Abstract
HIV can infiltrate the brain and lead to HIV-associated neurocognitive disorders (HAND). The pathophysiology of HAND is poorly understood, and there are no diagnostic biomarkers for it. Previously, an increase in inducible nitric oxide synthase levels and protein tyrosine nitration in the brain were found to correlate with the severity of HAND.1,2 In this study, we analyzed human brains from individuals who had HIV infection without encephalitis and with encephalitis/HAND and compared them to the brains of healthy individuals. We identified the nitrated proteins and determined the sites of modification using affinity enrichment followed by high-resolution and high-mass-accuracy nanoLC–MS/MS. We found that nitrated proteins were predominantly present in the HIV-infected individuals with encephalitis, and, interestingly, the modifications were predominantly located on immunoglobulin variable regions. Our molecular model indicated potential interactions with HIV envelope proteins and changes on the heavy and light chain interface upon the nitration and nitrohydroxylation of these residues. Therefore, our findings suggest a role for these modifications in the immune response, which may have implications in disease pathogenesis.
Keywords: HIV, nitroproteome, HIV-associated neurocognitive disorders, nitration, mass spectrometry, antibody, immune response
Introduction
Human immunodeficiency virus (HIV) infiltrates the blood-brain barrier and infects macrophages/microglia and astrocytes in the brain. During disease progression, patients show various levels of neurocognitive impairment, which is collectively termed HIV-associated neurocognitive disorders (HAND). Interestingly, there is no convincing evidence of HIV infection of the neurons. Thus, neuronal dysfunction has been linked to the indirect effects of factors released by HIV-infected or activated glial cells.3
Nitric oxide (NO), generated by nitric oxide synthase, is an important cellular messenger in signal transduction. In the HIV-infected brain, the expression of inducible nitric oxide synthase is increased, leading to elevated amounts of NO.1 Additionally, an imbalance between oxidants and reductants is observed.4,5 NO is known to react with superoxide to form peroxynitrite, which nitrates protein tyrosine and tryptophan residues through multiple mechanisms.6,7 Nitration of tyrosine can change the activity of enzymes and interfere with inter- and intramolecular interactions.8 Nitration of tryptophan was discovered more recently, and its functional consequences are not well-defined.7,9
Tyrosine nitration has been implicated in various neurodegenerative diseases such as Alzheimer’s disease,10 amyotrophic lateral sclerosis,11 and Parkinson’s disease.12 Moreover, it has been detected in vivo during an inflammatory challenge.13 The majority of patients with HAND show HIV encephalitis at autopsy,14 and elevated inducible nitric oxide synthase levels are correlated with the severity of HAND.1 The immunohistochemical analysis of brain tissues from demented and nondemented HIV-infected patients indicated an increased level of nitrotyrosine in the demented group.2 Nitrated l-prostaglandin d synthase with decreased activity upon modification was found in the cerebrospinal fluid of HIV-infected patients and was proposed as a HAND biomarker.15,16 However, other nitrated proteins in the brain have not been identified, and their role in HAND progression has not been investigated.
The two main methods used in nitrated protein identification are two-dimensional gel electrophoresis (2DE) separation and immunoprecipitation followed by mass spectrometric analysis. 2DE does not always have the resolving power to separate proteins with similar sizes and chemical properties. Thus, an immunoreactive gel spot detected by western blot analysis with the antinitrotyrosine antibody may not always correspond to a single protein. Additionally, hydrophobic proteins cannot be easily extracted from gel spots, biasing the method toward hydrophilic proteins. Another problem observed in both methods is the specificity of the antinitrotyrosine antibodies. Two antinitrotyrosine antibodies were shown to bind to tyrosine and tryptophan structures similar to 3-nitrotyrosine.17,18 Detecting low-abundance nitrated peptides on the identified proteins is essential for the identification of nitrated proteins. The nanoflow liquid chromatography has a higher sensitivity and enables the detection of low-abundance peptides in samples when the amounts are limited. A nanoflow liquid chromatography system coupled to a high-resolution tandem mass spectrometer is ideal for the detection of low-abundance nitrated peptides and identifying the modification sites. Determining the modification site ensures the correct identification of nitrated proteins and provides insight into the possible molecular and functional changes upon nitration.
In this study, we identified nitrated proteins and the sites of nitration in postmortem human brain tissues from individuals without HIV infection (control), with HIV infection but without encephalitis (HIV), or with HIV infection with encephalitis (HIV-E). Immunoprecipitation was utilized for the enrichment of nitrated proteins in each brain lysate. A nano-HPLC system coupled to high-resolution Orbitrap Velos mass spectrometer was used for analysis. We identified the nitrated proteins and the sites of modifications, and we used molecular modeling to predict the biological role of these post-translational modifications during HIV infection of the brain.
Experimental Methods
Sample Preparation
Frozen postmortem human brain tissues (parietal cortex) from individuals with HIV infection but without encephalitis (n = 2) and HIV infection with encephalitis (n = 2) were obtained from the University of California, Los Angeles (case IDs: 1093, 6081, 5007, and 5008). Frozen control human brains (n = 4) from a matching site of the brain were obtained from the University of Maryland, Baltimore (case IDs: 5125, 5343, 5346, and 5189) (Supporting Information Table 1). The tissues were washed with ice-cold buffer (20 mM Tris/HCl, pH 6.8, 100 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol containing Roche complete protease inhibitor cocktail) and then homogenized using a pestle in the same buffer (1:5 w/v) with the addition of 1% (v/v) Triton-X 100 and 0.1% (w/v) SDS. The lysate was centrifuged at 18 000g for 20 min to remove insoluble cellular components. The BCA assay (Pierce) was used according to the manufacturer’s directions to determine protein concentrations.
Immunoprecipitation of Nitrated Proteins
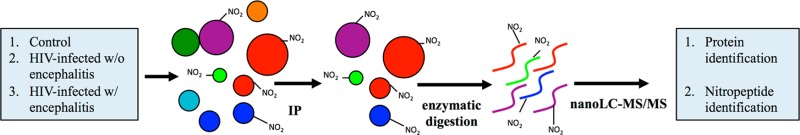
Two-hundred micrograms of protein from each brain sample was used. The samples were cleaned using detergent removal spin columns (Pierce), 0.5 mL, according to the manufacturer’s directions. The columns were initially equilibrated with Dulbecco’s phosphate buffered saline (PBS). The samples were incubated with monoclonal anti-3-nitrotyrosine antibody (clone 1A6) cross-linked to Protein G–agarose (Millipore) overnight at 4 °C. The following day, the flow through was collected using microcentrifuge spin filters (Pierce) with a 30 μm filter. The immunoprecipitate was washed three times with 1× PBS, and the proteins were eluted using 1, 2, and 5% formic acid (FA) (v/v). The anti-3-nitrotyrosine antibody Protein G–agarose conjugate was washed with 1× PBS. The immunoprecipitation procedure was repeated twice, and all of the eluents from a single sample were combined, their pH was neutralized, and they were dried (Scheme 1). We termed the proteins pulled down with the anti-3-nitroyrosine antibody protein G–agarose as immunoprecipitate. The immunoprecipitated proteins were called nitrated only if their nitrated peptides were detected in the MS/MS analysis.
