Abstract
Bacterial β-class carbonic anhydrases (CAs) are zinc metalloenzymes catalysing reversible hydration of CO2. They maintain the intracellular balance of CO2/bicarbonate required for biosynthetic reactions and represent a new group of antimicrobial drug targets. Genome sequence analysis of Pseudomonas aeruginosa PAO1, an opportunistic human pathogen causing life threatening infections, identified three genes, PAO102, PA2053 and PA4676, encoding putative β-CAs that share 28–45 % amino acid sequence identity and belong to clades A and B. The genes are conserved among all sequenced pseudomonads. The CAs were cloned, heterologously expressed and purified. Metal and enzymic analyses confirmed that the proteins contain Zn2+ and catalyse hydration of CO2 to bicarbonate. PAO102 (psCA1) was 19–26-fold more active, and together with PA2053 (psCA2) showed CA activity at both pH 7.5 and 8.3, whereas PA4676 (psCA3) was active only at pH 8.3. Circular dichroism spectroscopy suggested that psCA2 and psCA3 undergo pH-dependent structural changes. Taken together, the data suggest that psCA1 may belong to type I and psCA3 to type II β-CAs. Immunoblot analysis showed that all three CAs are expressed in PAO1 cells when grown in ambient air and at 5 % CO2; psCA1 appeared more abundant under both conditions. Growth studies of transposon mutants showed that the disruption of psCA1 impaired PAO1 growth in ambient air and caused a minor defect at high CO2. Thus, psCA1 contributes to the adaptation of P. aeruginosa to low CO2 conditions and will be further studied for its role in virulence and as a potential antimicrobial drug target in this organism.
Introduction
Carbonic anhydrases (CAs), EC 4.2.1.1, are metalloenzymes that catalyse the reversible hydration of CO2 to HCO3−. They are ubiquitous in all three domains of life: Bacteria, Archaea and Eukarya (Smith & Ferry, 2000). Based on sequence and structure characteristics, CAs are grouped into five evolutionarily distinct classes α, β, γ, δ and ζ (Elleuche & Pöggeler, 2010; Hewett-Emmett & Tashian, 1996; Smith & Ferry, 2000; Supuran, 2008; Tripp et al., 2001). Such diversity is reflected in the variety of physiological roles that these enzymes play in different organisms, including respiration, pH homeostasis, CO2/bicarbonate transport and carbon fixation (reviewed by Smith & Ferry, 2000). The most extensively studied α-class CAs are present in mammals, protozoa, prokaryotes, fungi, algae and plant cytoplasm. The β-class CAs are found in plant chloroplasts, bacteria, fungi and algae (Smith et al., 1999; Smith & Ferry, 2000). The γ-class CAs are common in archaea and bacteria and both the δ- and ζ-classes have been identified in marine diatoms (Supuran, 2011). Despite their differences, most CAs require a single Zn2+ ion for catalytic activity (Covarrubias et al., 2005; Guilloton et al., 1992; Smith & Ferry, 1999). Exceptions include γ-class CAs that may also contain Fe2+ or Co2+ (Iverson et al., 2000) and ζ-class CAs that may use Cd2+ for catalytic activity (Xu et al., 2008).
The continuing growth of genomic information and computational analysis has indicated that β-CAs, present in all three domains of life, are phylogenetically very diverse and can be grouped into two monophyletic groups representing monocot and dicot CAs and four clades (A–D) (Smith et al., 1999; Smith & Ferry, 2000). Clades A and D are exclusively prokaryotic, whereas clades B and C represent CAs from the Eukarya and Bacteria domains. In prokaryotes, β-CAs are widespread in metabolically diverse species, suggesting their importance in prokaryotic biology (Smith & Ferry, 2000). Functional characterization of several prokaryotic β-CAs demonstrated their major role in providing endogenous HCO3− during aerobic growth under low partial pressure of CO2 in Escherichia coli (Guilloton et al., 1993; Merlin et al., 2003), Corynebacterium glutamicum (Mitsuhashi et al., 2004), Streptococcus pneumoniae (Burghout et al., 2010), Ralstonia eutropha (Kusian et al., 2002) and cyanobacteria (Fukuzawa et al., 1992). They are also shown to play role in a broader spectrum of functions, for example cyanate degradation in E. coli (Guilloton et al., 1992), host colonization in Helicobacter pylori (Bury-Moné et al., 2008), survival in a host in Salmonella enterica serovar Typhimurium (Sa. Typhimurium; Valdivia & Falkow, 1997) and St. pneumoniae (Burghout et al., 2010), and growth during infection in a mouse model of tuberculosis in Mycobacterium tuberculosis (Covarrubias et al., 2005). Recently, prokaryotic β-class CAs that share no sequence similarity with human α-CA isoenzymes have been recognized as promising drug targets for antimicrobial treatments (Supuran, 2007b; Supuran & Scozzafava, 2007). They include CAs from E. coli (Cronk et al., 2006), He. pylori (Nishimori et al., 2007), Sa. Typhimurium (Nishimori et al., 2011), St. pneumoniae (Burghout et al., 2011), Haemophilus influenzae (Hoffmann et al., 2011), Brucella suis (Joseph et al., 2010, 2011; Supuran, 2007b; Winum et al., 2010) and My. tuberculosis (Nishimori et al., 2010).
Crystal structures of six β-class bacterial CAs (ECCA from E. coli; Cronk et al., 2001), HICA from Ha. influenzae (Cronk et al., 2006), Rv1284 and Rv3588c from My. tuberculosis (Covarrubias et al., 2005, 2006), stCA1 from Sa. Typhimurium (Brunzelle, 2011; Vullo et al., 2011) and carboxysomal CsoSCA from Halothiobacillus neapolitanus (Sawaya et al., 2006) have been determined. These bacterial β-CAs share a unique α-helix/β-sheet fold with at least four α-helices packed onto the β-sheet core and forming the surface of the protein (reviewed by Covarrubias et al., 2005; Rowlett, 2010). Most β-CAs coordinate catalytic Zn2+ in a pseudo-tetrahedral environment Cys2-His-X, where X is water, an anion or an Asp residue. Based on Zn2+coordination and the organization of the nearby residues, β-CAs can be divided into two distinct structural groups designated type I and type II, which are commonly identified based on crystal structure analyses (Joseph et al., 2011). These differences determine the catalytic properties of the enzymes. Type I β-CAs have a so-called ‘open active site’ with Zn2+ coordinated by two Cys, one His residue and a water molecule required for catalytic activity (Covarrubias et al., 2005, 2006; Smith & Ferry, 1999; Nishimori et al., 2010). These enzymes show catalytic activity at pH below and above 8. In contrast, type II β-CAs have a so-called ‘closed active site’ with Zn2+ coordinated by His, Asp and two Cys residues. These enzymes are not catalytically active at pH below 8, as no H2O is coordinated to the Zn2+. However, at pH above 8, the nearby Arg residue forms a salt bridge with the Asp residue coordinated to the Zn2+, which liberates the fourth Zn2+-coordinating position for an incoming H2O and enables catalytic activity (Covarrubias et al., 2005, 2006; Cronk et al., 2001, 2006). As first exemplified for mycobacterial Rv3588c, the type II proteins may undergo a large structural change, including a carboxylate shift, and gain the open active site (Covarrubias et al., 2006).
P. aeruginosa is an environmental bacterium with a remarkable capacity to cause disease in susceptible hosts. It is responsible for severe infections of lungs, heart, urinary tract and wounds, and represents a particular threat in cystic fibrosis patients as well as immunocompromised patients suffering from AIDS, cancer or severe burns (Kalai et al., 2005; Ohri & Plummer, 2004; Richard et al., 1994). It is also a major causative agent of nosocomial infections (Mesaros et al., 2007). These infections are becoming increasingly difficult to treat, as P. aeruginosa is resistant to most available antimicrobial agents (Mesaros et al., 2007). Since P. aeruginosa occupies a variety of niches ranging from soil to different tissues in a human body, it needs to adapt to a broad spectrum of environmental conditions including different levels of CO2. Such adaptations are particularly important for bacterial survival during lung infections, where the concentration of CO2 changes from 0.03 % as in the air to 5–6 % in the lungs respiratory zone (Høiby, 2006). However, little is known about the molecular mechanisms involved in P. aeruginosa response to changes in CO2 availability. Here we have identified three putative β-class CAs encoded in the P. aeruginosa genome. For the first time, the proteins were purified and proven to be functionally active. Their role in P. aeruginosa growth and survival at different CO2 levels was investigated.
Methods
Materials.
