There is great commercial interest in graphene, and annual production volumes are predicted to grow sharply.[1] The emerging graphene industry is not focused exclusively on monolayers, but is also pursuing related members of the graphene material family that include few-layer graphene (FLG), multi-layer graphene or ultrathin graphite (UTG), graphene oxide (GO), and three-dimensional materials assembled from these two-dimensional building blocks.[2] The manufacture and sale of graphene-based materials will inevitably be accompanied by human exposures, and an effort is underway to assess graphene safety through in vitro[3–6] and in vivo [4,7] methods. Many production methods for FLG and UTG include processing steps where the materials are handled as bulk dry powders, and dust formation and inhalation of these plate-like structures has been proposed as the most important likely exposure pathway.[4,6] As with other nanomaterials, the expense of in vivo toxicity testing and the desire to minimize the use of animals has led to an increased emphasis on mechanistic in vitro methods for screening assessment. These in vitro methods are especially useful in nanotoxicology where a single material may be produced in a large variety of formulations, which in the case of graphene includes materials with different layer numbers, lateral dimensions, and surface chemistries.
Most in vitro methods in toxicology were developed for chemicals rather than engineered nanomaterials, and there is significant potential for these small, high-surface-area materials to induce unanticipated artifacts in nanotoxicology assays. For example, carbon nanotubes have been reported to interfere with fluorescent and colorimetic probes used to assay for cell viability in vitro.[8,9] Nanotubes have also been reported to adsorb and deplete micronutrients and amino acids from cell culture media, giving rise to growth inhibition associated primarily with folic acid deficiency.[10] In general, when immersed in a biological environment, the solid-liquid interface will interact with many components of the surrounding medium, and may develop a layer of adsorbed small molecules, ions, natural organic matter,[11] proteins and/or amino acids,[3,12] which can influence their cytotoxicity or biocompatibility.
To our knowledge, there have been no studies of graphene-induced interferences with in vitro assays. Graphene materials typically have very high specific surface areas (theoretical value of 2630 m2 g−1 for two sides of the ideal monolayer) and many graphene-based materials are hydrophobic. We therefore hypothesize that physical adsorption of biological molecules or synthetic probe dyes will be a major source of artifacts in in vitro toxicity assays. There is an extensive literature on the adsorption of organic or biological molecules from aqueous solution onto graphene based surfaces.[13–15] The affinity of carbon surfaces for organic molecules coupled with the high surface area of graphene materials suggests this issue requires careful study.
We further hypothesize that the tendency of graphene materials to interfere with in vitro assays will vary systematically with the material type. For multilayer graphenes, the specific surface area scales approximately as A ∼ 2630/N m2 g−1, where N is the layer number. Though this does not account for surface roughness or internal microporosity, it is often close to the measured Brunauer–Emmett–Teller (BET) surface areas because graphene materials are relatively smooth and low in porosity. Additionally, graphene oxide is significantly more hydrophilic than pristine graphene or FLG, and will likely show reduced hydrophobic forces, which are often a major driving force for adsorption of organic molecules from aqueous phases. Although there have been no studies on graphene-induced artifacts, the general literature on the graphene-biological interface does give some useful information. Theoretical calculations have predicted the adsorption of the amino acid L-leucine onto graphene,[16] and the binding of four aromatic amino acids to both carbon nanotubes (CNTs) and graphene sheets.[17] Several studies have investigated dye quenching by graphene-based materials.[18,19]
We imagine four possible ways in which graphene materials may interfere with in vitro testing: (1) by adsorbing nutrients and reducing their biological availability; (2) by attenuating either excitation light or fluorescence emission light in biological assays, (3) by quenching fluorescent probes giving false positives or negatives in assays for cell viability or toxicity endpoints, and (4) by physically adsorbing the probe dyes and preventing them from interacting with their biomolecular target.
To study these issues, we assembled a set of three graphene-based nanomaterials of different surface area and surface chemistry (Table 1), and included carbon black as a reference material. These graphene samples were chosen to be representative of the range of properties and geometries in graphene-family materials now under development in industry. For the biochemical interaction study we carried out amino acid and micronutrient profiling in cell culture medium previously exposed to graphene materials and performed a detailed study of the interaction with a model probe, dichlorofluorescein (DCF). DCF and its analogs are commonly used as probes for reactive oxygen species (ROS) production during in vitro toxicity testing,[20] and there have been studies performed using this assay with graphene-family nanomaterials.[21,22,26] Procedures for both the cellular[23] and the acellular[24] assays are reported elsewhere. In brief, typical assays for oxidative stress start with an acetylated version of the compound, such as dichlorodihydrofluorescein diacetate (H2DCFDA), which is uncharged and able to permeate the cell membrane. Once inside, the acetate groups are cleaved by non-specific interactions with endogenous esterases to the charged dichlorodihydrofluorescein derivative, which is colorless and trapped inside by its membrane impermeability. Acellular experiments require that the acetate functional groups are cleaved off using a base such as sodium hydroxide to form the active version of the dye.[25] This version is then oxidized to DCF by the various reactive oxygen species present[25,26] that may be present in the cell; the more ROS present, the stronger the fluorescent signal from the probe. Here we use the fluorescent product (DCF) directly, to study adsorptive and quenching artifacts isolated from the complicating effects of cellular uptake, intracellular cleavage, and intracellular oxidation in complex redox environments that include heme proteins, cytochrome c, and peroxidases.[27]
Table 1.