Scheme 1. Experimental Design.
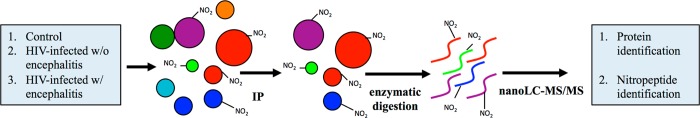
Brain samples from the parietal cortex of HIV-negative controls, HIV-infected individuals without encephalitis, and HIV-infected individuals with encephalitis were immunoprecipitated with anti-3-nitrotyrosine antibody conjugated to protein G–agarose. The elution fraction with enriched nitrated proteins was trypsinized. The peptides were separated on a reverse-phase nano-LC system and analyzed by high-resolution tandem mass spectrometry.
Protein Digestion
The immunoprecipitated samples were reconstituted in 100 mM ammonium bicarbonate, reduced with 5 mM dithiothreitol, and alkylated with 15 mM iodoacetamide. The proteins were digested with trypsin (Sigma) at a 1:20 trypsin-to-protein ratio (w/w) for 18 h at 37 °C.
LC–MS/MS Analysis
The tryptic peptides were enriched and separated on an Eksigent nanoflow LC system coupled to a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). A 2 cm long trap column (YMC gel ODS-A S-10 μm) and a 75 μm × 10 cm analytical column containing Magic AQ C18 material, 5 μm, 100 Å (Michrom Bioresources) were utilized. The peptides were separated on a 70 min linear gradient and directly introduced to the LTQ-Orbitrap Velos at a flow rate of 300 nL/min and a spray voltage of 2.0 kV. Data-dependent tandem MS analysis was employed in the Orbitrap, with a 30 000 resolution for MS and 7500 resolution for MS/MS. Full scans were acquired from m/z 300–2000 with up to the 15 most intense ions isolated using a 1.9 Da window. The peptide ions were fragmented using a collision energy of 35% in the HCD cell with a dynamic exclusion of 30 s. The first mass value was fixed at m/z 140, and the minimum signal for triggering an MS/MS scan was set to 2000. An ambient air-lock mass was set at m/z 371.10123 for real-time calibration.19 Unassigned and singly charged ion rejection was enabled.
Bioinformatics
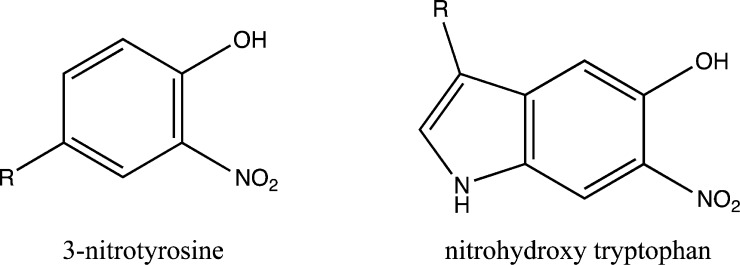
MS and MS/MS data were searched using Proteome Discoverer (v. 1.3.0.339) with the Mascot (v. 1.27) algorithm. Database searching of MS/MS spectra was performed using the National Center for Biotechnology Information nonredundant database (2012). Homo sapiens was selected for the taxonomy, and 230 236 protein sequences were searched. The scan event filter had the following criteria: MS2 for MS order, HCD for activation type, and full for scan type. Trypsin was selected as the enzyme with the allowance of a maximum of two missed cleavages. The anti-3-nitrotyrosine antibody, clone 1A6 (Millipore), has been reported to bind to nitrohydroxytryptophan also because of the similarity in the chemical structures of these modified amino acids.17 Thus, we included nitrohydroxylated tryptophan in the bioinformatics analysis. Carbamidomethylation (C, +57.02146) was set as a fixed modification. The variable modifications included oxidation (M, +15.99492), deamidation (N, Q, +0.98402), acetylation (protein N terminal, +42.01057), nitration (Y, +44.98508), and nitrohydroxylation (W, +60.97999) (Figure 1). The precursor mass tolerance was set to 20 ppm, and the fragment tolerance was set to 0.05 Da. Peptide validator was applied to the Mascot results to search against a decoy database and to obtain false discovery rates (strict, 0.01; relaxed, 0.05). The results were filtered for minimum medium peptide confidence for peptide identification and differentiable protein groups for protein group identification.
Figure 1.
Chemical structures of 3-nitrotyrosine and nitrohydroxy tryptophan. The exact location of nitro and hydroxyl groups on the tryptophan cannot be determined by mass spectrometric approaches. The anti-3-nitrotyrosine antibody has previously been shown to be capable of binding to both of these modified amino acids.17
Molecular Modeling
A comparative model of immunoglobulin light and heavy chains was obtained using the HIV-neutralizing antibodies (PDB identifier: 2CMR and 4JDT) as templates with the molecular modeling program MOE (Chemical Computing Group Inc., version 2012.10). In this model, the loop between the β4 and β5 strands that differs in size and has a disordered structure in the templates was not modeled. Because the modified residues were either on the complementarity-determining region (CDR) or were two amino acids away from the CDR, our model gives insights into the molecular changes that take place upon nitration and nitrohydroxylation as well as their potential effect on antibody structure and antigen recognition.