Restriction enzymes were purchased from New England Biolabs. E. coli Tuner BL21(DE3), E. coli NovaBlue Singles, pET15b and Clonables 2× ligation premix were purchased from Novagen. Oligonucleotide primers were obtained from Integrated DNA Technologies. Mini protease inhibitor cocktail was purchased from Roche. Immobilized metal affinity chromatography resin was purchased from Sigma Aldrich. Purified Pseudomonas fluorescens esterase was purchased from MP Biomedicals and Sigma Aldrich, respectively. All the other chemicals were of analytical grade.
Bacterial strains and media.
P. aeruginosa strain PAO1, the non-mucoid sequenced strain, was used in this study. Transposon mutants, PW1172, PW4542 and PW8872 with the corresponding genotypes: PAO102-E10 : : ISphoA/hah, PA2053-H07 : : ISphoA/hah and PA4676-B07 : : ISlacZ/hah, were obtained from the University of Washington Two-Allele library. The mutations were confirmed by two-step PCR: first, transposon-flanking primers were used to verify that the gene of interest was disrupted and second, transposon-specific primers were used to confirm the transposon insertion. The primer sequences are available at www.gs.washington.edu. PAO1 and transposon mutants were grown at 37 °C in biofilm minimal medium (BMM) medium, which contained (per litre): 9.0 mM sodium glutamate, 50 mM glycerol, 0.02 mM MgSO4, 0.15 mM NaH2PO4, 0.34 mM K2HPO4, 145 mM NaCl, 20 µl trace metals and 1 ml vitamin solution. Trace metal solution (per litre of 0.83 M HCl): 5.0 g CuSO4 . 5H2O, 5.0 g ZnSO4 . 7H2O, 5.0 g FeSO4 . 7H2O, 2.0 g MnCl2 . 4H2O). Vitamin solution (per litre): 0.5 g thiamine, 1 mg biotin. The pH of the medium was adjusted to 7.0. The cultures were grown in ambient air, 5 % CO2, or in the presence of 100 µM acetazolamide (AZ) and 6.4 mM DMSO (AZ solvent). The cultures treated with DMSO alone as a control showed no growth defect. For quantitative growth assays, cultures were first grown in tubes containing 5 ml BMM to mid-exponential phase, diluted to obtain equal optical densities, and used to inoculate 100 ml of fresh BMM medium in 250 ml flasks. Growth data were based on at least three biological replicates.
PAO1 was used to obtain genomic DNA for cloning CA-encoding genes. E. coli cells were grown at 37 °C in Luria–Bertani (LB) broth (10 g tryptone, 5 g yeast extract, 5 g NaCl, per litre) on a rotary shaker at 200 r.p.m. E. coli Nova Blue Singles Competent cells were used as a cloning host. E. coli Tuner BL21(DE3) cells were used for the heterologous expression of CAs.
Bioinformatic analysis.
blastp sequence alignments, using the National Centre for Biotechnology Information (NCBI) non-redundant database (GenBank release 160.1), as well as the conserved domain database (CDD) prediction and conserved residues analyses were used to predict CA-encoding genes in the P. aeruginosa PAO1 genome (www.pseudomonas.com). For phylogenetic analyses, amino acid sequences of the 11 best-studied β-class CAs were aligned with three P. aeruginosa CAs using clustal w (Larkin et al., 2007). The obtained multiple sequence alignment was then used to build an unrooted phylogenetic tree using the neighbour-joining algorithm in the Geneious 5.5.7 software. Microarray expression data were retrieved from the Gene Expression Omnibus data repository available at http://www.ncbi.nlm.nih.gov/geo.
Immunoblot analysis.
P. aeruginosa PAO1 cells were grown to mid-exponential phase and harvested. Cell pellets were washed twice with saline, resuspended in 20 mM Tris buffer (pH 8.3), containing 1 : 100 (v/v) complete Mini protease inhibitor cocktail (Roche), and disrupted using FastPrep bead beater (MP Biomedicals) in five 15 s cycles at maximum speed, with 5 min on ice between cycles. Cell extracts (5 µg) were separated by 12 % SDS-PAGE and electroblotted onto PVDF membranes (Immobilon; Millipore) using a Bio-Rad Trans-Blot SD Electrophoretic Transfer Cell unit following the manufacturer’s protocols. The membranes were probed with primary antiserum raised to β-class Cab from Methanobacterium thermoautotrophicum ΔH (Smith et al., 1999) (generously shared by Dr James Ferry, Pennsylvania State University). Goat anti-mouse immunoglobulin G, conjugated to Alexa Fluor 680 (Molecular Probes), was used as the secondary antibody. Antibody binding was detected by using an Odyssey infrared imager (LI-COR). Coomassie-stained gels were run in parallel as loading controls.
Cloning, protein expression and purification of PAO1 CAs.
CA-encoding genes PAO102, PA2053 and PA4676 (GenBank accession numbers NP_248792, NP_250743 and NP_253365, respectively) were amplified by PCR using Pfu polymerase, P. aeruginosa PAO1 genomic DNA as a template and primers containing NdeI and BamHI recognition sequences (Table 1). The resulting products were digested with BamHI and NdeI and ligated into similarly digested pET15b vector, and transformed into E. coli NovaBlue. The resultant plasmid sequences were verified by double stranded sequencing. The plasmids were then transformed into E. coli Tuner BL21(DE3) for production of the 6×His fusion proteins. E. coli TunerBL21(DE3) harbouring PAO102, PA2053 and PA4676 were grown at 37 °C in LB medium supplemented with 100 µg ampicillin ml−1 and 0.05 mM ZnSO4. At an OD600 of 0.6, fusion protein expression was induced by addition of IPTG to a final concentration of 1 mM. Simultaneously, to prevent possible Zn2+ depletion, 0.5 mM ZnSO4 was added and growth was continued for 3 h. Cells were harvested by centrifugation, resuspended in the lysis buffer containing 20 mM Tris, pH 7.9, 5 mM imidazole, 150 mM NaCl, and lysed by sonication using a 550 Sonic Dismembrator (Fisher Scientific) for 10 pulses of 10 s each at 1 min intervals. After centrifugation, the cell-free supernatants were loaded onto Ni2+ charged immobilized metal affinity chromatography columns. The loaded columns were washed with the lysis buffer, followed by the same buffer with 60 mM imidazole, and proteins were eluted with the lysis buffer containing 300 mM imidazole. The elution fractions appeared free of detectable contaminating proteins, as determined by SDS-PAGE followed by Coomassie blue R-250 staining. The fractions containing the 6×His-tagged proteins were pooled and dialysed in five steps (2 h first step and 1 h each of the following steps) against dialysis buffers with decreasing amounts of imidazole, NaCl and glycerol. The first dialysis buffer contained 20 mM Tris/HCl, pH 7.9, 150 mM imidazole, 100 mM NaCl and 10 % glycerol, the second buffer contained 20 mM Tris/HCl, pH 7.9, 50 mM imidazole, 50 mM NaCl and 5 % glycerol, and the final dialysis (storage) buffer contained 20 mM Tris/HCl, pH 7.5 or pH 8.3. Dialysis against the final dialysis buffer was repeated three times.
Table 1. PCR primers used in this study.
| Primer | Sequence (5′→3′) | Comments |
| PAO102_F* | AGAGAGCATATGCCAGACCGTATG | External psCA1 primer, forward |
| PAO102_R | AGAGAGGGATCCTCACGAGCTCAG | External psCA1 primer, reverse |
| PA2053_F | AGAGAGCATATGCGTGACATCATCG | External psCA2 primer, forward |
| PA2053_R | AGAGAGGGATCCTCAGGCGAC | External psCA2 primer, reverse |
| PA4676_F | AGAGAGCATATGAGCGACTTGCAG | External psCA3 primer, forward |
| PA4676_R | AGAGAGGGATCCTCAGCAGCAAC | External psCA3 primer, reverse |
The forward and the reverse primers include the recognition sites for NdeI and BamHI (underlined). Six bases (AGAGAG) were added to the start of each primer.
Protein identification.
To verify the identity of the high molecular mass protein bands detected by SDS-PAGE, the protein bands were excised and digested with trypsin. The resultant peptides were analysed by MALDI-TOF using a Voyager DE-PRO instrument operated in reflectron mode at the Protein Resource Facility at Oklahoma State University. Proteins were identified using Mascot (v2.2.2; Matrix Science) and a database generated based on the P. aeruginosa genome. Protein identifications were accepted if their Mascot probability based MOWSE scores (PBM) were statistically significant, and at least five peptides were identified.
Cross-linking analysis.
