d were plotted using linear least square fitting, with the square of the correlation coefficient R2 indicated.
In Fig. 5c, we applied the mPAM to characterize subcellular cell stiffness of drug-treated live single VSMCs. The whole-cell average Young’s modulus Eave of drug-treated live single VSMCs was significantly less than the value measured for untreated controls, confirming the critical involvements of actin microfilaments and myosin II-mediated cytoskeleton contractility in regulating mechanical stiffness of VSMCs. Interestingly, cell stiffness of VSMCs treated with blebbistatin and Y27632 decreased more significantly as compared to the cells treated with cytochalastin D, again highlighting the importance of intracellular cytoskeleton contractility in mediating cell stiffness. In Fig. 5d, we further examined how drug treatments of VSMCs to block myosin II and ROCK activities and actin polymerization could affect the correlations among cell stiffness, cellular traction force, and cell spread area. Our results in Fig. 5d suggested that the positive correlations among the whole-cell average Young’s modulus Eave, total cellular traction force, and cell spread area were retained even after drug treatments of VSMCs. Further, it appeared that as compared to untreated controls, the elastic modulus of drug-treated VSMCs became less sensitive to changes of total traction force and cell spread area, especially for the cells treated with blebbistatin and Y27632, as reflected by the significantly decreased slopes of the linear fittings for VSMCs treated with blebbistatin and Y27632. The total cellular traction force of drug-treated VSMCs also became less sensitive to changes of cell spread area, even though such changes were less obvious as compared to drug-treated cells in correlations between whole-cell average Young’s modulus Eave and total cellular traction force and whole-cell average Young’s modulus Eave and cell spread area.
Together, our data in Fig. 5 confirmed the important functional roles of actin polymerization and myosin II-mediated cytoskeleton contractility in regulating cell stiffness. Our observations suggested that cytoskeleton contractility, cell shape, and cell stiffness were tightly coupled cellular systems involved together in mediating complex biomechanical functions such as rigidity sensing and force responses.
Conclusion
In conclusion, here we reported for the first time a whole-cell cell stiffness measurement technique with a subcellular spatial resolution, by using a cell stretching device (i.e., the mPAM) that allowed for quantitative control and real-time measurements of mechanical stimuli and cellular biomechanical responses. We applied this mPAM cell stretch platform to exertequibiaxial stressing to single live cells attached on the PDMS micropost tops. The local cell stretching forces and cell deformations generated during the mPAM cell stretch assays could be recorded in real time using live-cell fluorescence microscopy and be converted to a subcellular spatial map of cell stiffness using our computational scheme using the finite element analysis and the iterative Levenberg-Marquardt nonlinear optimization algorithm. Using the mPAM cellstretch assays and our cell stiffness computation scheme, we could determine the whole-cell cell stiffness with a subcellular spatial resolution (a few micrometers), which is not possible with any existing cell stiffness measurment technique. To demonstrate the general application of the mPAM for cell mechanics studies, we performed detailed studies to examine the correlations among cell stiffness, cellular traction force, and cell spread area for live single VSMCs. Our results suggested strong positive correlations among the whole-cell average Young’s modulus Eave, total cellular traction force, and cell spread area for live single VSMCs, suggesting that cell morphology and cell mechanical properties might be strongly coupled cellular systems in mediating complex biomechanical functions and force-sensing mechanisms of cells. Using the mPAM, we further illustrated the important functional roles of actin polymerization and myosin II-mediated cytoskeleton contractility in regulating cell stiffness. Together, our study here demonstrated for the first time a new approach for whole-cell cell stiffness measurements with a subcellular spatial resolution, which would likely facilitate studies of cell mechanics and mechanobiology where real-time live-cell subcellular measurements of cell mechanical propeties such as cellular traction forces and cell stiffness are desired.
Insight, innovation, integration.
The important role of biophysical cues in determining cell behaviours is increasingly recognized. An integrated physical description of cell mechanics will be required to understand the mechano-sensing processes. However, current techniques for measurements of cell mechanics often can only provide single mechanical properties characterizing the whole cell body or local mechanical properties describing a small cell area. This paper reports a novel technology platform that allows for quantitative control and real-time measurements of mechanical stimuli and cellular biomechanical responses. Using this platform, real-time live-cell whole-cell cell stiffness can be mapped with a subcellular spatial resolution. Our research provides a unique research tool for cell mechanics and likely will help explain the complex biomechanical functions and force-sensing mechanisms of cells.
ACKNOWLEDGEMENTS
We acknowledge financial support from the National Science Foundation (CMMI 1129611 & CBET 1149401), the American Heart Association (Scientist Development Grant, 12SDG12180025), and the department of Mechanical Engineering at the University of Michigan, Ann Arbor. We thank P. Mao for his assistance in microfabrication of the silicon micropost array master. We thank M. Yang and C. Chen for sharing the MATLAB program for quantifying cellular traction forces using the PDMS micropost array. We thank N. Chronics for providing us access to PDMS microfabrication equipments in his research laboratory. We also thank J. Mann for the previous development of the mPAM and the CSD platform. We acknowledge the Massachusetts Institute Technology (MIT) Microsystems Technology Laboratories (MTL) for support in microfabrication.
