Abstract
In magnetic resonance imaging, complex-valued measurements are acquired in time corresponding to spatial frequency measurements in space generally placed on a Cartesian rectangular grid. These complex-valued measurements are transformed into a measured complex-valued image by an image reconstruction method. The most common image reconstruction method is the inverse Fourier transform. It is known that image voxels are spatially correlated. A property of the inverse Fourier transformation is that uncorrelated spatial frequency measurements yield spatially uncorrelated voxel measurements and vice versa. Spatially correlated voxel measurements result from correlated spatial frequency measurements. This paper describes the resulting correlation structure between voxel measurements when inverse Fourier reconstructing correlated spatial frequency measurements. A real-valued representation for the complex-valued measurements is introduced along with an associated multivariate normal distribution. One potential application of this methodology is that there may be a correlation structure introduced by the measurement process or adjustments made to the spatial frequencies. This would produce spatially correlated voxel measurements after inverse Fourier transform reconstruction that have artificially inflated spatial correlation. One implication of these results is that one source of spatial correlation between voxels termed connectivity may be attributed to correlated spatial frequencies. The true voxel connectivity may be less than previously thought. This methodology could be utilized to characterize noise correlation in its original form and adjust for it. The exact statistical relationship between spatial frequency measurements and voxel measurements has now been established.
Keywords: fMRI activation, Complex-valued data, Image reconstruction, Fourier transform, k-Space
1. Introduction
In functional magnetic resonance imaging (fMRI), we apply magnetic field gradients to encode then measure the complex-valued Fourier transformation (FT) of the effective proton spin density (PSD) in a real-valued physical object. In fMRI, complex-valued measurements are acquired in spatial frequency space (usually two-dimensional), also called k-space from the use of the k variables for its axes (kx, ky). These measurements are transformed into a complex-valued image by an image reconstruction method. The most common image reconstruction method is the inverse Fourier transform. These discrete complex-valued measurements, when placed at their proper spatial frequency location, are ideally the discrete FT of the PSD. A dis crete inverse Fourier transform (IFT) is applied to the discretely measured signal to reconstruct a discretely measured PSD or image. The original object or PSD is real-valued, but due to imperfections in the imaging process, a complex-valued image of PSDs is produced (Haacke et al., 1999). These complex-valued measurements are collected to yield a complex-valued time course in each voxel. Traditional methods to detect brain activation utilize magnitude-only voxel time courses (Bandettini et al., 1993; Friston et al., 1994). Work by Menon (2002) and others indicate that the generally discarded phase portion of the fMRI voxel time courses may contain information regarding the brain's vasculature and that the entire voxel time series either in the form of real–imaginary or magnitude-phase should be used. Recently complex-valued methods to detect brain activation have been introduced (Nan and Nowak, 1999; Rowe, 2005a,b; Rowe and Logan, 2004, 2005). Preliminary work with these methods indicate that they can be used in fMRI to postacquistion suppress venous BOLD (Nencka and Rowe, 2005, 2006; Rowe, 2005c; Rowe and Nencka, 2006). These complex-valued detection methods could be combined with the current methods that connect spatial frequency measurements to voxel measurements and a more natural representation of the noise utilized.
After Fourier (or non-Fourier) image reconstruction, images are complex-valued containing a matrix of real and imaginary components of the measured effective PSD. The real part of the complex-valued measurements in each image will be stacked on top of the imaginary part of the measurements to form a single real-valued vector of measurements. A one-to-one relationship will be described between the vector of complex-valued measurements in an image and the real-valued vector with twice the dimension of stacked measurements. This one-to-one relationship or correspondence is often called an isomorphism in the mathematical literature. It is known that image voxel measurements are spatially correlated, in measured fMRI data. A property of the inverse Fourier transformation is that uncorrelated spatial frequency measurements yield spatially uncorrelated voxel measurements and vice versa. Additionally, correlated voxel measurements result from correlated spatial frequency measurements. The thrust of this paper is to relate the signal and noise characteristics of spatial frequency measurements and Fourier reconstructed image measurements. It will be shown that the true spatial correlation between voxel measurements could be inflated by correlated noise of spatial frequency measurements and that this correlated noise between spatial frequency measurements could be adjusted to yield an estimate of the true spatial correlation between voxel measurements. This has many implications for fMRI including connectivity results and activation thresholding (Logan and Rowe, 2004). There has been much work on the physiological basis for temporal autocorrelations between spatially separated voxels. It has been reported that there are spontaneous fluctuations in regional cerebral blood flow (rCBF) from spatially distinct regions (Biswal et al., 1995). Further, time-dependent changes in effective connectivity during learning have been observed (Buchel et al., 1999).