Characterization of graphene and reference materials used in this study.
| Material Name | Abbreviation | Surface Area [m2 g−1]a) | Lateral Sizeb) | Layer Numberb) | C/O Atomic Ratio by XPSd) |
|---|---|---|---|---|---|
| Graphene Oxide | GO | 2600 | 1–2 um | 1 | 1.70 |
| Few-Layer Graphene | FLG | 670 | 0.8 um | 3–5 | 14.6 |
| Carbon Black | CB | 356 | 16 nmc) | 0 | 10.0 |
| Ultra Thin Graphite | UTG | 125 | 5 um | 20 | 31.6 |
Measured on dry powders by nitrogen vapor adsorption and BET analysis except for GO, which is approximated to have the same surface area as the theoretical value for a graphene monolayer;
Lateral size and layer numbers are typical values from scanning electron microscopy (SEM) analysis, except for GO, which is reported as primarily in monolayer form as determined from step heights by atomic force microscopy (AFM) analysis on deposited films;
Primary particle diameter;
X-ray photoelectron spectroscopy (XPS).
Exposure of RPMI cell culture medium to graphene materials caused only a modest perturbation in amino acid levels (not shown), but significant effects on micronutrients (Figure 1, Part A). Folic acid (vitamin B9), pyridoxine HCl (a form of Vitamin B6), and niacinamide (the amide form of Vitamin B3) are all depleted by exposure to graphene materials, with folic acid depletion occurring at very low graphene doses (<10 ug mL−1). All three micronutrients studied contain planar conjugated units, which favor adsorption by hydrophobic and π−π interactions.[28,29] The preferential adsorption of folic acid correlates with its very low water solubility (1.6 mg L−1) compared to pyridoxine HCl (2 × 105 mg L−1) and niacinamide (5 × 105 mg L−1). For organic compounds there is a crude correlation between water solubility and adsorption sometimes referred to as Lundelius' rule,[30] which reflects the important role hydrophobic interactions typically play in both phenomena. Folic acid in particular has an extended planar conjugated structure capable of π−π interactions on carbon surfaces.[10] Folic acid depletion at these levels has already been shown to lead to growth inhibition in human HepG2 cells, which are not capable of producing their own folic acid, leading to a ‘starvation’ toxicity mechanism.[10] As these effects are determined by folate levels and not by the direct action of nanomaterials on cells, the previously published study[11] together with the present Figure 1 provide sufficient information to identify the doses where these effects will occur (at graphene mass doses above 1–10 ug/mL depending on surface area). The authors are not aware of any studies of graphene or other nanomaterials inducing this effect in vivo, but expect that folic acid may be replenished by the systematic circulation, so depletion is likely to be a transient effect. In cell culture, in contrast, there is a fixed, finite pool of folic acid, and a single adsorption event proceeding to equilibrium can lead to permanent depletion and cell growth inhibition.[10] We believe this is best regarded as an in vitro artifact, since this irreversible depletion mechanism does not accurately model physiological behavior in vivo.
Figure 1.
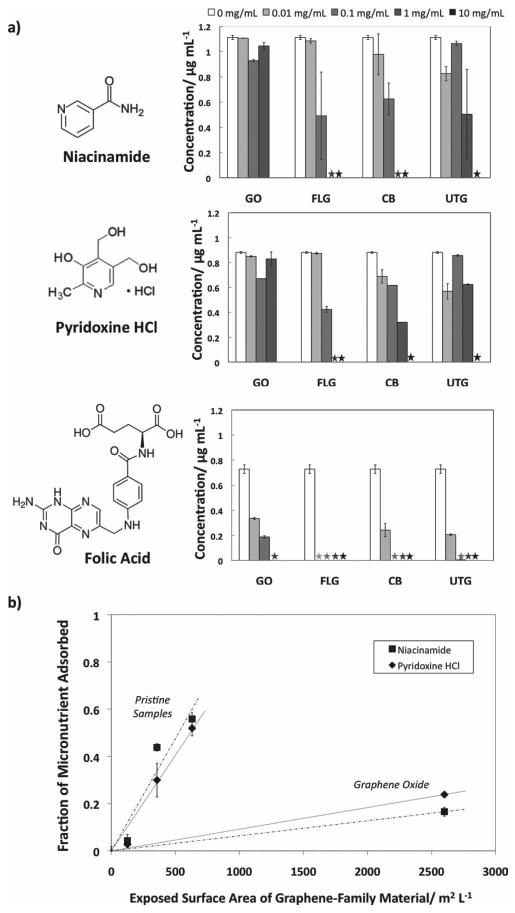
Interactions of graphene materials with essential micronutrients. A) Vitamin profiles of RPMI cell culture media after exposure to graphene and reference materials. Values are reported with a 95% confidence interval. Asterisks indicate that the concentration was below the detection limit. Folic acid becomes significantly depleted at low material doses typical of toxicology studies (∼10 ug mL−1), while pyridoxine HCl or niacinamide are depleted primarily at higher doses. B) Trends of micronutrient adsorption. Concentration of 1 mg mL−1 graphene material is used as an example. When looking at a single sample concentration of graphene material, it can be seen that there are two distinct trends. The pristine, more hydrophobic materials adsorb at a much higher rate than the highly oxidized GO material.