Results
Diverse Immunoglobulins Identified in the HIV-E Immunoprecipitate
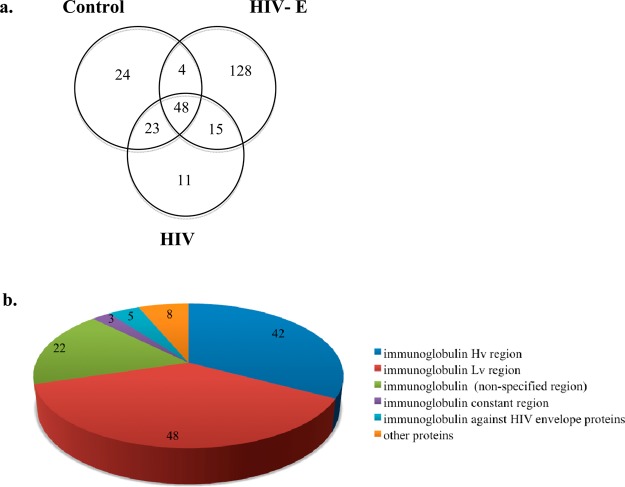
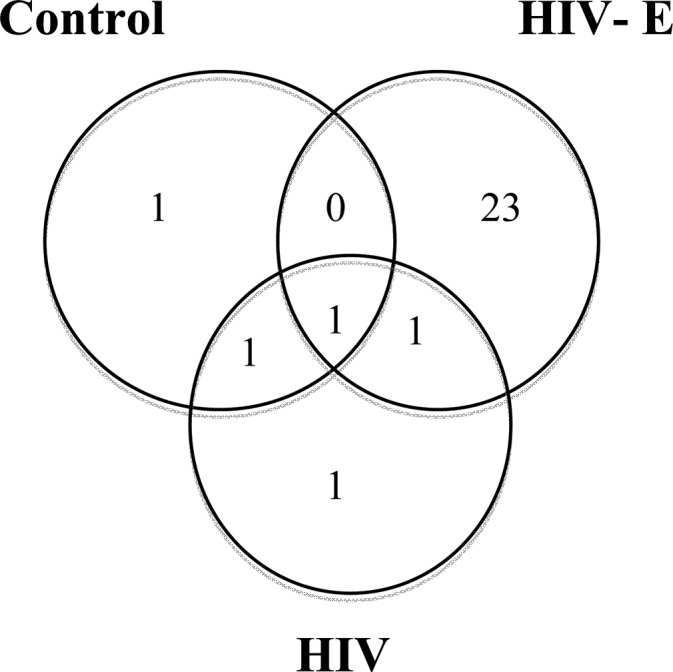
In the current study, a total of 253 differentiable proteins (Supporting Information Table 2) were identified in the immunoprecipitates of all samples with at least one unique peptide. Among these proteins, 24 were present only in the control, 11, only in HIV, and 128, only in the HIV-E sample sets (Figure 2). The number of proteins identified in all of our eight samples was 48. The number of proteins identified in both control and HIV-E samples was four, in both the control and HIV sample sets was 23, and in both the HIV-E and HIV sample sets was 15. Because the vast majority of the immunoprecipated proteins were in the HIV-E samples, it suggests that these protein modifications did not occur as a result of sample preparation but were specific for the underlying biological processes in HIV-E. The proteins identified in the control samples likely indicate either nonspecific binding to the anti-3-nitrotyrosine antibody or proteins that were not related with HIV infection. Thus, efforts were focused on the proteins that were identified only in the HIV sample sets (both the HIV and HIV-E sample sets) and only in the HIV-E sample set.
Figure 2.
Number of proteins identified in the sample sets and the distribution of proteins identified exclusively in the HIV-E brain. (a) Venn diagram representation of differentiable proteins in the parietal cortex brain tissue immunoprecipitates. The majority of the proteins are found exclusively in the HIV-E sample set. (b) Distribution of proteins found in the immunoprecipitates from HIV-E samples. Ninety proteins are immunoglobulin variable regions, of which five proteins are immunoglobulins against HIV envelope proteins.
The proteins that were identified only in the immunoprecipitates from the HIV sample set included small nuclear ribonucleoprotein Sm D1, proteolipid protein 1, NADPH dehydrogenase 1 alpha subcomplex 6, Apo-human serum transferrin, and Pitp-alpha complexed to phosphatidylinositol. The rest of the 11 proteins were immunoglobulins. Thirteen out of the 15 proteins that were identified in both HIV and HIV-E sample sets were also immunoglobulins. The other two proteins were cytochrome c oxidase subunit 5A and gamma-synuclein.
The NCBI nr database has a large number of immunoglobulin sequences predicted from the GenBank. A total of 128 proteins were identified only in the immunoprecipitate from the HIV-E sample set. Interestingly, 90 of these proteins were immunoglobulin variable regions, and five of them were immunoglobulins against HIV-1 envelope proteins (Figure 2). We also identified glyoxalase double mutant, carbonic anhydrase, cold agglutinin, and CD47. The other 36 proteins were either immunoglobulins without region specification or unnamed protein products. Next, we analyzed the nitrated peptides to confirm that the identified proteins were nitrated.
Nitrated Tyrosine and Nitrohydroxylated Tryptophan Are Found in the Immunoglobulin Variable Regions
Using Mascot, 18 peptides with nitrated tyrosines and 10 peptides with nitrohydroxylated tryptophan modifications were identified (Table 1). The lowest Mascot ion score was 38, and the highest E-value was 5.09 × 10–2. An investigation of murine and human immunoglobulin heavy chain CDR3 repertoire has shown that they exhibit differences in amino acid sequence and structure.20 The data was searched using Rodentia taxonomy as well to ensure that the modified peptides were not coming from the monoclonal anti-3-nitrotyrosine antibody used during the immunoprecipitation. The modified peptide Qac,deYNO2NASVSVPDSSGPER could be part of an unnamed protein product in Mus musculus as well as heterogeneous nuclear ribonucleoprotein K in H. sapiens. The rest of the modified peptides reported were found in the H. sapiens database but not in Rodentia.
Table 1. Nitrated Peptides from the Anti-3-Nitrotyrosine Antibody–Agarose-Immunoprecipitated Brain Proteinsa.
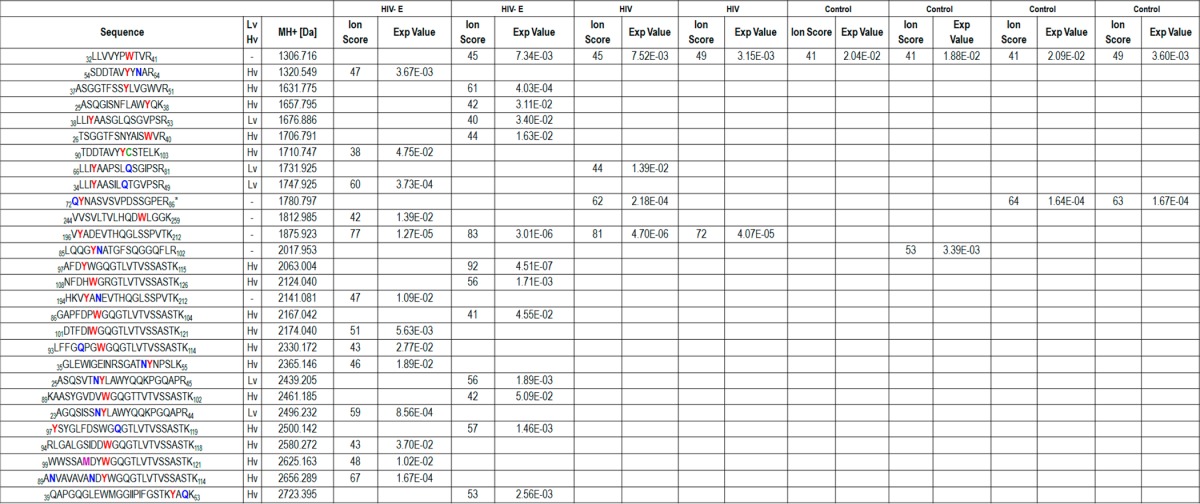
HIV, brain samples from individuals with HIV infection without encephalitis; HIV-E, brain samples from individuals with HIV infection and encephalitis; PSM, peptide spectrum match; Hv, heavy chain variable region; and Lv, light chain variable region on immunoglobulins. On the peptide sequences, nitrotyrosines and nitrohydroxytryptophans are shown in red, deamidated residues are indicated in green, carbamidomethylated cysteines are colored green, and the oxidized methionine is shown in purple. The peptide with an asterisk has N-terminus acetylation. The complete peptide identification information can be found in Supporting Information Table 4.