The purified proteins were dialysed in 20 mM citrate phosphate buffers, pH 7.5 and 8.3, and subjected to cross-linking using dithiobis(succinimidyl propionate) (DSP), a thiol-containing homobifunctional cross-linker, at a protein to DSP molar ratio of 1 : 6. After 30 min incubation at room temperature, the reaction was quenched with 3× SDS-PAGE sample buffer. Then the reaction mixtures were split into two tubes, one tube with no DTT, and the other receiving DTT to a final concentration of 1 mM to promote cleavage of the cross-linker. The proteins were separated using SDS-PAGE followed by Coomassie blue staining.
Enzyme activity.
Carbonic anhydrase activity was assayed using the modified colorimetric method described by Covarrubias et al. (2005). The buffer contained 25 mM TAPS (pH 7.5 or pH 8.3), 80 µM m-cresol purple and 100 mM sodium sulfate that was added to maintain the ionic strength of the reaction mixture. The reaction was initiated by addition of 0.5 ml CO2-saturated water to 0.5 ml of a 2× buffer solution containing protein sample. Changes in absorbance at 578 nm (A578) were measured at 25 °C at 1 s intervals using a Shimadzu UV-Vis spectrophotometer with time t = 0 coinciding with manual addition of substrate. A buffer and BSA were used as negative controls. BSA showed no CA activity. One unit of enzyme activity was defined as the amount of enzyme (mg) required to change pH from 7.5/8.3 to 6.3 min−1 at 25 °C.
Esterase activity was measured as described by Covarrubias et al. (2005) with slight modifications. The reaction was initiated by adding 100 µl of the protein sample to 800 µl 50 mM Tris/HCl, pH 7.5, and 100 µl 5 mM p-nitrophenylacetate solution. The substrate solution was prepared freshly for every measurement by dissolving p-nitrophenylacetate in DMSO. Changes in absorbance at 406 nm (A406) were measured at 25 °C at 1 s intervals using a Shimadzu UV-Vis spectrophotometer with time t = 0 coinciding with manual addition of protein. A buffer and BSA were used as negative controls. BSA showed no esterase activity. Recombinant P. fluorescens esterase was used as a positive control and showed specific activity of 5.5±0.6 U mg−1. One unit of enzyme activity was calculated as the amount of enzyme (mg) required to hydrolyse p-nitrophenylacetate (µmol) min−1 at 25 °C.
Elemental analysis.
Quantitative analysis of the elemental content of the purified CAs was performed using a Thermo Jarrell-Ash Enviro 36 Inductively Coupled Argon Plasma (ICAP) instrument with simultaneous 20-element capability at the Chemical Analysis Laboratory, University of Georgia, Athens. Prior to the analyses, CAs were dialysed in 20 mM Tris buffer at pH 7.5 and 8.5 at 4 °C. To prevent zinc contamination, all the solutions were prepared using plasticware and deionized water (18 MΩ) produced by a Barnsted-thermolyne deionization system. Furthermore, buffer samples were subjected to the ICAP analysis and the detected amounts of zinc were subtracted as blanks from the corresponding measurements in the protein samples. Protein concentration was calculated based on A280 (Mach et al., 1992). To avoid possible fold interference, the proteins were first denatured in 6 M guanidine hydrochloride.
Circular dichroism (CD) spectroscopy.
Far-UV CD spectroscopy analysis was performed using a Jasco J815 spectropolarimeter equipped with a Peltier temperature controller (Jasco). Spectra were acquired using a 0.1 cm path length cuvette at 10 °C at a scan rate of 50 nm min−1 with a resolution of 1.0 nm and data integration time of 2 s (Mukherjee et al., 2009).
Results and Discussion
P. aeruginosa PAO1 genome encodes three putative β-class CAs
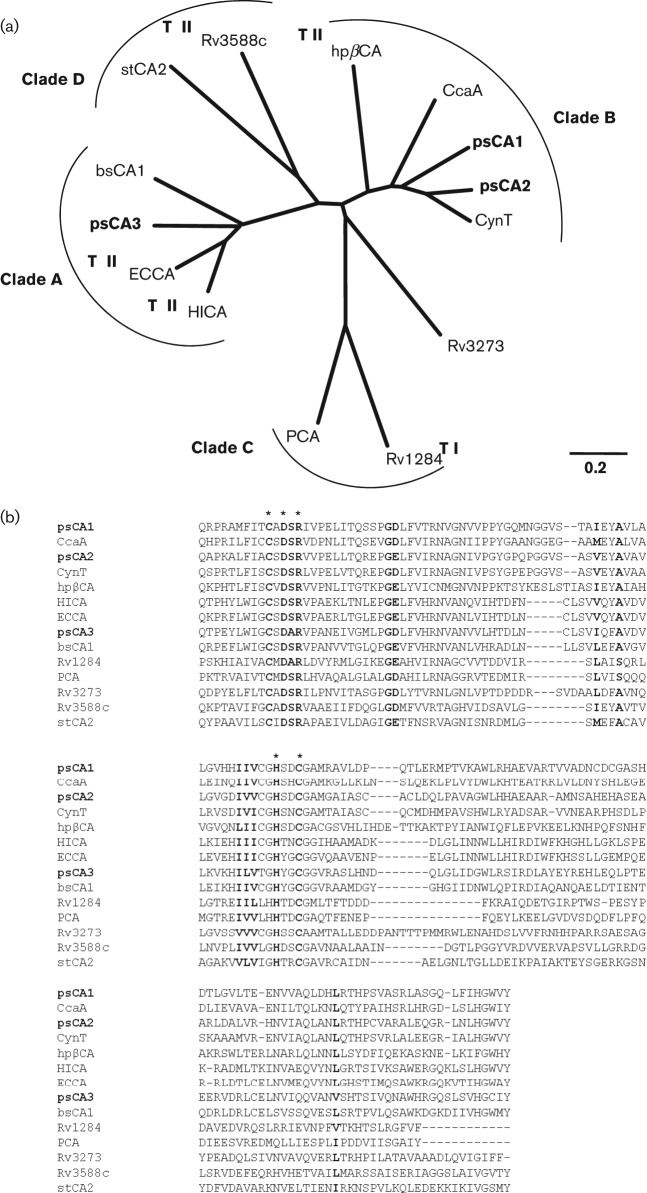
Sequence analysis of the P. aeruginosa PAO1 genome identified three genes (PAO102, PA2053 and PA4676) encoding putative β-class cytoplasmic CAs. The genes are present in all sequenced pseudomonads and share 23–100 % amino acid sequence identity. We designated them pseudomonad CAs, psCA1, psCA2 and psCA3, correspondingly. Comparative amino acid sequence analyses showed that the predicted CAs share 28–45 % amino acid sequence identity and cluster with distinct sets of homologues. The CDD prediction placed psCA1 and psCA2 in clade B, but psCA3 in clade A, of β-class CAs. Clade A represents primarily CAs from Gram-negative bacterial species that are more closely related to plant CAs than are the other clades, whereas clade B includes CAs from Gram-negative bacterial and eukaryotic species (Smith & Ferry, 2000). Phylogenetic analysis using 11 functionally and/or structurally characterized bacterial β-CAs confirmed that the PAO1 CAs belong to distinct clades and showed that psCA1 and psCA2 are related to clade B CynT homologues from Synechocystis (So & Espie, 1998), E. coli (Guilloton et al., 1992) and He. pylori (Supuran, 2007a), whereas psCA3 is related to clade A HICA from Ha. influenza (Cronk et al., 2006) and ECCA from E. coli (Cronk et al., 2001) (Fig. 1a). To further support the hypothesis that the identified genes encode functional CAs, the amino acid sequences of their active sites as predicted by the CDD sequence analysis were aligned with the active sites of 11 characterized β-class CAs (Fig. 1b). The active sites share 12–54 % sequence identity and contain 14 highly conserved amino acid residues. These include five residues that are characteristic for β-CAs. The first four residues that are required for the coordination of Zn2+ (Cys42, Asp44, His98 and Cys101 in ECCA from E. coli; Cronk et al., 2001) include Cys60, Asp62, His119 and Cys122 (psCA1); Cys39, Asp41, His98 and Cys101 (psCA2); Cys42, Asp44, His98 and Cys101 (psCA3), while the fifth characteristic residue (Arg46 in ECCA) is involved in the formation of an Asp–Arg catalytic dyad (salt bridge): Arg64 (psCA1), Arg43 (psCA2) and Arg46 (psCA3). Additionally, Gly51 (mtCA1), known to be involved in the formation of a dimer interface (Covarrubias et al., 2005), aligns with Gly76 (psCA1), Gly55 (psCA2) and Gly58 (psCA3). The remaining conserved residues have not been assigned a functional role. Overall, the sequence analyses strongly suggested that psCA1, psCA2 and psCA3 encode functional CAs.
Fig. 1.