Micronutrient depletion depends on specific material properties (Figure 1B). Among the pristine (unfunctionalized) nanocarbons (FLG, UTG, and carbon black) the extent of adsorption correlates well with total BET surface area. Adsorption on graphene oxide is much less despite its very high total surface area (Figure 1). This is likely due to oxygen-containing functional groups (C/O ∼ 1.7)[31,32] that reduce the total area of hydrophobic surface, and break the remaining hydrophobic surface into small patches that may or may not allow dye adsorption. Water clusters that are hydrogen bonded to those functional groups can also partially restrict solute access to the hydrophobic domains.[31,33] It has also been proposed that acidic oxygen groups remove electrons from the π-electron system of the basal plane, which interferes and weakens the dispersion interactions between the aromatic rings and the π electrons of the graphene surface.[34] Hydrogen bonding is sometimes cited as a mechanism for physical adsorption from aqueous solution,[29] but the lower adsorptive capacity of GO indicates that this is not the predominant mechanism here. It has been previously noted that CNTs with larger inner diameters and therefore flatter graphitic sidewalls allow for better π-stacking of aromatic molecules and will therefore be better adsorbents.[35] It is interesting that the micronutrient absorptivity of graphene in this study is higher than that reported previously for single-walled CNTs.[10] The ratio of the two slopes in Figure 1B is approximately 10:1; suggesting that when compared to FLG, GO surfaces contain approximately 10% hydrophobic surfaces in patches large enough to adsorb the two smaller micronutrients.
Graphene materials may also cause optical interferences with in vitro assays. Part A of Figure 2 shows the potential of graphene materials to extinguish light at an optical path length of 1 cm. GO has photoluminescent behavior[36] and preferentially absorbs light in the blue region. The FLG sample has different opto-electronic properties and, when dispersed uniformly in solution using serum proteins, is optically grey in the visible spectrum (Figure 2).
Figure 2.
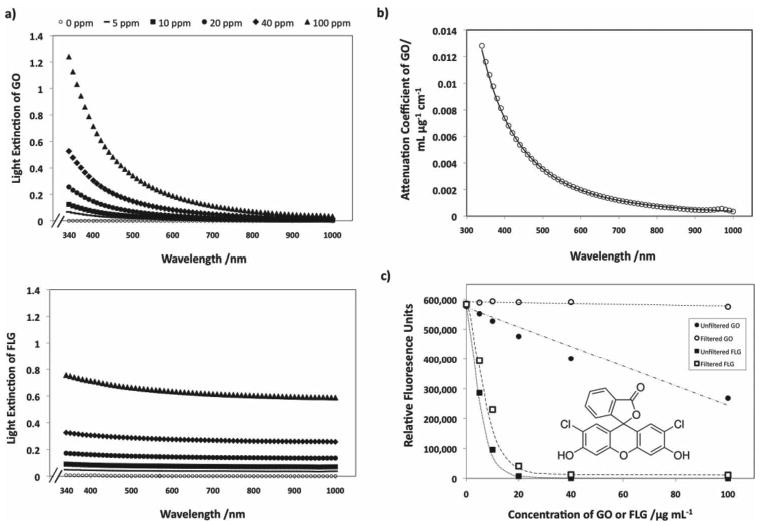
Capability of graphene materials to interfere with light during in vitro assays. A) Dose dependent optical absorption spectra for FLG (above) and GO (below) samples in PBS at a path length of 1 centimeter. GO is naturally hydrophilic and readily disperses, FLG was predispersed using 0.5% fetal bovine serum (FBS) by volume. B) The wavelength-dependent adsorptivity coefficient for GO, with units of mL ug−1 cm−1, as fit by the function 2 × 106λ−3.27, where λ is the wavelength in nm. FLG was found to have a specific attenuation coefficient of 0.0073 ± 0.0013 mL ug−1 cm−1, approximately independent of wavelength. C) Fluorescence quenching of the ROS molecular probe, dichlorofluorescein (DCF) by two graphene materials. DCF was prepared at a concentration of 10 ug mL−1 in PBS buffer (pH 7.4). The fluorescence of DCF dye is only affected when measured in the presence of GO; upon removal, the signal is restored to that of the control. There is only a partial recovery of fluorescent signal in the case of DCF, indicating that most of the quenching is caused by physical adsorption of the dye, which is removed along with the FLG by filtration.
The wavelength-dependent attenuation coefficient for GO is shown in Part B of Figure 2 and is well described by AC = 2 × 106λ−3.27, with λ in nm and AC in mL ug−1 cm−1. The FLG data is adequately described by a mass attenuation coefficient of 0.0073 ± 0.0013 mL ug−1 cm−1. Even at low concentrations (<20 ug−1 mL−1) these effects are potentially significant. It is important to note that these effects can vary greatly with experimental configuration. Extinction is the sum of absorption and scattering, which are strongly dependant on optical path length, and the extent of scattered light captured also depends on the distance between the sample and detector.
Graphene may also interfere with optical assays through specific adsorption or quenching of dyes. To study these effects we chose dichlorofluorescein (DCF) as a model fluorescent dye, whose analogues are commonly used to assay for reactive oxygen species (ROS), and studies in the literature have used this assay with graphene-family nanomaterials.[21,22] Potential artifacts that can be induced in this assay by engineered nanoparticles have been studied elsewhere,[26,37] but did not include graphene materials. Experiments were done using GO and FLG, chosen with the intent to bracket the panel of materials as the strongest and weakest adsorbent.