The 32LLVVYPWNO2OHTVR41 peptide from beta globin was detected in seven of the samples except for one of the HIV brain samples (Table 1). The 85LQQGYNO2NdeATGFSQGGFLR102 peptide from palmitoyl-protein thioesterase 1 chain A was present only in one of the four control samples, whereas the 72Qac,deYNO2NASVSVPDSSGPER86 peptide from human heterogeneous nuclear ribonucleoprotein K was found in two of the control samples and one HIV brain sample (Figure 3).
Figure 3.
Venn diagram representation of nitrated and nitrohydroxylated peptides in the sample sets. The majority of the nitrotyrosine and nitrohydroxytryptophan containing peptides were identified in the tryptic digests of the immunoprecipitated proteins from the HIV-infected with encephalitis sample set.
The nitrated peptides that were identified only in the immunoprecipitates from HIV and/or HIV-E sample sets may reflect the effect of HIV infection on the human proteins. The 196VYNO2ADEVTHQGLSSPVTK212 peptide from antitubulin IgG1 kappa VL chain was found in both HIV and HIV-E samples and not in the controls. The 66LLIYNO2AAPSLQdeSGIPSR81 peptide from immunoglobulin kappa light chain variable region was present in one HIV brain sample. Twenty three of the nitrated and nitrohydroxylated peptides were found only on peptides from HIV-E brain immunoprecipitates, and 21 of them were on immunoglobulin variable regions (Table 1). Among these peptides, 108NFDHWO2OHGRGTLVTVSSASTK126 may belong to anti-HIV-1 gp41 immunoglobulin heavy chain (gi: 212675107) and 23AGQSISSNdeYNO2LAWYQQKPGQAPR44 may belong to anti-HIV-1 gp120 immunoglobulin kappa chain variable region (gi: 299742). The high amino acid level variation observed in immunoglobulin variable regions and the sequence homology in the immunoglobulin protein family made protein identification challenging by shotgun proteomic methods. The MS/MS spectra of the modified peptides can be found in Supporting Information Figures 1–14.
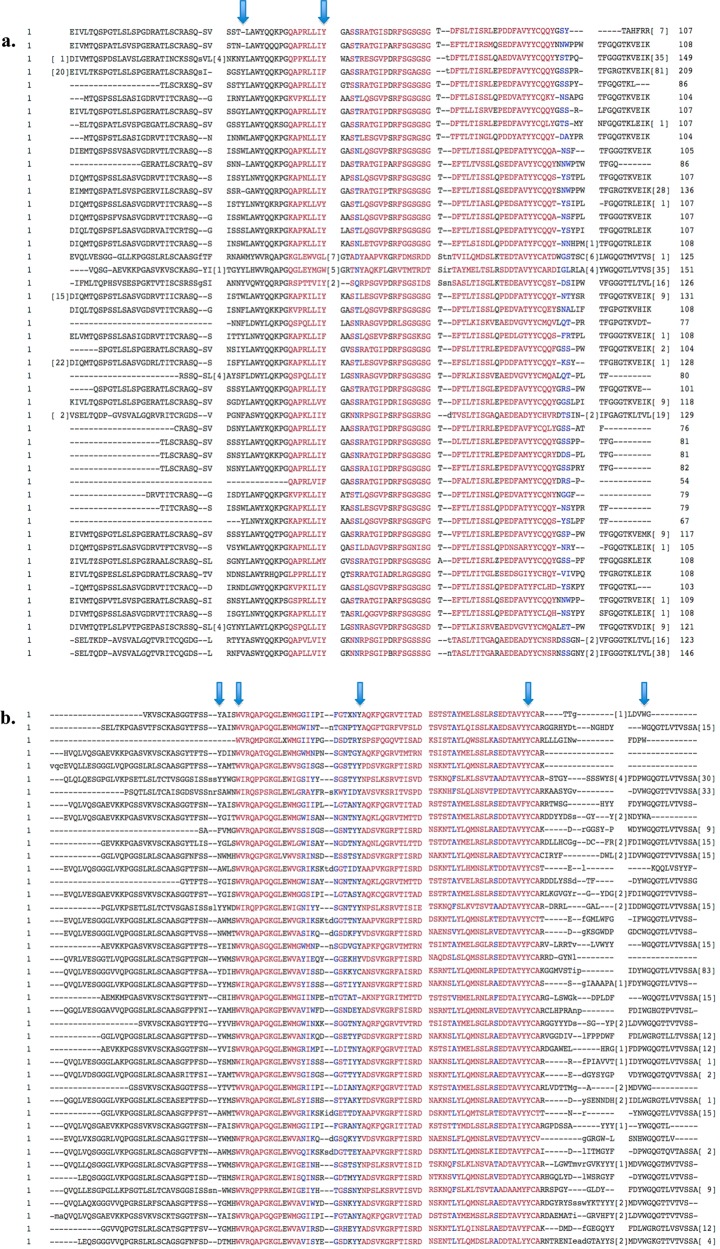
The MS/MS spectra for m/z 1600.75139, 1660.78826, 2069.02292, 2102.96147, 2155.08268, 2307.07646, 2423.1778, and 2552.19048 were matched to more than one peptide sequence (Supporting Information Table 4); therefore, the identity of these peptides was ambiguous. These eight spectra were found only in the HIV-E sample set, and the peptide sequences belonged to immunoglobulin variable regions (five belonged to the heavy chain and three belonged to the light chain). Despite the ambiguity in the exact sequence, the identified human protein sequences had high sequence homology, and the modified residues were mostly conserved (Figure 4).
Figure 4.
Amino acid sequence alignment of immunoglobulin variable regions identified only in the HIV-E immunoprecipitates. The National Center for Biotechnology constraint-based multiple alignment tool (COBALT) was used. Highly conserved residues are in red, and mostly conserved residues are in blue. The nitrated residues, shown with an arrow, are conserved among different immunoglobulin variable regions. (a) Immunoglobulin light chain variable region sequences. (b) Immunoglobulin heavy chain variable region sequences.