Sequence analyses of psCA1, psCA2 and psCA3. (a) Unrooted bootstrap neighbour-joining phylogenetic tree calculated from an alignment of the amino acid sequences of the PAO1 CAs and the following bacterial β-class CAs: gi|78099988| CynT E. coli, gi|2493490| CcaA Synecocystis, gi|6014888| hpβCA He. pylori, gi|47606320| ECCA E. coli, gi|1175500| HICA Ha. influenza, gi|23500522| bsCA 1 B. suis, gi|54042506| Rv1284 M. tuberculosis, gi|332075681| PCA St. pneumoniae, gi|15610409| Rv3273 M. tuberculosis, gi|15610724| Rv3588c M. tuberculosis, gi|16445318| stCA 2 Sa. Typhimurium. TI and TII represent type I and type II β-CAs, respectively. Bar, 0.2 changes per nucleotide position. (b) Amino acid sequence alignment of the predicted active sites of the PAO1 CAs and the eleven bacterial β-CAs used in the phylogenetic analyses. The active site residues of the CAs were predicted by using the CDD algorithms in the NCBI (Marchler-Bauer et al., 2011). The conserved amino acids are shown in bold. Five residues involved in Zn2+ coordination and catalytic activity are shown with an asterisk (Joseph et al., 2010).
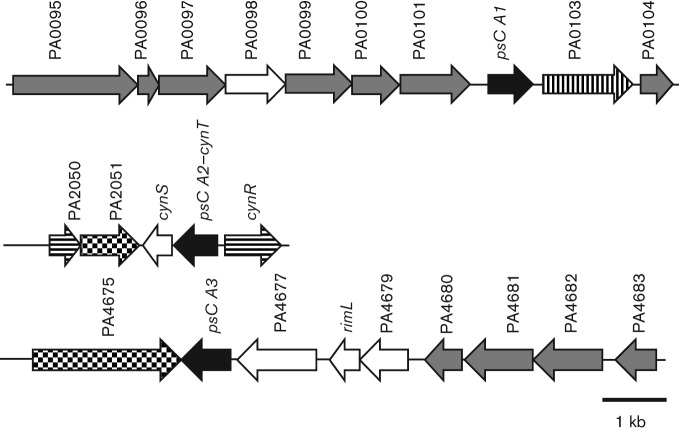
Despite sequence similarity, the genetic neighbourhoods of the CA-encoding genes are dissimilar (Fig. 2). psCA1 is clustered with PAO103 encoding a putative sulfate permease of the major facilitator superfamily (MFS), which is likely involved in bicarbonate transport as described for other members of this family (Felce & Saier, 2004). So, psCA1-PAO103 may be responsible for CO2/bicarbonate homeostasis in PAO1. psCA2 shares 65 % amino acid sequence identity with CynT from E. coli and is closely clustered with cynS, a cyanate lyase, and transcriptional regulator cynR. Cyanate lyase catalyses the conversion of cyanate into ammonia and carbon dioxide. We predict that psCA2 may catalyse the hydration of CO2 generated by cyanase into HCO3−, thereby preventing depletion of the HCO3− required for further degradation of cyanate or for other metabolic processes as suggested by Guilloton et al. (1992). Finally, psCA3 is clustered with a predicted peptidase (PA4677) and TonB receptor (PA4675), and may be functionally related to protein modification or degradation. Altogether, this suggests that the PAO1 CA enzymes may have distinct functional roles in the physiology of this organism. Multiple CAs in a single organism are not uncommon; for example, two out of three CAs in My. tuberculosis also showed distinct characteristics and were suggested to have different functional duties (Covarrubias et al., 2005).
Fig. 2.
The genetic organization of CA-encoding genes in the P. aeruginosa PAO1 genome. The numbers above the arrows indicate CDS (PA) numbers from the PAO1 genome assembly (http://www.pseudomonas.com). Carbonic anhydrases psCA1, psCA2 and psCA3 are shown in black. Putative transporters are shown in vertical lines, putative transcriptional regulator and sigma factor are shown in horizontal lines, predicted receptors are shown in tiled squares, putative enzymes are shown in white and hypothetical proteins are shown in grey.
P. aeruginosa CAs were heterologously expressed and purified
To study the catalytic activity of the predicted P. aeruginosa β-CAs, the proteins were first cloned, heterologously expressed and purified. psCA1, psCA2 and psCA3 contain 242, 220 and 215 amino acids, yielding molecular monomeric masses of 27, 23 and 24 kDa, respectively. SDS-PAGE of the purified proteins followed by MALDI-TOF-based protein identification showed that psCA1, psCA2 and psCA3 formed dimers with the corresponding molecular masses of ~55, 50 and 50 kDa (Fig. 3). The fact that the dimers could be detected by SDS-PAGE indicates a strong interaction between the monomers. Chemical cross-linking using DSP, a homo-bifunctional reagent specifically reacting with primary amine groups, confirmed dimerization of the proteins; however, it did not yield as much dimers (data not shown). Dimers are considered essential structural units of β-CAs, as once formed, they complete the active site environment (Covarrubias et al., 2006). Further oligomerization of psCAs is also possible as was shown for a number of β-CAs forming tetramers or octamers (Rowlett, 2010), that may be associated with gaining a catalytically active form of the enzyme (Covarrubias et al., 2006).
Fig. 3.
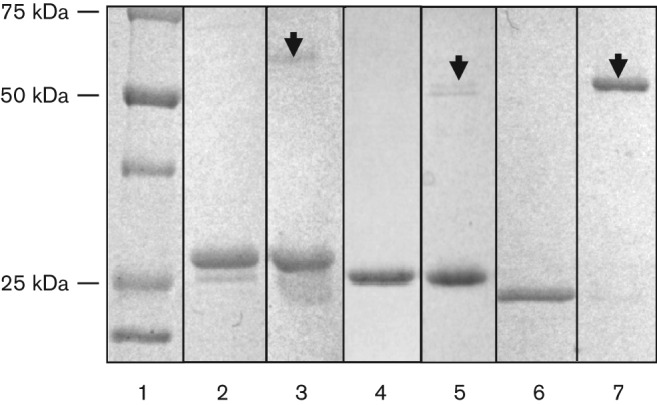
Coomassie-blue-stained gel of the His-Tag-purified CAs from P. aeruginosa PAO1. Lanes: 1, protein ladder; 2 and 3, psCA1 boiled and not boiled; 4 and 5, psCA2 boiled and not boiled; 6 and 7, psCA3 boiled and not boiled. Arrowheads show protein dimerization. The identity of the proteins was confirmed by MALDI-TOF.
P. aeruginosa CAs contain zinc and undergo pH-dependent structural changes
Since zinc is required for catalytic activity in β-CAs (Supuran, 2011), the purified proteins were analysed for zinc content using ICAP analyses. As expected, all three proteins contained zinc. The calculated zinc/protein molar ratios were 0.5 for psCA1 and psCA2, and 1.1 for psCA3. It is not clear why the zinc/protein molar ratios in psCA1 and psCA2, but not psCA3, were 0.5, suggesting one zinc atom per dimer. Zinc is commonly present in β-CAs as one atom per protein monomer (Covarrubias et al., 2005; Guilloton et al., 1992; Smith & Ferry, 1999). Lower zinc/protein ratios have been correlated with a lack of catalytic activity (Covarrubias et al., 2005). Although the catalytic activities of the purified proteins reported below did not correlate with the zinc content, it is possible that providing a 1 : 1 zinc/protein ratio may further increase the measured catalytic activities of psCA1 and psCA2. Since the His tag may chelate zinc, His tag-free purification may be needed to prevent zinc depletion in psCA1 and psCA2.
To estimate a possible effect of pH on the secondary structure, the proteins were dialysed at pH 7.5 and 8.3 and subjected to far-UV CD spectroscopy. The collected spectra of all three proteins showed discrete minima at 208 nm and 222 nm (Fig. 4), which is characteristic of a protein with α-helical character and agrees with a characteristic α-helix/β-sheet fold of β-CAs (Cronk et al., 2001; Smith et al., 2000). Interestingly, the CD spectra of psCA3 and psCA2, but not psCA1, exhibited a shift at pH 7.5 versus pH 8.3, suggesting that these two proteins undergo a conformational change as a function of pH. This may relate to the pH-dependent structural changes yielding an accessible active site shown in a type II mycobacterial CA Rv3588c, and thus presenting a regulatory mechanism for β-CAs catalytic activity (Covarrubias et al., 2006).
Fig. 4.