2. Statistical theory
In this section, the statistical properties of the complex-valued spatial frequency measurements are described for a single time point image. The statistical properties of the complex-valued image measurements from a complex-valued inverse Fourier transformation of the complex-valued spatial frequency measurements are described. This is done for a one-dimensional image where the characteristics of the transformation in terms of mean and covariance are easier to understand then generalized for a two-dimensional image.
2.1. One dimension
Consider a one-dimensional horizontal complex-valued magnetic resonance image with px complex-valued voxels. To obtain this image, we must measure px spatial frequencies corresponding to the ky = 0 center line. In this one-dimensional magnetic resonance image, complex-valued k-space measurements are taken in time but correspond to specific spatial frequencies. We will assume that the k-space measurements are acquired from left to right.
Let sC = (sC1, . . ., sCpx)T be these px measured complex-valued spatial frequencies stacked into a px × 1 complex-valued vector sC that is the sum of s0C, a vector of true noiseless complex-valued spatial frequencies and ∈C, a vector of complex-valued measurement error as in Appendix A where “T” denotes transposition. As shown in Appendix A, the measurements can be represented as a single real-valued vector by stacking the px real measurements upon the px imaginary measurements to yield the vector s, that is the sum of a vector of true noiseless complex-valued spatial frequencies s0, and a vector of complex-valued measurement error ∈. Since the vector s is what is measured with error, it is assumed to be characterized as having a multivariate normal distribution with mean s0 and covariance matrix Λ as described in Appendix A.
The Fourier image reconstruction process to generate a complex-valued measured image ρC consists of pre-multiplying the measured spatial frequencies sC by the Fourier matrix ΩCx in Eq. (A.2). As shown in Appendix A, this is equivalently represented as the pre-multiplication if the real-valued vector of measured spatial frequencies s by the real-valued matrix Ωx to arrive at the real-valued representation of the measured image ρ. The real-valued representation of the measured image ρ is a linear transformation of the real-valued representation of the measured spatial frequencies and thus normally distributed with mean ρ0 = Ωxs0 and covariance matrix .
An example of this methodology might be useful. Although explicit analytic expressions exist for the mean and covariance of the complex-valued transformed one-dimensional images measurements given the mean and covariance of the one-dimensional spatial frequency measurements, simulations were carried out to verify the analytic results in addition to determining those for magnitude-only image measurements where closed form analytic solutions do not exist due to the nonlinear and non one-to-one mapping. The simulations were performed under known conditions. These can be used to precisely characterize the signal and noise of the transformed measurements. All computations utilized Matlab (The Mathworks, Natick, MA, USA). Data was generated to mimic a one-dimensional magnetic resonance imaging experiment. Although this simulation is a mathematical ideal and possibly unrealistic, its results are useful in understanding the properties of the described methodology. Random complex-valued error vectors of dimension px were generated in the form of the real-valued representation. A large number, L, of random vectors of dimension 2px for the px real measurements stacked upon the px imaginary measurements denoted by s1, . . ., sL were generated from a normal distribution with mean s0 and covariance . Without loss of generality, s0 = 0 while Λ1 and Λ2 are taken to be unit variance correlation matrices. The 2 × 2 correlation matrix Λ1 is taken to have an off diagonal correlation of ϱ1 = 0.5 while the px × px correlation matrix Λ2 is taken to be an AR(1) correlation matrix with (i, j)th element where ϱ2 = 0.25. The number of randomly generated vectors was selected to be L = 106.