Nitrated Tyrosines and Nitrohydroxylated Tryptophan Residues Are Conserved in Immunoglobulin Light and Heavy Chain Variable Regions
Among the nitrated and nitrohydroxylated peptides that were observed in the HIV-E sample set, 17 belonged to immunoglobulin heavy chain variable regions and four belonged to immunoglobulin light chains (Table 1). Although a higher number of nitrated tyrosine- and nitrohydroxylated tryptophan-containing peptides were found in the HIV-E sample set, the same modified peptide sequences were not present in both HIV-E samples. We observed that despite the variation in the peptide sequences from immunoglobulin variable regions the modified tyrosine and tryptophan residues were conserved among the nitrated peptides. Thus, we aligned the amino acid sequences of the 42 immunoglobulin heavy chain variable region proteins and 48 immunoglobulin light chain variable region proteins that were identified in the immunoprecipitates (Figure 4). Again, these residues were mostly conserved in the human immunoglobulin variable regions, which suggests that these modifications may be of functional significance. Additionally, two tyrosines are found on the CDR of the immunoglobulin heavy chain variable region, and four of the modified residues are two amino acids away from the CDR, further suggesting biological significance of the nitration at these sites.
Molecular Model of Immunoglobulin Light and Heavy Chains Suggest Potential Changes in Inter- and Intramolecular Interactions upon Nitration
We modeled immunoglobulin light and heavy chains using the crystal structures of neutralizing antibody with gp41 innercore21 and gp12022 (PDB code: 2CMR and 4JDT) to determine the potential molecular changes that take place upon the nitration of the tyrosine and tryptophan residues (Figure 5a). Although we do not know the antigens of the modified antibodies and cannot determine the number of modifications on a single antibody using shotgun proteomics methods, this model demonstrates the potential interactions that the modified residues may be involved in. For convenience, we used the residue numbers from the neutralizing antibody from the 2CMR crystal structure. The nitration of heavy chain CDR residues Y32 and Y59 may affect antigen recognition. In this case, nitration of Y32 could lead to H-bonding with the H199 of the gp41 innercore (Figure 5b). Similarly, nitrated Y59 may have a favorable intermolecular interaction with R214 of the gp41 innercore (Figure 5c). Conserved W36 is located one residue away from the CDR in the β strand of the immunoglobulin heavy chain, and the nitrohydroxylated form can hydrogen bond with the Q6 and C22 (Supporting Information Figure 15a). However, modified W105 is found at the interface between the heavy and the light chains of the immunoglobulin. This region is hydrophobic and cannot accommodate the nitro group (Supporting Information Figure 15b). Thus, the nitrohydroxylation of this residue may interfere with the intramolecular interactions of the two chains. Nitrated Y91 is also located at the interface between the heavy and the light chains, but its nitration would have minor effects on antibody shape (data not shown). Interestingly, the nitrated immunoglobulin light chain Y36 fits well into the interface between the two chains. The nitro group can be well-accommodated by the H-bonds with heavy chain W105 and a backbone amine (Figure 5d). Finally, light chain Y49 does not interact directly with the antigen but is found below the flexible loop that binds to the antigen (Figure 5e). Thus, by interacting with the loop residues, it may change the shape of CDR3. The model of immunoglobulin with gp120 did not show any significant change in antigen recognition upon nitration of tyrosine and tryptophan, although the changes in intramolecular interactions were similar (data not shown). As explained above, the modification of each residue may have unique effects on antigen binding and chain interactions.
Figure 5.
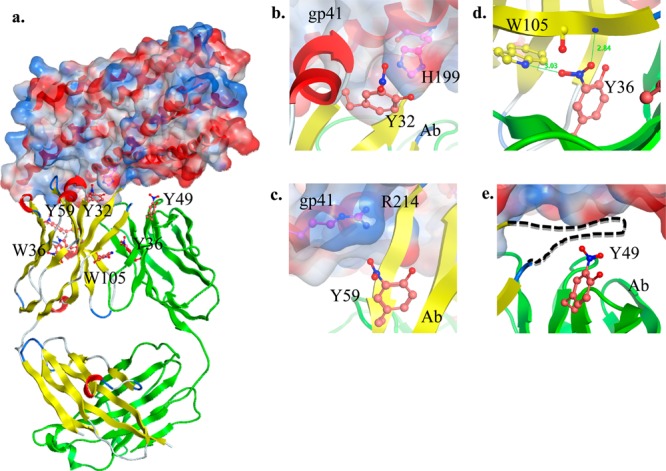
Model of the complex between immunoglobulin light and heavy chains and the HIV envelope protein gp41 innercore obtained using the crystal structure of the neutralizing antibody as templates. (a) The cartoon of the heavy and light chains of the immunoglobulin are shown in yellow and green, respectively. The solvent-accessible surface of gp41 is drawn semitransparent and is colored according to the electrostatic potential (red, negative; blue, positive) around the cartoon of the receptor (colored in red). Nitrated Y32, Y36, Y49 and nitrohydroxylated W36 and W105 of the immunoglobulin as well as H199 and R214 of the receptor are drawn in a ball-and-stick representation. The positions of the nitro- and hydroxyl-group substitutions were chosen to minimize steric conflicts with the immunoglobulin. (b) Nitrated heavy chain Y32 may interact with H199 of gp41. (c) Heavy chain Y59 is also solvent-exposed, and its nitration may have a positive effect on gp41 binding through the favorable interaction between the nitro group and R214. (d) Light chain Y36 resides at the interface between the heavy and the light chains. Interestingly, the nitro group can be accommodated well at this position through H-bonds with heavy chain W105 and a backbone amine. (e) Light chain Y49 resides below the disordered loop (dashed line) between the heavy chain β4 and β5 strands, and nitration of this residue may affect this loop, which contains CDR3 and is critical for antigen binding.
Discussion
In this study, we identified nitrated proteins in the brain of HIV-infected individuals with neurocognitive disorders. The majority of the nitrated peptides belong to immunoglobulin heavy and light chain variable regions. Despite the sequence variation, the nitrated tyrosine and nitrohydroxylated tryptophan residues were conserved in the human immunoglobulins detected in these samples and the previously reported HIV-1-neutralizing antibodies that bind to the HIV envelope glycoproteins gp41 and gp120. The potential roles of tyrosine and tryptophan and their modified forms in antibody–antigen binding are addressed here.