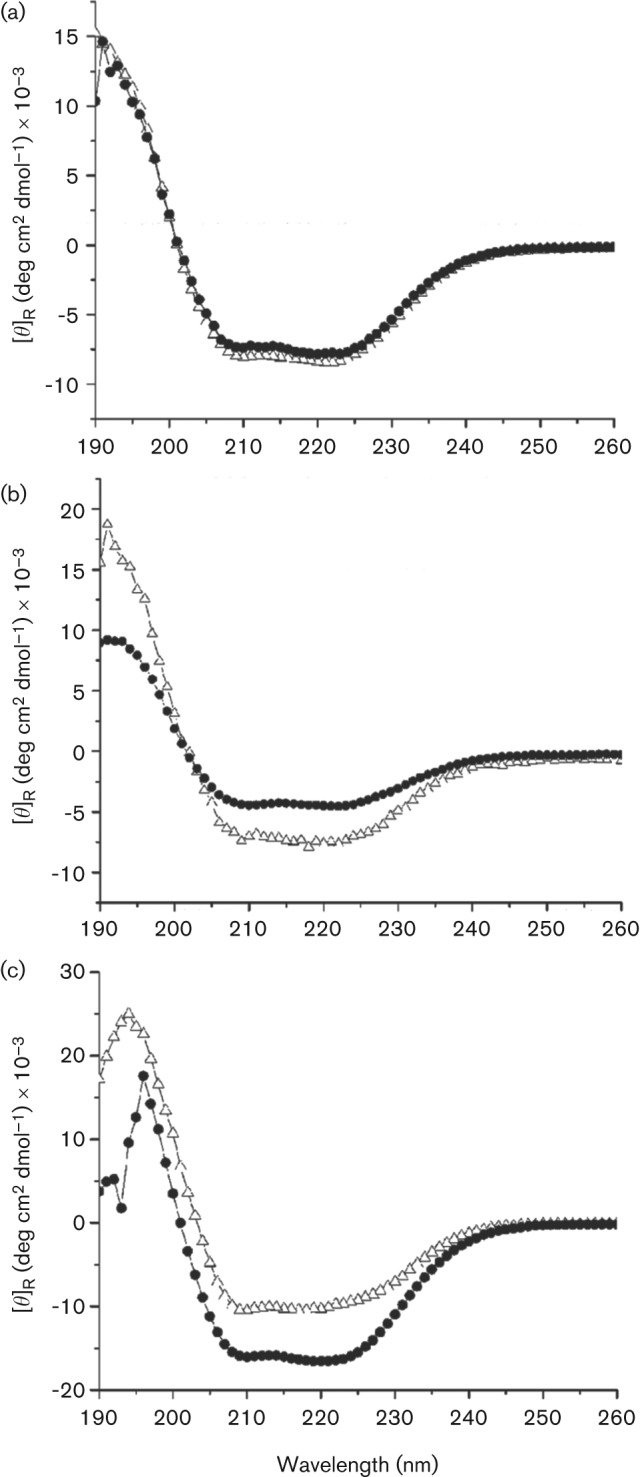
CD spectroscopy of the purified CAs. Far-UV spectra of the recombinant proteins psCA1 (a), psCA2 (b) and psCA3 (c) were recorded at 10 °C in 20 mM citrate phosphate buffer at pH 7.5 (Δ) and pH 8.3 (•). The means of three scans were graphed.
psCA1, psCA2 and psCA3 are functionally active CAs
To confirm that the purified proteins are functionally active CAs, their specific carbonic anhydrase activities were measured at pH 7.5 and 8.3. The results place PAO1 CAs in the range of catalytic activities of several previously reported β-CAs with psCA1 being most similar to the CA from Bacillus subtilis SA3 and psCA2 with psCA3 being more similar to Cab from Me. thermoautotrophicum (Table 2). psCA1 and psCA2 were active at both pH; however, the activity of psCA1 was at least 19-fold higher. psCA3 showed catalytic activity only at pH 8.3. Since α-class CAs are also known to catalyse the reverse hydrolysis of esters (Şentürk et al., 2011), we tested the CAs for specific esterase activity as well. However, none of the purified enzymes exhibited detectable esterase activity with p-nitrophenylacetate as a substrate (data not shown). A similar lack of esterase activity was detected in several β-CAs including hpβCA from He. pylori, Cab from Me. thermoautotrophicum, bsCA 1 from Br. suis (Innocenti & Supuran, 2010), and Rv1284 and Rv3588c from My. tuberculosis (Covarrubias et al., 2005).
Table 2. Carbonic anhydrase specific activities of the recombinant proteins measured at pH 7.5 and pH 8.3.
| Protein | Carbonic anhydrase specific activity (U mg−1) | ||
| pH 7.5 | pH 8.0 | pH 8.3 | |
| psCA1 | 401±39* | 750±14 | |
| psCA2 | 21±2 | 29±2 | |
| psCA3 | nd† | 26±3 | |
| Caba‡ | 39 | ||
| acCAb | 5236 | ||
| CsoSCAc | 3.41±0.08 | ||
| baCAd§ | 737.9 | ||
sd (n = 3).
No activity was detected.
Published records include β-class CAs from: a, Me. thermoautotrophicum (Smith & Ferry, 1999); b, Acetobacterium woodii (BrausStromeyer et al., 1997); c, Thimicrospira srunogenam (Dobrinski et al., 2010); d, Ba. subtilis SA3 (Ramanan et al., 2009).
Proteins were named in this study.
The catalytic CA activities and the CD spectra of the enzymes showed that psCA1 displays no effect of pH on the secondary structure and is active at both pH 7.5 and 8.3. This suggests that psCA1 may belong to the type I β-CAs together with Rv1284 from My. tuberculosis (Nishimori et al., 2010). These proteins have an ‘open’ or ‘accessible’ catalytic site and are active at pH below and above 8. In contrast, psCA3 was active only at pH 8.3 and showed a pH-dependent shift in the CD spectra. These data indicate that psCA3 may belong to the type II β-CAs, known to possess a ‘closed’ or ‘blocked’ catalytic site containing a four-residue Cys2-His-Asp coordination sphere for Zn2+ and no Asp–Arg dyad (Rowlett, 2010). These enzymes are not active at pH <8. However, at pH >8, the Asp–Arg dyad is reformed and liberates the fourth coordinating position required for the catalytic water molecule, as shown in ECCA from E. coli (Cronk et al., 2001), Rv3588c from My. tuberculosis (Covarrubias et al., 2005, 2006) and HICA from Ha. influenza (Cronk et al., 2006). The phylogenetic analysis also supports this prediction by grouping psCA3 closely with the type II ECCA and HICA. For psCA2, sequence alignment and phylogenetic analyses confidently grouped this protein with psCA1 and CynT orthologues into clade B of β-CAs. In agreement, psCA2 showed a low-level activity at both pH, but displayed a pH-dependent change in the spectral characteristics. However, the analysis of thermal unfolding together with the secondary structure content estimation for the protein (data not shown) suggested its partial unfolding under the conditions tested, which may explain the low protein stability in solutions and its low catalytic activity. Further studies are needed to classify this protein into a structural type.
psCA1, psCA2 and psCA3 are expressed in P. aeruginosa
Since CAs are known to be required for sequestering cellular CO2 under the CO2-poor conditions in several micro-organisms (Burghout et al., 2010), we first tested whether the expression of the psCAs is affected in response to PAO1 growth at different CO2 levels: low, as in ambient air (0.03 % CO2) and high, in the presence of 5 % CO2. For this, immunoblot analysis of PAO1 cell extracts was used. All three proteins were detected in PAO1 cells as recognized by the primary antiserum raised against β-class Cab from Me. thermoautotrophicum ΔH (Fig. 5). At low partial pressure of CO2, psCA1 appeared more abundant, followed by psCA3 and psCA2. At high CO2, the apparent abundance of psCA1 slightly increased, whereas the abundance of psCA2 and psCA3 did not change and reduced, respectively (Fig. 5). The expression of several heterotrophic β-CAs has been reported to be induced by low CO2 (Aguilera et al., 2005; Amoroso et al., 2005; Kaur et al., 2009), whereas the expression of others is regulated by additional or alternative factors including growth rate, pH or stress (Kaur et al., 2009; Merlin et al., 2003). The differential expression of psCA proteins suggests their independent regulation and further supports the hypothesis that they play different physiological roles in P. aeruginosa.
Fig. 5.