Figure 2 Part C shows that graphene materials suppress DCF fluorescence at doses as low as 5 ug mL−1. Fluorescence intensities were measured after 30 min of incubation with the graphene and both before and after the materials were removed using a PTFE filter. Kinetics studies (not shown) indicated that at least 90% of the total physical adsorption occurs within the first 30 min of incubation. At equal mass dose, the FLG sample is a much more effective fluorescence quencher for DCF dye than GO.
GO is only able to affect fluorescence if it is in solution during the measurement. Removal of GO by filtration restores the fluorescence back to the original intensity, indicating that the amount of DCF dye adsorbed by the graphene oxide is below the detectible limit. The signal loss in the GO experiment must therefore be dominated by optical extinction or by fluorescence quenching of solution -phase molecules within Förster resonance energy transfer (FRET) distances of suspended GO. This FRET-based mechanism of quenching is shown to be unimportant at these material doses (Supporting Information). In contrast, optical absorption is estimated to be of the same order of magnitude as the observed effect, based on the mass attenuation coefficients reported in Figure 2a applied to both the excitation and emission wavelengths for DCF used in this study (485 and 525 nm, respectively). We thus propose that the combined absorption of excitation and emission light by GO gives rise to the signal loss shown in Figure 2.
As can be seen in Figure 2C, FLG is a much more effective inhibitor of the fluorescence signal than GO at an equal mass dose. The mechanism, however, is not the same; removal of FLG recovers only a fraction of the fluorescence signal suggesting that the unrecovered portion of the fluorescence is from dye that adsorbed on the material (which is consequently removed with the graphene during the filtering process). Dye quenching in contact with graphene surfaces is well established.[8,18,38] Quenching due to adsorption of the dye is responsible for more than two-thirds the signal loss seen at each of the concentrations studied, and can therefore be considered the dominant effect. The remaining quenching effect can be predicted using the light attenuation coefficient for FLG reported in Figure 2A. The loss of signal predicted due to these optical interferences is within 5% of the fluorescence quenching measured that cannot be attributed to the adsorbed dye. Quenching of dye molecules by long-range resonance energy transfer from the FLG sheets was also considered as a contributing method,[19] but was not found to be significant (a more detailed explanation is included in Supporting Information).
Physical adsorption not only leads to dye quenching, but is also important in its own right, since it removes fluorescent probes from solution and can prevent their intended interaction with the biological target molecule and subsequent conversion to fluorescent form. Investigators frequently wash the cells prior to adding a molecular probe,[23] an effort which can help remove the graphene nanomaterials from the system and avoid these adsorptive artifacts. However, in some cases, the chemical target for the probe is in the extracellular medium, and may be removed with washing, or, washing steps will not remove any of the graphene nanomaterials that have been internalized by the cells during the exposure time, nor any that have accumulated on the cellular membrane, as described by others.[22] We therefore measured the extent of physical adsorption of DCF onto FLG as a function of material dose and in the presence of potentially competing adsorbates (Figure 3). Three cases were considered, the first of which was adsorption of DCF onto FLG in PBS buffer, without any competing organic solutes (bottom curve).
Figure 3.
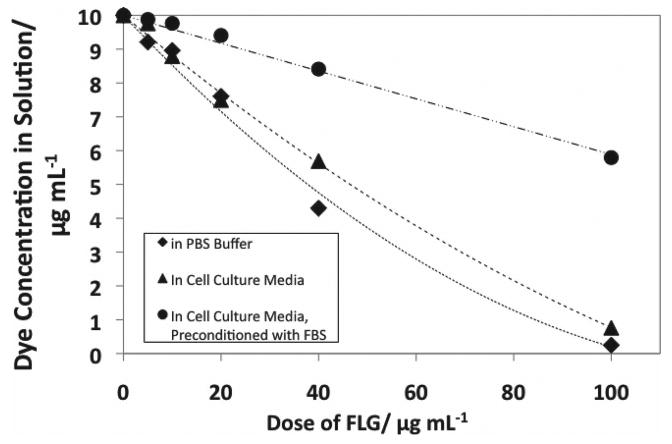
Depletion of DCF dye as a function of material dose by physical adsorption onto graphene materials. Dye was prepared at a concentration of 10 ug mL−1 in PBS buffer (pH 7.4) or cell culture media (pH 7.4) and concentration was calculated from the measured extinction at 500 nm wavelength of light, after filtering out the FLG samples.
We hypothesized that pre-incubation of FLG with cell culture medium or serum- supplemented cell culture medium would decrease physical adsorbtion of DCF dye. FLG was incubated in (1) cell culture medium or (2) cell culture medium supplemented with 10 v-% fetal bovine serum (FBS) overnight. The pre-conditioned FLG was then diluted into cell culture medium containing the DCF dye. As can be seen in Figure 3, pre-conditioning significantly reduces dye adsorption by FLG, by 9% for media pre-conditioning and by 60% for serum pre-conditioning. Increasing the amount of FBS in the cell culture media beyond 10% had a negligible effect on the amount of dye adsorbed (data not shown). Pre-conditioning graphene-based nanomaterials with FBS has been reported to mitigate cytotoxicity[3] and has the added benefit of increasing dispersion stability. Colloidal stability can also contribute to biocompatibility,[39] as well as lessening reaggregation and gravitational settling of the graphene platelets during experiments, allowing for more consistency in the delivered dose. Supplmentation using FBS is used to improve dispersion stability, and similar effects have been observed for the use of surfactants to disperse CNTs.[40]
We can also use our results to explore the effects of intracellular adsorption which may occur when DCFDA is used to probe intracellular ROS production. Cellular uptake increases the concentration of the dye to several hundred times that in the extracellular medium,[41] and elevated concentrations lead to larger fractions of adsorbed dye and potentially more severe adsorptive artifacts (see analysis in Supporting Information). At the elevated intracellular concentrations, adsorption on graphene may approach saturation, and this amount can be estimated if the adsorbed dye is limited to a monolayer. A rough estimate of the space-filling 2D area of the DCF molecule can be made by assuming it to be equivalent to the area of 9 benzene rings (the amount needed to overlay and completely cover one DCF molecule), or approximately 5.5 × 10−19 m2 per molecule or 830 m2 g−1 of DCF. Our FLG sample has a surface area of 670 m2 g−1, giving an estimated maximum adsorption capacity of over 800 mg of DCF dye per gram of FLG adsorbent. This high adsorption capacity could lead to a significant reduction in the effective concentration of dye within the cell and underestimation of the intracellular ROS activity.