Selectivity for Protein Nitration in HIV-Infected Individuals with Encephalitis
Nitrated proteins have been previously detected in various inflammatory conditions and in multiple tissues. Thus, it is not surprising to detect the highest number of nitrated peptides in HIV-infected individuals with encephalitis. During the infection, reactive oxygen and nitrogen species are released from activated macrophages and microglia within the brain.23 Peroxynitrite is the key species involved in the nitration of tyrosine6 and tryptophan24 residues. Its conjugated acid can diffuse through membranes, and its anionic form can be transported by anion channels.25 How nitrohydroxylation takes place in the setting of HIV infection is an interesting question that needs to be addressed in future studies. Interestingly, we did not find any of the peptides from the variable region of the immunoglobulins listed in Table 1 with only hydroxylation or nitration (data not shown). Thus, in our samples, these modifications were always present together on tryptophan residues. A likely possibility is that both of these modifications occur either simultaneously or in quick succession. Peroxynitrite has been shown to both nitrate and hydroxylate tryptophan24 and may be part of the nitrohydroxylation process. In vitro nitration of tryptophan, however, yields a mixture of products depending on the type of nitrating agent, its concentration, and the reaction conditions.7 We cannot exclude the possibility of artificial hydroxylation of nitrated tryptophan in the postmortem brain; however, its presence in only the HIV-E sample set indicates relevance to the disease condition and is in agreement with previous reports on oxidative and nitrosative stress in HIV infection and HAND progression.23
Unlike enzymatic post-translational modifications such as phosphorylation, N-glycosylation, and ubiquitination, which have consensus sequences on their substrates, post-translational modifications based on radical reactions have not been found to have such a consensus sequence. Interestingly, nitration selectivity has been linked to the proximity to the site where the nitrating agent is produced, the abundance of the protein and its primary sequence, the abundance of tyrosine, and the residue exposure.26 Among the nitrated peptides listed in Table 1, 22 out of 28 are from immunoglobulin variable chains. Although, this modification does not seem to be specific to only immunoglobulin variable chains in the brains from individuals with HIV-E, they seem to be favored. Our observation is consistent with the previous reports of nitration because (1) oxidative and nitrosative species are present at HIV-infected cells,23 (2) immunoglobulins are expressed at elevated levels and are diversified during infection,27 (3) tyrosine and tryptophan residues are found at a high frequency in the CDR of immunoglobulins on the variable regions,28−30 and (4) tyrosines and tryptophan residues are predominantly located on the solvent-exposed antigen-binding surface of immunoglobulins.29−31 Additionally, lack of methionine and cysteine residues in the close proximity to tyrosine has been determined to be important for nitration reactions. Only three nitrated peptides out of the 22 (Table 1) have cysteine and methionine residues in the sequence. Thus, although nitration is not specific to immunoglobulin variable regions in the HIV-infected brain, it seems to be favored.
Role of Tyrosine and Tryptophan on Immunoglobulins in Inter- and Intramolecular Interactions
The structure, function, and properties of immunoglobulin binding fragments are widely studied because of their potential applications in vaccine design and therapeutics based on synthetic antibodies or peptides. The sequence similarities between antibodies isolated from patients and synthetic antibody libraries have been determined previously.29,30,32 Both tyrosine and tryptophan were predicted to be ideal residues within antibody binding regions because of the following physical and chemical properties: hydrogen-bond formation, hydrophobic interactions, electrostatic interactions between positively charged groups and the ring structure, amphipathicity, large size for maximized intermolecular interaction, and enhanced mobility when adjacent to small residues such as glycine, alanine, and serine.29 The importance of tyrosine in synthetic antibody affinity and specificity has been reviewed.31 Also, tyrosine accounts for 25% of the immunoglobulin side chains that interact with antigens in isolated antibodies.29 In our study, the majority of the proteins identified in the immunoprecipitate from HIV-E samples were immunoglobulin variable chains. Immunoglobulins are produced by B-cells and may enter the brain either by the cerebrospinal fluid33 or through the blood-brain barrier, which may be disrupted during HIV infection.34−36 Once in the brain, they can bind to viral antigens. In accordance with previous studies, the alignment of immunoglobulin light variable region and heavy variable region sequences of the identified proteins revealed that the modified tyrosine and tryptophan residues are mostly conserved (Figure 4).
The structural basis of broad neutralizing antibodies for HIV envelope proteins gp41 and gp120 has been investigated previously.22,37 Often, the molecular effect of a modification depends on where the residue is located on the protein and the interacting residues in the milieu. In our structural model using two neutralizing-antibody crystal structures, we characterized potential antigen-binding effects of the immunoglobulin heavy chain Y32 and Y59 of the CDR. Similarly, immunoglobulin light chain Y49 may influence antigen interactions by affecting the flexible loop that contains the immunoglobulin CDR3. The rest of the modified residues are located on the framework region (FR) of the variable region, internal to the heavy chain β barrel (W36), or at the interface between the light and heavy chains (L, Y36; H, W105). Their nitration could modify the fold of each chain and the interaction between heavy and light chains. Whereas the CDR residues are involved directly in antigen binding, the FR residues can influence the overall structure of the variable region, affecting the recognition.38
Tyrosine Modification on Immunoglobulins in HIV Infection
During immune activation, variation is introduced to the immunoglobulins by VDJ recombination. In response, the viral genes mutate to escape antibody neutralization. A previous study found tyrosine sulfation on the amino terminus of CCR5, which was shown to be critical for recognition of gp120.39 Furthermore, antibodies that mimic the coreceptor through tyrosine sulfation have been detected. This modification was suggested as a secondary level of antibody variation to enhance antigen recognition.40
The in vitro nitration of a monoclonal human immunoglobulin and its mass spectrometric analysis revealed tyrosine nitration on both light and heavy chains.41 An antibody-based breast cancer drug has been reported to contain tyrosine nitration.42 Although the oxidation of a critical tryptophan residue on an humanized antibody was shown to lead to a loss of its binding activity to its antigen,43 the functional consequences of nitration and nitrohydroxylation on immunoglobulins are largely unknown. To our knowledge, our study is the first report of nitration on human immunoglobulin variable regions with site-specific information. Because of structural similarity, nitrohydroxy tryptophan may serve similar functional roles as nitrated tyrosine. Our data indicates a role of nitration in the immune system, and it may be a marker of encephalitis, which is commonly observed as a pathological correlate in patients with HAND. Our findings may have important implications in elucidating the immune response during HIV infection and in the development of novel therapeutics.
Implications of Immunoglobulin Tyrosine and Tryptophan Modifications and Future Directions
To date, all HIV vaccine strategies have failed. There is widespread interest in antibodies for vaccine design and antibody-based therapeutics development as antibody–drug conjugates.44 Antibodies either are isolated from patients and characterized or synthetic antibody/peptide libraries are screened for affinity and specificity for antigens of interest. Tyrosine has been identified as a critical residue in both isolated and synthetic antibodies. Tyrosine sulfation was discovered on the CDR3 region of CCR5 mimetic gp9 and gp16 broadly neutralizing antibodies against HIV envelope proteins.45 Furthermore, sulfated tyrosine containing CCR5 and CD4 mimetic peptide fused to a dimeric antibody constant domain showed enhanced potency.46 Investigation of the role of nitration and nitrohydroxylation of the tyrosine and tryptophan residues on immunoglobulins is necessary to determine how they alter the function of immunoglobulins when compared to the previously reported role of tyrosine sulfation for HIV envelope protein binding.