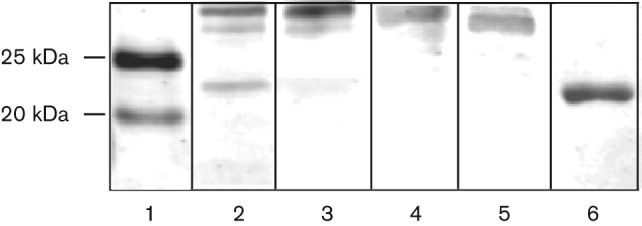
Immunoblot analysis of cytosolic cell extract from P. aeruginosa PAO1 grown in BMM at ambient air or 5 % CO2. Lanes: 1, protein ladder; 2, PAO1 cell extract at ambient air; 3, PAO1 cell extract at 5 % CO2; 4, 5 and 6, purified psCA1, psCA2 and psCA3, respectively. The CAs were probed with the primary antiserum raised against β-class CA, Cab from Me. thermoautotrophicum (gift from Dr James Ferry, Pennsylvania State University; Smith et al., 1999).
psCA1 contributes to the adaptation of P. aeruginosa to low CO2
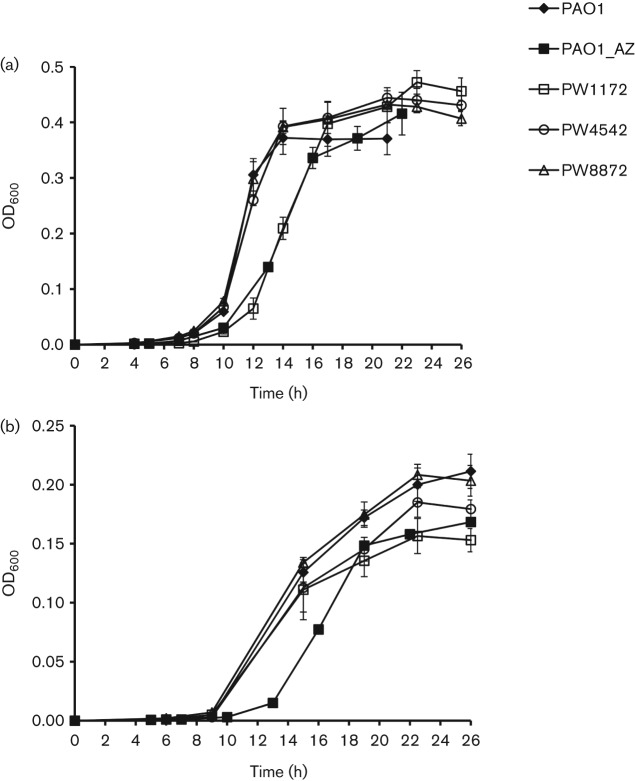
To test if the CAs play a role in PAO1 growth at different CO2 levels, PAO1 and the transposon mutants PW1172, PW4542 and PW8872 with disrupted psCA1, psCA2 and psCA3, respectively, were grown and monitored in ambient air or in the presence of 5 % CO2. At low partial pressure of CO2, the disruption of psCA2 and psCA3 had no effect, whereas the lack of psCA1 delayed growth for at least 2 h and decreased growth rate from 0.36 h−1 in PAO1 to 0.26 h−1 in PW1172 (Fig. 6a). A similar growth inhibition was observed in the presence of AZ, one of the most potent permeant inhibitors for bacterial α-, β- and γ-CAs (Burghout et al., 2010; Joseph et al., 2010; Nishimori et al., 2009, 2011). This suggested that the growth defect in the mutant was most likely due to the lack of CA activity. This also indicated that the other two CAs, psCa2 and psCA3, expressed at lower levels and showing low CA activity, did not complement the lack of psCA1 under the conditions tested. Thus, psCA1, the most abundant PAO1 β-CA with the highest CA activity, plays a role in PAO1 growth at low partial pressure of CO2 in ambient air, as was also shown for several other β-CAs including PCA in St. pneumonia (Burghout et al., 2010) and Can in E. coli (Merlin et al., 2003).
Fig. 6.
Growth characteristics of strain PAO1 and transposon mutants PW1172, PW4542 and PW8872 with the corresponding genotypes: PAO102-E10 : : ISphoA/hah, PA2053-H07 : : ISphoA/hah and PA4676-B07 : : ISlacZ/hah, obtained from the University of Washington Two-Allele library. The cultures were grown in BMM in ambient air (a), or at 5 % CO2 (b). To inhibit CA activity, 100 µM AZ in 50 % DMSO was added. All graphs represent the means of three independent growth experiments.
The low atmospheric partial pressure of CO2 together with its rapid diffusion from the cell causes depletion of cellular bicarbonate, which is needed for a number of bicarbonate-dependent carboxylation reactions in central metabolism and biosynthesis of small molecules and fatty acids (Aguilera et al., 2005; Burghout et al., 2010; Merlin et al., 2003). Therefore, growth in air requires sufficient CA activity for endogenous supply of bicarbonate via enzymic conversion of CO2 (Merlin et al., 2003). This explains the impaired growth of PW1172 in ambient air. The recovery of growth in the PW1172 mutant as well as in AZ-treated PAO1 suggests an alternative source of endogenous CO2/bicarbonate, which may include additional CAs that are not inhibited by AZ. Sequence analysis retrieved no α-class and three homologues of γ-class cytosolic CAs in the PAO1 genome. These included PAO066, PA3754 and PA5540 that share 21–56 % amino acid sequence identity with functional γ-class CAs, Cam in Methanosarcina thermophila and YrdA in E. coli (Alber & Ferry, 1994; Park et al., 2012), and potentially could contribute to the total CA activity in PAO1. Overall, the presence of multiple CAs indicates their high physiological significance in P. aeruginosa and a robustness of adaptations to low environmental CO2 in this organism.
Interestingly, at 5 % CO2, AZ showed a similar growth inhibition as in ambient air (Fig. 6b), suggesting that CA activity is important for PAO1 growth under this condition as well. However, PW1172 and PW4542 showed only a small defect in growth. This is noteworthy, since the addition of CO2 was shown to often overcome the need for CA activity (Kusian et al., 2002; Merlin et al., 2003). It is also noteworthy that PAO1 showed at least twofold decrease in growth rate and maximum yield at elevated CO2. Based on sensitivity to CO2, heterotrophs range from those whose growth requires CO2 under low CO2 conditions (Kusian et al., 2002; Brown & Howitt, 1969; Bury-Moné et al., 2008; Kempner & Schlayer, 1942; Merlin et al., 2003) to those whose growth is inhibited by elevated CO2 (Enfors & Molin, 1980; Gill & Tan, 1979), including organisms like P. aeruginosa that may exhibit both characteristics. In the former case, the need for CO2 is attributed to CO2 fixation in biosynthetic reactions, and CAs, as discussed above, are shown to be commonly required for survival of these organisms at low levels of CO2 (Kusian et al., 2002; Burghout et al., 2010; Hashimoto & Kato, 2003; Merlin et al., 2003). Whereas CO2 inhibition, the phenomenon used in food preservation (Dixon & Kell, 1989), is consistent with its inhibitory effect on respiratory metabolism (Dixon & Kell, 1989), enzymic activity (King & Nagel, 1975) and/or alterations in cell membrane permeability (Sears & Eisenberg, 1961), the role of CAs in the organisms inhibited by elevated CO2 has not been specifically addressed. One possible mechanism may relate to the role of CAs in the maintenance of intracellular pH homeostasis through the CO2/HCO3− buffer system.
Finally, based on the microarray expression data (GDS2869) available at the Gene Expression Omnibus data repository, the transcription of psCA1 and psCA2, but not psCA3, was induced about three to four times in P. aeruginosa isolates from cystic fibrosis lung sputa grown both in vitro and in vivo (Son et al., 2007). This suggests a potential role of these proteins in the ability of P. aeruginosa to survive in a host, as has been shown for β-CAs in He. pylori (Bury-Moné et al., 2008), Sa. Typhimurium (Valdivia & Falkow, 1997), St. pneumoniae (Burghout et al., 2010) and My. tuberculosis (Covarrubias et al., 2005).
Conclusions
Three functionally active CAs were identified in P. aeruginosa PAO1. According to sequence analyses, these proteins are highly conserved among pseudomonads and represent clades A and B of bacterial β-class CAs. The high conservation and multiple occurrence of the proteins suggests their fundamental physiological significance in pseudomonads. Our results indicate that psCA1 belongs to type I, and psCA3 to type II β-CAs. All three CAs are expressed in PAO1 cells at different CO2 levels, while psCA1, the most active and abundant β-CA, plays a role in PAO1 survival at low CO2. The enzymes will be further studied for structural features as well as their possible role in P. aeruginosa virulence.
Acknowledgements
This work was supported by the Oklahoma State University Start-Up funding and Grant-in-Aid from American Heart Association (award 09BGIA2330036). We thank Dr James Ferry (Pennsylvania State University) for providing primary antiserum. Mass spectrometry analyses were performed in the DNA/Protein Resource Facility at Oklahoma State University, using resources supported by the NSF MRI and EPSCoR programs (award DBI/0722494). Transposon mutants were obtained from the University of Washington Two-Allele library (grant NIH P30 DK089507).