In summary, graphene-based materials have significant potential to interfere with in vitro toxicity testing methods through both optical and adsorptive effects at toxicologically relevant doses (less than 10–100 ug/mL) (Figure 4). Optical interferences can occur for both few-layer graphene and graphene oxide. The specific attenuation coefficients for each in units of mL ug−1 cm−1 are 0.0073 ± 0.0013 for few layer graphene and 2 × 106λ−3.266, where λ is the wavelength in nm, for graphene oxide. The effects of light extinction by graphene materials can be very significant and in general will be more important in long path length (large volume) experiments than in short-path-length microplate experiments. Removal of the nanomaterial by washing and/or centrifugation can remove optical interferences, and would lead to accurate data here for GO since it adsorbs the DCF dye to only a limited extent. FLG in contrast adsorbs and quenches the DCF dye in a manner that depends on surface area (1/layer-number) and can lead to underreporting of ROS whether the material is removed or not. Intracellular concentrations of dye can be significantly higher than in the extracellular space, and the natural scaling laws for adsorption suggest the intracellular artifacts will be more pronounced. We show that soaking the FLG in cell culture media with serum prior to testing may be a practical method for reducing these adsorptive artifacts.
Figure 4.
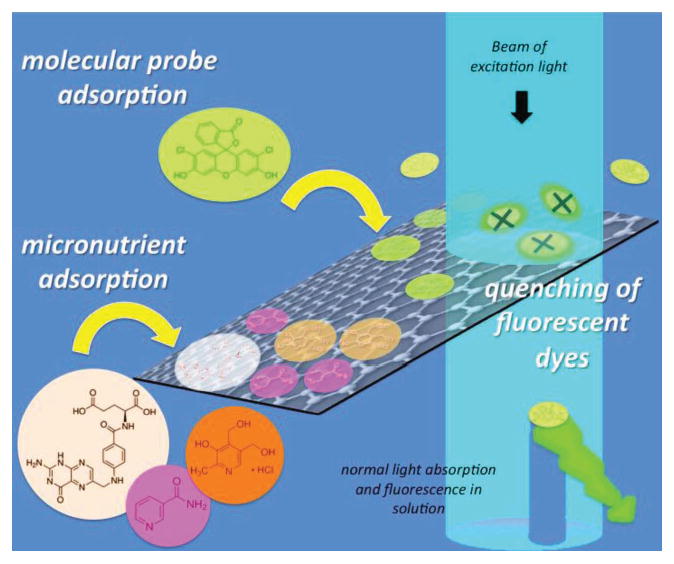
Summary of interferences and potential artifacts caused by graphene materials in cell culture experiments. Physical adsorption on abundant hydrophobic surfaces of graphene materials causes depletion of micronutrients and indirect toxicity, and the static fluorescence quenching of molecular probes can give false positives or negatives.
Folic acid depletion of cell culture medium is observed here for a 3–5 layer FLG sample at doses lower than 10 ug mL−1, which makes FLG more active than single-walled carbon nanotubes reported previously.[10] Folate adsorption at these levels has already been shown to lead to apparent toxicity in the form of growth inhibition of human liver cells in culture.[11] Similar material doses also lead to strong adsorption of the ROS probe, DCF, which in turn lead to fluorescence quenching and underreporting of ROS activity. Data are provided here on four biologically important solutes (folic acid, pyridoxine, niacinimide, and DCF) and in all cases we observe a much stronger adsorption on FLG than on the hydrophilic graphene oxide. This suggests the dominance of hydrophobic forces and π-interactions on adsorption in these systems. Among the three non-functionalized (pristine) graphenic materials, the extent of adsorption correlates with total surface area. The extreme aspect ratio of graphene materials makes the edge contributions to total area insignificant, and thus total area scales as A ∼ 1/N, with a theoretical maximum of 2630 m2 g−1 for N equal to unity. Graphene materials with fewer layers and more hydrophobic (pristine) surfaces will likely be the most problematic in biological experiments. It is important to note that the adsorptive and optical phenomena documented here are not the only mechanisms through which graphene materials can drive cellular responses. Graphene materials of small lateral dimension are known to enter cells[6] and may initiate toxicity pathways through separate mechanisms such as membrane disruption or oxidative stress. It is difficult to apply the present findings to previously published studies, and thus we can make no claims that any particular published dataset has been influenced by adsorptive or optical artifacts. The present study does, however, document artifacts in a systematic way that may allow other researchers to avoid them through intelligent use of controls or protein pre-coating of graphene surfaces by preconditioning in serum.[3] In general, great care must be taken to validate assays and characterize media alterations if we are to obtain an accurate picture of the potential of graphene materials to produce adverse biological responses.