Antibodies are one of the major sensors of the body, especially during an infection. After the introduction of antiretroviral therapy, HAND nosology has changed with the addition of more prevalent, milder forms of HAND.47 Recognition of these forms of HAND requires extensive neuropsychological testing, which may not be easily accomplished in a clinical practice setting. There is a need for developing biomarkers that correlate with the neurocognitive state of the HIV-infected patients, follow medication effects, and help to provide appropriate therapy. The exclusive presence of nitration on immunoglobulins in the HIV-E sample set makes them potential biomarkers if these findings can be confirmed in either cerebrospinal fluid or serum. There has been increased interest in post-translational modifications in biomarker studies because protein expression alone does not always reflect the change in the system during disease states.3 Targeted and sensitive assays for detecting post-translational modifications on abundant proteins such as hemoglobin and albumin in biological fluids have been designed.48,49 Cerebrospinal fluid, which is rich in immunoglobulins, can be analyzed without depletion of abundant proteins for the validation of nitrated tyrosine- or nitrohydroxylated tryptophan-containing peptides as HAND biomarkers.
However, our study has several limitations. We used autopsy tissue; hence, some of the changes may have occurred terminally or postmortem. The study used a small sample size, and the samples were not perfectly matched for age and gender. Despite these limitations, the differences between HIV-E and controls were quite striking, suggesting that the differences were most likely driven by the underlying biological processes associated with HIV-E, as described above.
Conclusions
In this study, we identified nitrated proteins in control, HIV, and HIV-E brain lysate using an affinity-enrichment protocol followed by LC–MS/MS analysis. The predominant presence of these modified peptides on immunoglobulin variable regions and on conserved tyrosine and tryptophan residues suggests potential functional roles. Furthermore, the high frequencies of tyrosine on the antigen-binding fragments of antibodies and the function of tryptophan in inter- and intramolecular interactions make the modification of these residues significant. Our results provide unique insight into HIV neuropathogenesis and have opened the door for future structure–function studies. This may also have implications for further development of biomarker assays.
Acknowledgments
R.J.C, M.A.B., and L.U. were supported by grant R01NS039253 (to R.J.C.) from the National Institute of Neurological Disorders and Stroke. A.N. is supported by intramural NIH funds. We thank the Johns Hopkins School of Medicine Mass Spectrometry and Proteomics Facility for technical assistance and Drs. Stefani Thomas and Ronald Schnaar for the critical reading of the manuscript.
Glossary
Abbreviations
- CCR5
CC chemokine receptor 5
- CD47
cluster of differentiation 47
- CDR
complementarity-determining region
- COBALT
constraint-based multiple alignment
- FA
formic acid
- FR
framework region
- HAND
HIV-associated neurocognitive disorders
- HCD
high-energy collision dissociation
- HIV
human immunodeficiency virus or sample from individuals with HIV infection without encephalitis
- HIV-E
sample from individuals with HIV infection and encephalitis
- Hv
immunoglobulin heavy chain variable region
- IgG
immunoglobulin G
- LC
liquid chromatography
- Lv
immunoglobulin light chain variable region
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PBS
phosphate buffered saline
- PSM
peptide spectrum match
- PMSF
phenylmethylsulfonyl fluoride
- SDS
sodium dodecyl sulfate
- TFA
trifluoroacetic acid
Supporting Information Available
Sample information, protein and peptide identifications, additional molecular model figures and MS/MS spectra of modified peptide. This material is available free of charge via the Internet at http://pubs.acs.org. The MSF files associated with this manuscript can be downloaded from Peptide Atlas at the following address: ftp://PASS00386:PJ9873yh@ftp.peptideatlas.org/.
The authors declare no competing financial interest.
Dedication
∥ Deceased November 12, 2012.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Adamson D. C.; McArthur J. C.; Dawson T. M.; Dawson V. L. Mol. Med. 1999, 2, 98–109. [PMC free article] [PubMed] [Google Scholar]
- Boven L. A.; Gomes L.; Hery C.; Gray F.; Verhoef J.; Portegies P.; Tardieu M.; Nottet H. S. J. Immunol. 1999, 7, 4319–4327. [PubMed] [Google Scholar]
- Uzasci L.; Nath A.; Cotter R. J. Neuroimmune Pharmacol. 2013, 5, 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagna A.; Le Grazie C.; Accordini A.; Giulidori P.; Cavalli G.; Bottiglieri T.; Lazzarin A. Neurology 1995, 9, 1678–1683. [DOI] [PubMed] [Google Scholar]
- Choi J.; Liu R. M.; Kundu R. K.; Sangiorgi F.; Wu W.; Maxson R.; Forman H. J. J. Biol. Chem. 2000, 5, 3693–3698. [DOI] [PubMed] [Google Scholar]
- Radi R. Proc. Natl. Acad. Sci. U.S.A. 2004, 12, 4003–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakura F.; Ikeda K. Nitric Oxide 2006, 2, 152–161. [DOI] [PubMed] [Google Scholar]
- Szabo C.; Ischiropoulos H.; Radi R. Nat. Rev. Drug Discovery 2007, 8, 662–680. [DOI] [PubMed] [Google Scholar]
- Nuriel T.; Hansler A.; Gross S. S. J. Proteomics 2011, 11, 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield D. A.; Reed T.; Sultana R. Free Radical Res. 2011, 1, 59–72. [DOI] [PubMed] [Google Scholar]
- Franco M. C.; Ye Y.; Refakis C. A.; Feldman J. L.; Stokes A. L.; Basso M.; Fernandez Melero; de Mera R. M.; Sparrow N. A.; Calingasan N. Y.; Kiaei M.; Rhoads T. W.; Ma T. C.; Grumet M.; Barnes S.; Beal M. F.; Beckman J. S.; Mehl R.; Estevez A. G. Proc. Natl. Acad. Sci. U.S.A. 2013, 12, E1102–E1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson S. R.; Held J. M.; Schilling B.; Oo M.; Gibson B. W.; Andersen J. K. Anal. Chem. 2009, 18, 7823–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulak K. S.; Miyagi M.; Yan L.; West K. A.; Massillon D.; Crabb J. W.; Stuehr D. J. Proc. Natl. Acad. Sci. U.S.A. 2001, 21, 12056–12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherner M.; Cysique L.; Heaton R. K.; Marcotte T. D.; Ellis R. J.; Masliah E.; Grant I. J. NeuroVirol. 2007, 1, 23–28. [DOI] [PubMed] [Google Scholar]