Abbreviations:
- AZ
acetazolamide
- CA
carbonic anhydrase
- CD
Circular Dichroism
- CDD
conserved domain database
- ICAP
inductively coupled argon plasma
References
- Aguilera J., Petit T., de Winde J. H., Pronk J. T. (2005). Physiological and genome-wide transcriptional responses of Saccharomyces cerevisiae to high carbon dioxide concentrations. FEMS Yeast Res 5, 579–593. 10.1016/j.femsyr.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Alber B. E., Ferry J. G. (1994). A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc Natl Acad Sci U S A 91, 6909–6913. 10.1073/pnas.91.15.6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso G., Morell-Avrahov L., Müller D., Klug K., Sültemeyer D. (2005). The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol Microbiol 56, 549–558. 10.1111/j.1365-2958.2005.04560.x [DOI] [PubMed] [Google Scholar]
- Braus-Stromeyer S. A., Schnappauf G., Braus G. H., Gössner A. S., Drake H. L. (1997). Carbonic anhydrase in Acetobacterium woodii and other acetogenic bacteria. J Bacteriol 179, 7197–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown O. R., Howitt H. F. (1969). Growth inhibition and death of Escherichia coli from CO2 deprivation. Microbios 3, 241–246. [Google Scholar]
- Brunzelle, J. S., Wawrzak, Z., Onopriyenko, O., Anderson, W. F. & Savchenko, A. (2011). 1.54a resolution crystal structure of a beta-carbonic anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. lt2.
- Burghout P., Cron L. E., Gradstedt H., Quintero B., Simonetti E., Bijlsma J. J. E., Bootsma H. J., Hermans P. W. M. (2010). Carbonic anhydrase is essential for Streptococcus pneumoniae growth in environmental ambient air. J Bacteriol 192, 4054–4062. 10.1128/JB.00151-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghout P., Vullo D., Scozzafava A., Hermans P. W. M., Supuran C. T. (2011). Inhibition of the β-carbonic anhydrase from Streptococcus pneumoniae by inorganic anions and small molecules: Toward innovative drug design of antiinfectives? Bioorg Med Chem 19, 243–248. 10.1016/j.bmc.2010.11.031 [DOI] [PubMed] [Google Scholar]
- Bury-Moné S., Mendz G. L., Ball G. E., Thibonnier M., Stingl K., Ecobichon C., Avé P., Huerre M., Labigne A. & other authors (2008). Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect Immun 76, 497–509. 10.1128/IAI.00993-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias A., Larsson A. M., Högbom M., Lindberg J., Bergfors T., Björkelid C., Mowbray S. L., Unge T., Jones T. A. (2005). Structure and function of carbonic anhydrases from Mycobacterium tuberculosis. J Biol Chem 280, 18782–18789. 10.1074/jbc.M414348200 [DOI] [PubMed] [Google Scholar]
- Covarrubias A. S., Bergfors T., Jones T. A., Högbom M. (2006). Structural mechanics of the pH-dependent activity of β-carbonic anhydrase from Mycobacterium tuberculosis. J Biol Chem 281, 4993–4999. 10.1074/jbc.M510756200 [DOI] [PubMed] [Google Scholar]
- Cronk J. D., Endrizzi J. A., Cronk M. R., O’neill J. W., Zhang K. Y. J. (2001). Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci 10, 911–922. 10.1110/ps.46301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk J. D., Rowlett R. S., Zhang K. Y., Tu C., Endrizzi J. A., Lee J., Gareiss P. C., Preiss J. R. (2006). Identification of a novel noncatalytic bicarbonate binding site in eubacterial beta-carbonic anhydrase. Biochemistry 45, 4351–4361. 10.1021/bi052272q [DOI] [PubMed] [Google Scholar]
- Dixon N. M., Kell D. B. (1989). The inhibition by CO2 of the growth and metabolism of micro-organisms. J Appl Bacteriol 67, 109–136. 10.1111/j.1365-2672.1989.tb03387.x [DOI] [PubMed] [Google Scholar]
- Dobrinski K. P., Boller A. J., Scott K. M. (2010). Expression and function of four carbonic anhydrase homologs in the deep-sea chemolithoautotroph Thiomicrospira crunogena. Appl Environ Microbiol 76, 3561–3567. 10.1128/AEM.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuche S., Pöggeler S. (2010). Carbonic anhydrases in fungi. Microbiology 15, 23–29. 10.1099/mic.0.032581-0 [DOI] [PubMed] [Google Scholar]
- Enfors S. O., Molin G. (1980). Effect of high concentrations of carbon dioxide on growth rate of Pseudomonas fragi, Bacillus cereus and Streptococcus cremoris. J Appl Bacteriol 48, 409–416. 10.1111/j.1365-2672.1980.tb01029.x [DOI] [PubMed] [Google Scholar]
- Felce J., Saier M. H., Jr (2004). Carbonic anhydrases fused to anion transporters of the SulP family: evidence for a novel type of bicarbonate transporter. J Mol Microbiol Biotechnol 8, 169–176. 10.1159/000085789 [DOI] [PubMed] [Google Scholar]
- Fukuzawa H., Suzuki E., Komukai Y., Miyachi S. (1992). A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci U S A 89, 4437–4441. 10.1073/pnas.89.10.4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill C. O., Tan K. H. (1979). Effect of carbon dioxide on growth of Pseudomonas fluorescens. Appl Environ Microbiol 38, 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloton M. B., Korte J. J., Lamblin A. F., Fuchs J. A., Anderson P. M. (1992). Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J Biol Chem 267, 3731–3734. [PubMed] [Google Scholar]
- Guilloton M. B., Lamblin A. F., Kozliak E. I., Gerami-Nejad M., Tu C., Silverman D., Anderson P. M., Fuchs J. A. (1993). A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol 175, 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Kato J. (2003). Indispensability of the Escherichia coli carbonic anhydrases YadF and CynT in cell proliferation at a low CO2 partial pressure. Biosci Biotechnol Biochem 67, 919–922. 10.1271/bbb.67.919 [DOI] [PubMed] [Google Scholar]
- Hewett-Emmett D., Tashian R. E. (1996). Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Mol Phylogenet Evol 5, 50–77. 10.1006/mpev.1996.0006 [DOI] [PubMed] [Google Scholar]
- Hoffmann K. M., Samardzic D., Heever K., Rowlett R. S. (2011). Co(II)-substituted Haemophilus influenzae β-carbonic anhydrase: spectral evidence for allosteric regulation by pH and bicarbonate ion. Arch Biochem Biophys 511, 80–87. 10.1016/j.abb.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N. (2006). P. aeruginosa in cystic fibrosis patients resists host defenses, antibiotics. Microbe 1, 571–577. [Google Scholar]
- Innocenti A., Supuran C. T. (2010). Paraoxon, 4-nitrophenyl phosphate and acetate are substrates of α- but not of β-, γ- and ζ-carbonic anhydrases. Bioorg Med Chem Lett 20, 6208–6212. 10.1016/j.bmcl.2010.08.110 [DOI] [PubMed] [Google Scholar]
- Iverson T. M., Alber B. E., Kisker C., Ferry J. G., Rees D. C. (2000). A closer look at the active site of γ-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39, 9222–9231. 10.1021/bi000204s [DOI] [PubMed] [Google Scholar]
- Joseph P., Turtaut F., Ouahrani-Bettache S., Montero J. L., Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Köhler S. & other authors (2010). Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Brucella suis. J Med Chem 53, 2277–2285. 10.1021/jm901855h [DOI] [PubMed] [Google Scholar]
- Joseph P., Ouahrani-Bettache S., Montero J. L., Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Winum J. Y., Köhler S., Supuran C. T. (2011). A new β-carbonic anhydrase from Brucella suis, its cloning, characterization, and inhibition with sulfonamides and sulfamates, leading to impaired pathogen growth. Bioorg Med Chem 19, 1172–1178. 10.1016/j.bmc.2010.12.048 [DOI] [PubMed] [Google Scholar]
- Kalai S., Achour W., Abdeladhim A., Bejaoui M., Ben Hassen A. (2005). [Pseudomonas aeruginosa isolated in immunocompromised patients: antimicrobial resistance, serotyping, and molecular typing]. Med Mal Infect 35, 530–535 (in French). 10.1016/j.medmal.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Kaur S., Mishra M. N., Tripathi A. K. (2009). Regulation of expression and biochemical characterization of a β-class carbonic anhydrase from the plant growth-promoting rhizobacterium, Azospirillum brasilense Sp7. FEMS Microbiol Lett 299, 149–158. 10.1111/j.1574-6968.2009.01736.x [DOI] [PubMed] [Google Scholar]