Experimental Section
Materials and Characterization
Graphene oxide was synthesized by a modified Hummer's method and characterized by atomic-force microscopy and SEM as described previously.[42] In aqueous suspension it exists primarily as monolayers with mean lateral dimensions between 1 and 2 μm.[42] Few-layer graphene (3–5 layers), ultrathin graphite ∼20 layers), and carbon black reference samples were obtained commercially and characterized. Available surface area was determined by application of BET theory to nitrogen vapor adsorption (Autosorb-1, Quantachrome, Boynton Beach, FL). Nominal size was determined by using a LEO 1530 scanning electron microscope. Elemental analysis was determined using an Elemental Combustion System (COSTECH instruments, Valencia, CA). Surface characterization was done by X-ray photoelectron spectroscopy using a Perkin Elmer 5500 Surface Analyzer. Cell culture media (RPMI medium 1640–11875) and phosphate buffered saline (PBS) were obtained from Invitrogen Corporation (Carlsbad, CA).
Amino Acid and Vitamin Profiling
Graphene materials or reference materials were added to serum-free RPMI 1640–11875 (Invitrogen Co) at doses ranging from 0.01 to 10 mg mL−1. Once added, the mixtures were protected from light, sonicated for 30 min to ensure dispersion, and left on lab rollers for 2 h to ensure equilibrium had been reached. Nanomaterials were separated from cell culture media by centrifugal ultrafiltration using Amicon Ultra-15 filter tubes. Samples were then frozen at −20 °C and spent media analysis was conducted at Irvine Scientific (Santa Ana, CA). Amino acid concentrations were determined by high-performance liquid chromatography with UV detection. All samples were tested in triplicate.
Interference with Molecular Probe Dyes
Graphene nanomaterials were prepared in a stock solution with PBS buffer, cell culture media, or cell culture media supplemented with 10% FBS by volume and allowed to equilibrate for at least 12 h. The graphene nanomaterials were then diluted into a working solution with a final concentration 10 ug mL−1 dichlorofluorescein dye (Sigma Aldrich, St. Louis, MO) in PBS buffer or cell culture medium (pH 7.4). Fluorescence was measured using a Spectramax M2 (Molecular Devices, Dowington PA) microplate reader, and UV-visible absorbance was measured using a Jasco V-630 spectrophotometer (Jasco, Oklahoma City, OK). Measurements were made both with carbon nanomaterials in solution and after removal using a Millex 0.2 μm PTFE syringe filter (Millipore, Billerica, MA). Carbon nanomaterials could not be detected in the filtrate when using UV-visible absorbance. Conversion of fluorescence or absorption to dye concentration was done using a calibration curve constructed from the measurements of known dye concentrations.
Supplementary Material
Acknowledgments
Acknowledgments are made to the NIEHS R01 ES016178 Grant, NSF CBET-1132446, NIEHS Superfund Research Program P42 ES013660, US Department of Education through the GAANN Award, and CONACYT-Estancias Posdoctorales y Sabáticas al Extranjero para la Consolidación de Grupos de Investigación (propuesta:165215), and to NSF (DMR 0955612). The authors are grateful for technical assistance from Dr. Indrek Kulaots and Dr. Vanesa Sanchez of Brown University.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Megan A. Creighton, School of Engineering, Brown University, Providence, RI 02912, USA
Prof. J. Rene Rangel-Mendez, División de Ciencias Ambientales, Instituto Potosino de Investigación Científica y Tecnológica, San Luis Potosí, 78216, Mexico
Prof. Jiaxing Huang, Department of Materials Science and Engineering, Northwestern University, Evanston, IL 60208, USA
Prof. Agnes B. Kane, Department of Pathology and Laboratory Medicine, Brown University, Providence, RI 02912, USA
Prof. Robert H. Hurt, Email: Robert_Hurt@Brown.edu, School of Engineering, Brown University, Providence, RI 02912, USA
References
- 1.Segal M. Nat Nanotechnol. 2009;4:611. doi: 10.1038/nnano.2009.161. [DOI] [PubMed] [Google Scholar]
- 2.a) Lee SH, Kim HW, Hwang JO, Lee WJ, Kwon J, Bielawski CW, Ruoff RS, Kim SO. Angew Chem Int Ed. 2010;49:10084. doi: 10.1002/anie.201006240. [DOI] [PubMed] [Google Scholar]; b) Lee SH, Lee DH, Lee WJ, Kim SO. Adv Funct Mater. 2011;21:1338. [Google Scholar]; c) Zhou WW, Zhu JX, Cheng CW, Liu JP, Yang HP, Cong CX, Guan C, Jia XT, Fan HJ, Yan QY, Li CM, Yu T. Energ Environ Sci. 2011;4:4954. [Google Scholar]