REFERENCES
- 1.Janmey PA, McCulloch CA. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 2.Bao G, Suresh S. Nat Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 3.Lo C-M, Wang H-B, Dembo M, Wang Y.-l. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Developmental Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connelly JT, Gautrot JE, Trappmann B, Tan DWM, Donati G, Huck WTS, Watt FM. Nat Cell Biol. 2010;12:711–U177. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 8.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingber DE. Ann Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 12.Lee GYH, Lim CT. Trends Biotech. 2007;25:111–118. doi: 10.1016/j.tibtech.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Mills JP, Diez-Silva M, Quinn DJ, Dao M, Lang MJ, Tan KSW, Lim CT, Milon G, David PH, Mercereau-Puijalon O, Bonnefoy S, Suresh S. Proc Natl Acad Sci U S A. 2007;104:9213–9217. doi: 10.1073/pnas.0703433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai M, Kita A, Leach J, Rounsevell R, Huang JN, Moake J, Ware RE, Fletcher DA, Lam WA. J Clin Invest. 2012;122:408–418. doi: 10.1172/JCI58753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamara K. B. a. C. D. W. a. I. S. Physics in Medicine and Biology. 2005;50:81. [Google Scholar]
- 16.Wirtz D, Konstantopoulos K, Searson PC. Nature Reviews Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic A, Lynch CD, Sheetz MP. PLoS One. 2009;4:e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Käs J, Ulvick S, Bilby C. Biophys J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nat Nanotech. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 21.Remmerbach TW, Wottawah F, Dietrich J, Lincoln B, Wittekind C, Guck J. Cancer Res. 2009;69:1728–1732. doi: 10.1158/0008-5472.CAN-08-4073. [DOI] [PubMed] [Google Scholar]
- 22.Gavara N, Roca-Cusachs P, Sunyer R, Farré R, Navajas D. Biophys J. 2008;95:464–471. doi: 10.1529/biophysj.107.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trepat X, Deng L, An SS, Navajas D, Tschumperlin DJ, Gerthoffer WT, Butler JP, Fredberg JJ. Nature. 2007;447:592–595. doi: 10.1038/nature05824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Na S, Trache A, Trzeciakowski J, Sun Z, Meininger G, Humphrey J. Ann Biomed Eng. 2008;36:369–380. doi: 10.1007/s10439-008-9438-7. [DOI] [PubMed] [Google Scholar]
- 25.Evans E, Yeung A. Biophys J. 1989;56:151–160. doi: 10.1016/S0006-3495(89)82660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid-Schönbein GW, Sung KL, Tözeren H, Skalak R, Chien S. Biophys J. 1981;36:243–256. doi: 10.1016/S0006-3495(81)84726-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldmann WH. Biotech Lett. 2000;22:431–435. [Google Scholar]
- 28.Thoumine O, Ott A. J Cell Sci. 1997;110:2109–2116. doi: 10.1242/jcs.110.17.2109. [DOI] [PubMed] [Google Scholar]
- 29.Desprat N, Guiroy A, Asnacios A. Rev Sci Instrum. 2006;77:055111–055119. [Google Scholar]
- 30.Mitrossilis D, Fouchard J, Pereira D, Postic F, Richert A, Saint-Jean M, Asnacios A. Proc Natl Acad Sci U S A. 2010;107:16518–16523. doi: 10.1073/pnas.1007940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dao M, Lim CT, Suresh S. Journal of the Mechanics and Physics of Solids. 2003;51:2259–2280. [Google Scholar]
- 32.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Journal of Biomechanics. 2001;34:1545–1553. doi: 10.1016/s0021-9290(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 33.Alenghat FJ, Fabry B, Tsai KY, Goldmann WH, Ingber DE. Biochemical and Biophysical Research Communications. 2000;277:93–99. doi: 10.1006/bbrc.2000.3636. [DOI] [PubMed] [Google Scholar]
- 34.Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ. Nat Mater. 2005;4:557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- 35.Puig-De-Morales M, Grabulosa M, Alcaraz J, Mullol J, Maksym GN, Fredberg JJ, Navajas D. J Applied Physiol. 2001;91:1152–1159. doi: 10.1152/jappl.2001.91.3.1152. [DOI] [PubMed] [Google Scholar]
- 36.Van Vliet KJ, Bao G, Suresh S. Acta Materialia. 2003;51:5881–5905. [Google Scholar]
- 37.Heidemann SR, Wirtz D. Trends in Cell Biology. 2004;14:160–166. doi: 10.1016/j.tcb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saez A, Buguin A, Silberzan P, Ladoux B. Biophys J. 2005;89:L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galbraith CG, Sheetz MP. Proc Natl Acad Sci U S A. 1997;94:9114–9118. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, Kirschner SB, Liu Y, Reich DH, Chen CS. Proc Natl Acad Sci U S A. 2007;104:14553–14558. doi: 10.1073/pnas.0611613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann JM, Lam RH, Weng S, Sun Y, Fu J. Lab Chip. 2012:12. doi: 10.1039/c2lc20896b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagayama K, Adachi A, Matsumoto T. J Biomech. 2011;44:2699–2705. doi: 10.1016/j.jbiomech.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 44.Levenberg K. Quarterly of Applied Mathematics. 1944;2:164–168. [Google Scholar]
- 45.Yang MT, Fu J, Wang YK, Desai RA, Chen CS. Nat Protoc. 2011;6:187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bathe K-J. Finite Element Procedures (Part 1-2) Prentice Hall; 1995. [Google Scholar]
- 47.Wang N, Tytell JD, Ingber DE. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 48.Nekouzadeh A, Pryse KM, Elson EL, Genin GM. J Biomech. 2008;41:2964–2971. doi: 10.1016/j.jbiomech.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu HJ, Lee CF, Locke A, Vanderzyl SQ, Kaunas R. PLoS One. 2010;5:e12470. doi: 10.1371/journal.pone.0012470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shroff SG, Saner DR, Lal R. Am J Physiol. 1995;269:C286–292. doi: 10.1152/ajpcell.1995.269.1.C286. [DOI] [PubMed] [Google Scholar]
- 51.Tee SY, Fu J, Chen CS, Janmey PA. Biophys J. 2011;100:L25–27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]