- Kempner W., Schlayer C. (1942). Effect of CO2 on the growth rate of the Pneumococcus. J Bacteriol 43, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. D., Nagel C. W. (1975). Influence of carbon dioxide upon the metabolism of Pseudomonas aeruginosa. J Food Sci 40, 362–366. 10.1111/j.1365-2621.1975.tb02202.x [DOI] [Google Scholar]
- Kusian B., Sültemeyer D., Bowien B. (2002). Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO2 concentrations. J Bacteriol 184, 5018–5026. 10.1128/JB.184.18.5018-5026.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A. & other authors (2007). clustal w and clustal_x version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Mach H., Middaugh C. R., Lewis R. V. (1992). Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal Biochem 200, 74–80. 10.1016/0003-2697(92)90279-G [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C. & other authors (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39 (Database issue), D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C., Masters M., McAteer S., Coulson A. (2003). Why is carbonic anhydrase essential to Escherichia coli? J Bacteriol 185, 6415–6424. 10.1128/JB.185.21.6415-6424.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesaros N., Nordmann P., Plésiat P., Roussel-Delvallez M., Van Eldere J., Glupczynski Y., Van Laethem Y., Jacobs F., Lebecque P. & other authors (2007). Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13, 560–578. 10.1111/j.1469-0691.2007.01681.x [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S., Ohnishi J., Hayashi M., Ikeda M. (2004). A gene homologous to β-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl Microbiol Biotechnol 63, 592–601. 10.1007/s00253-003-1402-8 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Saha B., Das A. K. (2009). Differential chemical and thermal unfolding pattern of Rv3588c and Rv1284 of Mycobacterium tuberculosis – a comparison by fluorescence and circular dichroism spectroscopy. Biophys Chem 141, 94–104. 10.1016/j.bpc.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Nishimori I., Minakuchi T., Kohsaki T., Onishi S., Takeuchi H., Vullo D., Scozzafava A., Supuran C. T. (2007). Carbonic anhydrase inhibitors: the beta-carbonic anhydrase from Helicobacter pylori is a new target for sulfonamide and sulfamate inhibitors. Bioorg Med Chem Lett 17, 3585–3594. 10.1016/j.bmcl.2007.04.063 [DOI] [PubMed] [Google Scholar]
- Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Innocenti A., Supuran C. T. (2009). Carbonic anhydrase inhibitors. Cloning, characterization, and inhibition studies of a new beta-carbonic anhydrase from Mycobacterium tuberculosis. J Med Chem 52, 3116–3120. 10.1021/jm9003126 [DOI] [PubMed] [Google Scholar]
- Nishimori I., Minakuchi T., Maresca A., Carta F., Scozzafava A., Supuran C. T. (2010). The β-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr Pharm Des 16, 3300–3309. 10.2174/138161210793429814 [DOI] [PubMed] [Google Scholar]
- Nishimori I., Minakuchi T., Vullo D., Scozzafava A., Supuran C. T. (2011). Inhibition studies of the β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium with sulfonamides and sulfamates. Bioorg Med Chem 19, 5023–5030. 10.1016/j.bmc.2011.06.038 [DOI] [PubMed] [Google Scholar]
- Ohri L. K., Plummer S. (2004). Prevention and control of nosocomial infections, 4th edition. Ann Pharmacother 38, 515–516. 10.1345/aph.1D430 [DOI] [Google Scholar]
- Park H.-M., Park J. H., Choi J. W., Lee J., Kim B. Y., Jung C. H., Kim J. S. (2012). Structures of the γ-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch. Acta Crystallogr D Biol Crystallogr 68, 920–926. 10.1107/S0907444912017210 [DOI] [PubMed] [Google Scholar]
- Ramanan R., Kannan K., Vinayagamoorthy N., Ramkumar K., Sivanesan S., Chakrabarti T. (2009). Purification and characterization of a novel plant-type carbonic anhydrase from Bacillus subtilis. Biotechnol Bioprocess Eng 14, 32–37. 10.1007/s12257-008-0099-z [DOI] [Google Scholar]
- Richard P., Le Floch R., Chamoux C., Pannier M., Espaze E., Richet H. (1994). Pseudomonas aeruginosa outbreak in a burn unit: role of antimicrobials in the emergence of multiply resistant strains. J Infect Dis 170, 377–383. 10.1093/infdis/170.2.377 [DOI] [PubMed] [Google Scholar]
- Rowlett R. S. (2010). Structure and catalytic mechanism of the β-carbonic anhydrases. Biochim Biophys Acta 1804, 362–373. 10.1016/j.bbapap.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Sawaya M. R., Cannon G. C., Heinhorst S., Tanaka S., Williams E. B., Yeates T. O., Kerfeld C. A. (2006). The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem 281, 7546–7555. 10.1074/jbc.M510464200 [DOI] [PubMed] [Google Scholar]
- Sears D. F., Eisenberg R. M. (1961). A model representing a physiological role of CO2 at the cell membrane. J Gen Physiol 44, 869–887. 10.1085/jgp.44.5.869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şentürk M., Gülçin İ., Beydemir Ş., Küfrevioğlu Ö. İ., Supuran C. T. (2011). In vitro inhibition of human carbonic anhydrase I and II isozymes with natural phenolic compounds. Chem Biol Drug Des 77, 494–499. 10.1111/j.1747-0285.2011.01104.x [DOI] [PubMed] [Google Scholar]
- Smith K. S., Ferry J. G. (1999). A plant-type (beta-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J Bacteriol 181, 6247–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. S., Ferry J. G. (2000). Prokaryotic carbonic anhydrases. FEMS Microbiol Rev 24, 335–366. 10.1111/j.1574-6976.2000.tb00546.x [DOI] [PubMed] [Google Scholar]
- Smith K. S., Jakubzick C., Whittam T. S., Ferry J. G. (1999). Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci U S A 96, 15184–15189. 10.1073/pnas.96.26.15184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. S., Cosper N. J., Stalhandske C., Scott R. A., Ferry J. G. (2000). Structural and kinetic characterization of an archaeal beta-class carbonic anhydrase. J Bacteriol 182, 6605–6613. 10.1128/JB.182.23.6605-6613.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A. K. C., Espie G. S. (1998). Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol Biol 37, 205–215. 10.1023/A:1005959200390 [DOI] [PubMed] [Google Scholar]
- Son M. S., Matthews W. J., Jr, Kang Y., Nguyen D. T., Hoang T. T. (2007). In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75, 5313–5324. 10.1128/IAI.01807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran C. T. (2007a). Novel targets against Helicoblacter pylori: a bioinformatic approach. Future Microbiol 2, 111–114. 10.2217/17460913.2.2.111 [DOI] [Google Scholar]
- Supuran C. T. (2007b). Carbonic anhydrases as drug targets–an overview. Curr Top Med Chem 7, 825–833. 10.2174/156802607780636690 [DOI] [PubMed] [Google Scholar]
- Supuran C. T. (2008). Carbonic anhydrases–an overview. Curr Pharm Des 14, 603–614. 10.2174/138161208783877884 [DOI] [PubMed] [Google Scholar]
- Supuran C. T. (2011). Bacterial carbonic anhydrases as drug targets: towards novel antibiotics? Front Pharmacol 2, 1–6. 10.3389/fphar.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran C. T., Scozzafava A. (2007). Carbonic anhydrases as targets for medicinal chemistry. Bioorg Med Chem 15, 4336–4350. 10.1016/j.bmc.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Tripp B. C., Smith K., Ferry J. G. (2001). Carbonic anhydrase: new insights for an ancient enzyme. J Biol Chem 276, 48615–48618. 10.1074/jbc.R100045200 [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Falkow S. (1997). Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277, 2007–2011. 10.1126/science.277.5334.2007 [DOI] [PubMed] [Google Scholar]
- Vullo D., Nishimori I., Minakuchi T., Scozzafava A., Supuran C. T. (2011). Inhibition studies with anions and small molecules of two novel β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium. Bioorg Med Chem Lett 21, 3591–3595. 10.1016/j.bmcl.2011.04.105 [DOI] [PubMed] [Google Scholar]
- Winum J. Y., Köhler S., Supuran C. T. (2010). Brucella carbonic anhydrases: new targets for designing anti-infective agents. Curr Pharm Des 16, 3310–3316. 10.2174/138161210793429850 [DOI] [PubMed] [Google Scholar]
- Xu Y., Feng L., Jeffrey P. D., Shi Y., Morel F. M. M. (2008). Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 452, 56–61. 10.1038/nature06636 [DOI] [PubMed] [Google Scholar]