- 3.Hu W, Peng C, Lv M, Li X, Zhang Y, Chen N, Fan C, Huang Q. ACS Nano. 2011;5:3693. doi: 10.1021/nn200021j. [DOI] [PubMed] [Google Scholar]
- 4.Schinwald A, Murphy FA, Jones A, MacNee W, Donaldson K. ACS Nano. 2012;6:736. doi: 10.1021/nn204229f. [DOI] [PubMed] [Google Scholar]
- 5.a) Chang Y, Yang ST, Liu JH, Dong E, Wang Y, Cao A, Liu Y, Wang H. Toxicol Lett. 2011;200:201. doi: 10.1016/j.toxlet.2010.11.016. [DOI] [PubMed] [Google Scholar]; b) Sasidharan A, Panchakarla LS, Sadanandan AR, Ashokan A, Chandran P, Girish CM, Menon D, Nair SV, Rao CN, Koyakutty M. Small. 2012;8:1251. doi: 10.1002/smll.201102393. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez VC, Jachak A, Hurt RH, Kane AB. Chem Res Toxicol. 2012;25:15. doi: 10.1021/tx200339h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Yang K, Wan J, Zhang S, Zhang Y, Lee ST, Liu Z. ACS Nano. 2011;5:516. doi: 10.1021/nn1024303. [DOI] [PubMed] [Google Scholar]; b) Zhang XY, Yin JL, Peng C, Hu WQ, Zhu ZY, Li WX, Fan CH, Huang Q. Carbon. 2011;49:986. [Google Scholar]
- 8.Casey A, Herzog E, Davoren M, Lyng FM, Byrne HJ, Chambers G. Carbon. 2007;45:1425. doi: 10.1016/j.tiv.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.a) Monteiro-Riviere NA, Inman AO, Zhang LW. Toxicol Appl Pharmacol. 2009;234:222. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]; b) Davoren M, Herzog E, Casey A, Cottineau B, Chambers G, Byrne HJ, Lyng FM. Toxicol In Vitro. 2007;21:438. doi: 10.1016/j.tiv.2006.10.007. [DOI] [PubMed] [Google Scholar]; c) Worle-Knirsch JM, Pulskamp K, Krug HF. Nano Lett. 2006;6:1261. doi: 10.1021/nl060177c. [DOI] [PubMed] [Google Scholar]
- 10.Guo L, Bussche AV, Buechner M, Yan AH, Kane AB, Hurt RH. Small. 2008;4:721. doi: 10.1002/smll.200700754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 12.a) Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. ACS Nano. 2011;5:7503. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]; b) Xia XR, Monteiro-Riviere NA, Mathur S, Song X, Xiao L, Oldenberg SJ, Fadeel B, Riviere JE. ACS Nano. 2011;5:9074. doi: 10.1021/nn203303c. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. ACS Nano. 2010;4:3623. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]; d) Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc Natl Acad Sci USA. 2008;105:14265. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Zhang WJ, Zhou CJ, Zhou WC, Lei AH, Zhang QL, Wan Q, Zou BS. Environ Contam Tox. 2011;87:86. doi: 10.1007/s00128-011-0304-1. [DOI] [PubMed] [Google Scholar]; b) Yang ST, Chen S, Chang YL, Cao AN, Liu YF, Wang HF. J Colloid Interf Sci. 2011;359:24. doi: 10.1016/j.jcis.2011.02.064. [DOI] [PubMed] [Google Scholar]; c) Liu T, Li Y, Du Q, Sun J, Jiao Y, Yang G, Wang Z, Xia Y, Zhang W, Wang K, Zhu H, Wu D. Colloids Surf B—Biointerfaces. 2012;90:197. doi: 10.1016/j.colsurfb.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, Cai X, Tan S, Li H, Liu J, Yang W. Chem Eng J. 2011;173:144. [Google Scholar]
- 15.a) Gao Y, Li Y, Zhang L, Huang H, Hu J, Shah SM, Su X. J Colloid Interf Sci. 2011;368:540. doi: 10.1016/j.jcis.2011.11.015. [DOI] [PubMed] [Google Scholar]; b) Li YH, Zhang P, Du QJ, Peng XJ, Liu TH, Wang ZH, Xia YZ, Zhang W, Wang KL, Zhu HW, Wu DH. J Colloid Interf Sci. 2011;363:348. doi: 10.1016/j.jcis.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Qin W, Li X, Bian WW, Fan XJ, Qi JY. Biomaterials. 2010;31:1007. doi: 10.1016/j.biomaterials.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Rajesh C, Majumder C, Mizuseki H, Kawazoe Y. J Chem Phys. 2009;130:124911. doi: 10.1063/1.3079096. [DOI] [PubMed] [Google Scholar]
- 18.a) Matte HSSR, Subrahmanyam KS, Rao KV, George SJ, Rao CNR. Chem Phys Lett. 2011;506:260. [Google Scholar]; b) Kim J, Cote LJ, Kim F, Huang JX. J Am Chem Soc. 2010;132:260. doi: 10.1021/ja906730d. [DOI] [PubMed] [Google Scholar]; c) Artiles MS, Rout CS, Fisher TS. Adv Drug Deliv Rev. 2011;63:1352. doi: 10.1016/j.addr.2011.07.005. [DOI] [PubMed] [Google Scholar]; d) Shao YY, Wang J, Wu H, Liu J, Aksay IA, Lin YH. Electroanal. 2010;22:1027. [Google Scholar]; e) Kuila T, Bose S, Khanra P, Mishra AK, Kim NH, Lee JH. Biosens Bioelectron. 2011;26:4637. doi: 10.1016/j.bios.2011.05.039. [DOI] [PubMed] [Google Scholar]; f) Liu Y, Liu CY, Liu Y. Appl Surf Sci. 2011;257:5513. [Google Scholar]