- Li W.; Malpica-Llanos T. M.; Gundry R.; Cotter R. J.; Sacktor N.; McArthur J.; Nath A. Neurology 2008, 19, 1753–1762. [DOI] [PubMed] [Google Scholar]
- Beasley A.; Anderson C.; McArthur J.; Sacktor N.; Nath A.; Cotter R. Clin. Proteomics 2010, 1, 29–41. [Google Scholar]
- Rebrin I.; Bregere C.; Kamzalov S.; Gallaher T. K.; Sohal R. S. Biochemistry 2007, 35, 10130–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B. A.; Ulrich M.; Stumbaum M.; Bernevic B.; Moise A.; Doring G.; Przybylski M. J. Am. Soc. Mass Spectrom. 2012, 11, 1831–1840. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; de Godoy L. M.; Li G.; Macek B.; Mortensen P.; Pesch R.; Makarov A.; Lange O.; Horning S.; Mann M. Mol. Cell. Proteomics 2005, 12, 2010–2021. [DOI] [PubMed] [Google Scholar]
- Zemlin M.; Klinger M.; Link J.; Zemlin C.; Bauer K.; Engler J. A.; Schroeder H. W. Jr.; Kirkham P. M. J. Mol. Biol. 2003, 4, 733–749. [DOI] [PubMed] [Google Scholar]
- Luftig M. A.; Mattu M.; Di Giovine P.; Geleziunas R.; Hrin R.; Barbato G.; Bianchi E.; Miller M. D.; Pessi A.; Carfi A. Nat. Struct. Mol. Biol. 2006, 8, 740–747. [DOI] [PubMed] [Google Scholar]
- Scharf L.; West A. P. Jr.; Gao H.; Lee T.; Scheid J. F.; Nussenzweig M. C.; Bjorkman P. J.; Diskin R. Proc. Natl. Acad. Sci. U.S.A. 2013, 15, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J.; Haughey N.; Li W.; Venkatesan A.; Anderson C.; Reid R.; Malpica T.; Pocernich C.; Butterfield D. A.; Nath A. Antioxid. Redox Signaling 2006, 11–12, 2089–2100. [DOI] [PubMed] [Google Scholar]
- Alvarez B.; Rubbo H.; Kirk M.; Barnes S.; Freeman B. A.; Radi R. Chem. Res. Toxicol. 1996, 2, 390–396. [DOI] [PubMed] [Google Scholar]
- Denicola A.; Souza J. M.; Radi R. Proc. Natl. Acad. Sci. U.S.A. 1998, 7, 3566–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischiropoulos H. Biochem. Biophys. Res. Commun. 2003, 3, 776–783. [DOI] [PubMed] [Google Scholar]
- Perreau M.; Levy Y.; Pantaleo G. Curr. Opin. HIV AIDS 2013, 4, 333–340. [DOI] [PubMed] [Google Scholar]
- Ohno S.; Mori N.; Matsunaga T. Proc. Natl. Acad. Sci. U.S.A. 1985, 9, 2945–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian I. S.; Bradwell A. R.; Olson A. J. J. Mol. Biol. 1991, 1, 133–151. [DOI] [PubMed] [Google Scholar]
- Fellouse F. A.; Wiesmann C.; Sidhu S. S. Proc. Natl. Acad. Sci. U.S.A. 2004, 34, 12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S.; Sidhu S. S. ACS Chem. Biol. 2009, 5, 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan S.; Zhang Y.; Fellouse F. A.; Shao L.; Schaefer G.; Sidhu S. S. J. Mol. Biol. 2008, 5, 1518–1528. [DOI] [PubMed] [Google Scholar]
- Burkala E. J.; He J.; West J. T.; Wood C.; Petito C. K. AIDS 2005, 7, 675–684. [DOI] [PubMed] [Google Scholar]
- Toborek M.; Lee Y. W.; Flora G.; Pu H.; Andras I. E.; Wylegala E.; Hennig B.; Nath A. Cell. Mol. Neurobiol. 2005, 1, 181–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin E. A.; Clements J. E.; Zink M. C.; Berman J. W. J. Neurosci. 2011, 26, 9456–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A.; Dormeyer W.; Wortelkamp S.; Woitalla D.; Kuhn W.; Meyer H. E. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2002, 1–2, 167–196. [DOI] [PubMed] [Google Scholar]
- Huang J.; Ofek G.; Laub L.; Louder M. K.; Doria-Rose N. A.; Longo N. S.; Imamichi H.; Bailer R. T.; Chakrabarti B.; Sharma S. K.; Alam S. M.; Wang T.; Yang Y.; Zhang B.; Migueles S. A.; Wyatt R.; Haynes B. F.; Kwong P. D.; Mascola J. R.; Connors M. Nature 2012, 7424, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K.; Sakamoto K.; Kojima M.; Aburatani T.; Ueda T.; Ueda H. FEBS J. 2006, 10, 2184–2194. [DOI] [PubMed] [Google Scholar]
- Farzan M.; Mirzabekov T.; Kolchinsky P.; Wyatt R.; Cayabyab M.; Gerard N. P.; Gerard C.; Sodroski J.; Choe H. Cell 1999, 5, 667–676. [DOI] [PubMed] [Google Scholar]
- Huang C. C.; Venturi M.; Majeed S.; Moore M. J.; Phogat S.; Zhang M. Y.; Dimitrov D. S.; Hendrickson W. A.; Robinson J.; Sodroski J.; Wyatt R.; Choe H.; Farzan M.; Kwong P. D. Proc. Natl. Acad. Sci. U.S.A. 2004, 9, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Gaza-Bulseco G.; Chumsae C.; Radziejewski C. H. Rapid Commun. Mass Spectrom. 2008, 1, 1–10. [DOI] [PubMed] [Google Scholar]
- Wan J.; Csaszar E.; Chen W. Q.; Li K.; Lubec G. PLoS One 2012, 4, e34511-1–e34511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.; Feng J.; Lin H. Y.; Mullapudi S.; Bishop E.; Tous G. I.; Casas-Finet J.; Hakki F.; Strouse R.; Schenerman M. A. Anal. Chem. 2007, 7, 2797–2805. [DOI] [PubMed] [Google Scholar]
- Schrama D.; Reisfeld R. A.; Becker J. C. Nat. Rev. Drug Discovery 2006, 2, 147–159. [DOI] [PubMed] [Google Scholar]
- Pejchal R.; Walker L. M.; Stanfield R. L.; Phogat S. K.; Koff W. C.; Poignard P.; Burton D. R.; Wilson I. A. Proc. Natl. Acad. Sci. U.S.A. 2010, 25, 11483–11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong J. A.; Dorfman T.; Quinlan B. D.; Chiang J. J.; Ahmed A. A.; Choe H.; Farzan M. J. Virol. 2011, 15, 7563–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A.; Arendt G.; Becker J. T.; Brew B. J.; Byrd D. A.; Cherner M.; Clifford D. B.; Cinque P.; Epstein L. G.; Goodkin K.; Gisslen M.; Grant I.; Heaton R. K.; Joseph J.; Marder K.; Marra C. M.; McArthur J. C.; Nunn M.; Price R. W.; Pulliam L.; Robertson K. R.; Sacktor N.; Valcour V.; Wojna V. E. Neurology 2007, 18, 1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. Y.; Lee B. W.; Kim D.; Lee Y. H.; Kim K. J.; Kang E. S.; Cha B. S.; Lee E. J.; Lee H. C. Acta Diabetol. 2011, 2, 167–172. [DOI] [PubMed] [Google Scholar]
- Chen H. J.; Chen Y. C. Anal. Chem. 2012, 18, 7881–7890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.