- 19.Swathi RS, Sebastian KL. J Chem Phys. 2009;130:086101. doi: 10.1063/1.3077292. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ID, Haugland RP, Spence MTZ. The Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 11th. Life Technologies; Carlsbad, CA: 2010. [Google Scholar]
- 21.a) Li Y, Liu Y, Fu YJ, Wei TT, Le Guyader L, Gao G, Liu RS, Chang YZ, Chen CY. Biomaterials. 2012;33:402. doi: 10.1016/j.biomaterials.2011.09.091. [DOI] [PubMed] [Google Scholar]; b) Zhang YB, Ali SF, Dervishi E, Xu Y, Li ZR, Casciano D, Biris AS. ACS Nano. 2010;4:3181. doi: 10.1021/nn1007176. [DOI] [PubMed] [Google Scholar]
- 22.Sasidharan A, Panchakarla LS, Sadanandan AR, Ashokan A, Chandran P, Girish CM, Menon D, Nair SV, Rao CNR, Koyakutty M. Small. 2012;8:1251. doi: 10.1002/smll.201102393. [DOI] [PubMed] [Google Scholar]
- 23.Rosenkranz AR, Schmaldienst S, Stuhlmeier KM, Chen WJ, Knapp W, Zlabinger GJ. J Immunol Methods. 1992;156:39. doi: 10.1016/0022-1759(92)90008-h. [DOI] [PubMed] [Google Scholar]
- 24.Keston AS, Brandt R. Analyt Biochem. 1965;11:1. doi: 10.1016/0003-2697(65)90034-5. [DOI] [PubMed] [Google Scholar]
- 25.Jakubowski W, Bartosz G. Cell Biol Int. 2000;24:757. doi: 10.1006/cbir.2000.0556. [DOI] [PubMed] [Google Scholar]
- 26.Doak SH, Griffiths SM, Manshian B, Singh N, Williams PM, Brown AP, Jenkins GJ. Mutagenesis. 2009;24:285. doi: 10.1093/mutage/gep010. [DOI] [PubMed] [Google Scholar]
- 27.a) Halliwell B, Whiteman M. Brit J Pharmacol. 2004;142:231. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bonini MG, Rota C, Tomasi A, Mason RP. Free Radical Bio Med. 2006;40:968. doi: 10.1016/j.freeradbiomed.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 28.Radovic L, editor. Chemistry and Physics of Carbon. Vol. 27. Marcel Dekker; New York: 2000. [Google Scholar]
- 29.Yang K, Xing B. Chem Rev. 2010;110:5989. doi: 10.1021/cr100059s. [DOI] [PubMed] [Google Scholar]
- 30.Lundelius EF. Kolloid Z. 1920;26:145. [Google Scholar]
- 31.Erickson K, Erni R, Lee Z, Alem N, Gannett W, Zettl A. Adv Mater. 2010;22:4467. doi: 10.1002/adma.201000732. [DOI] [PubMed] [Google Scholar]
- 32.Cote LJ, Kim J, Tung VC, Luo JY, Kim F, Huang JX. Pure Appl Chem. 2011;83:95. [Google Scholar]
- 33.Moreno-Castilla C. Carbon. 2004;42:83. [Google Scholar]
- 34.Coughlin RW, Ezra FS. Environ Sci Technol. 1968;2:291. [Google Scholar]
- 35.a) Liu Z, Sun XM, Nakayama-Ratchford N, Dai HJ. ACS Nano. 2007;1:50. doi: 10.1021/nn700040t. [DOI] [PubMed] [Google Scholar]; b) Gotovac S, Honda H, Hattori Y, Takahashi K, Kanoh H, Kaneko K. Nano Lett. 2007;7:583. doi: 10.1021/nl0622597. [DOI] [PubMed] [Google Scholar]
- 36.Eda G, Lin YY, Mattevi C, Yamaguchi H, Chen HA, Chen IS, Chen CW, Chhowalla M. Adv Mater. 2010;22:505. doi: 10.1002/adma.200901996. [DOI] [PubMed] [Google Scholar]
- 37.Pal AK, Bello D, Budhlall B, Rogers E, Milton DK. Dose Response. 2012;10:308. doi: 10.2203/dose-response.10-036.Pal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.a) Treossi E, Melucci M, Liscio A, Gazzano M, Samori P, Palermo V. J Am Chem Soc. 2009;131:15576. doi: 10.1021/ja9055382. [DOI] [PubMed] [Google Scholar]; b) Sheng L, Ren J, Miao Y, Wang J, Wang E. Biosens Bioelectron. 2011;26:3494. doi: 10.1016/j.bios.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 39.Duch MC, Budinger GRS, Liang YT, Soberanes S, Urich D, Chiarella SE, Campochiaro LA, Gonzalez A, Chandel NS, Hersam MC, Mutlu GM. Nano Lett. 2011;11:5201. doi: 10.1021/nl202515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Xia T, Ntim SA, Ji Z, George S, Meng H, Zhang H, Castranova V, Mitra S, Nel AE. ACS Nano. 2010;4:7241. doi: 10.1021/nn102112b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. J Immunol. 1983;130:1910. [PubMed] [Google Scholar]
- 42.a) Guo F, Kim F, Han TH, Shenoy VB, Huang J, Hurt RH. ACS Nano. 2011;5:8019. doi: 10.1021/nn2025644. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim F, Luo JY, Cruz-Silva R, Cote LJ, Sohn K, Huang JX. Adv Funct Mater. 2010;20:2867. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