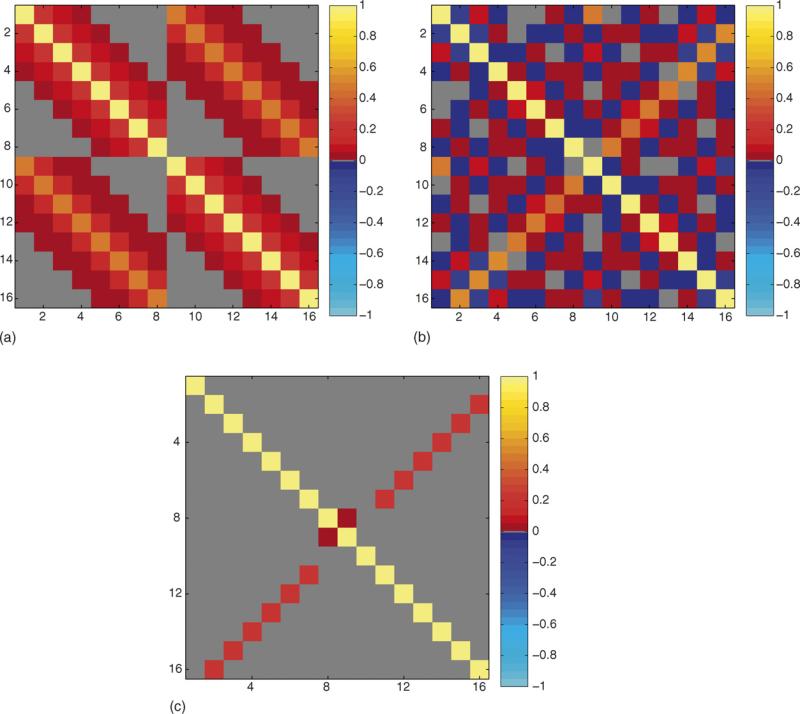
A value of px was chosen to be 8. Although the methodology equally applies to larger values, they are not shown to maintain the clarity of presentation. The sample correlation matrix from the L randomly generated one-dimensional spatial frequency vectors was computed as displayed in Fig. 1a. Further, each random one-dimensional spatial frequency vector was pre-multiplied by the appropriate inverse Fourier transform matrix Ωx given in Eq. (A.3) to produce random one-dimensional images. The sample correlation matrix of the real-valued representation ρ of the complex-valued one-dimensional image measurements ρC was computed as displayed in Fig. 1b. The sample spatial frequency correlation matrix matched its theoretical population correlation matrix in Eq. (A.4) and the sample image correlation matrix matched the population value in Eq. (A.5) utilizing the previously described theory. Further, since an analytic expression for the theoretical covariance or correlation matrix for magnitude-only image quantities can not be found, the L vectors containing real and imaginary image measurements of dimension 2px were converted to L vectors of dimension px con taining magnitude-only image quantities. The sample correlation matrix of the magnitude-only image vectors was computed for the as displayed in Fig. 1c. Note that both complex-valued voxels and real-valued magnitude-only voxels are correlated.
Fig. 1.
Correlation maps, px = 8, L = 106. (a) Correlation between complex spatial frequency measurements, Corr(s, s). (b) Correlation between complex image voxel measurements, Corr(r, r). (c) Correlation between magnitude image voxel measurements, Corr(m, m).
2.2. Two dimensions
In a two-dimensional echo planar magnetic resonance image, complex-valued measurements are taken in time but correspond to specific spatial frequencies on a Cartesian (kx, ky) grid. In a standard echo planar imaging (EPI) experiment, the measurements are taken in a “zig-zag” pattern. For example, with positive phase encode steps, the pattern starts at the bottom left of the grid with negative (kx, ky) values and moves from left to right, then right to left and so on, while going from bottom to top. The left–right direction is called the frequency encode direction while the top–bottom direction is called the phase encode direction.
We will assume that the data is collected according to this standard EPI trajectory.
Let SC be a py × px complex-valued matrix of two-dimensional measured spatial frequencies that is the sum of S0C, a matrix of true noiseless complex-valued spatial frequencies and EC, a matrix of complex-valued measurement error as in Appendix B. As shown in Appendix B, the matrix of spatial frequency measurements can be represented as a single real-valued vector by stacking the rows to form:
where Re( ) and Im( ) denote the operators that return the real and imaginary parts of their arguments and vec( ) denotes the the vectorization operator that stacks the columns of its matrix argument. This vector s, is the sum of a vector of true noiseless complex-valued spatial frequencies, s0, and a vector of complex-valued measurement error, ∈. Since the vector s is what is measured with error, it is assumed to be characterized as having a multivariate normal distribution with mean s0 and covariance matrix Φ as described in Appendix B.
The Fourier image reconstruction process to generate a complex-valued measured image RC consists of pre-multiplying the measured spatial frequencies SC by the Fourier matrix ΩCy in Eq. (B.1) and post-multiplying it by in Eq. (B.1). As shown in Appendix B, this is equivalently represented as the pre-multiplication of the real-valued vector of measured spatial frequencies s by the real-valued matrix Ω as in Eq. (B.6) to arrive at the real-valued representation of the measured image ρ. The real-valued representation of the measured image ρ is a linear transformation of the real-valued representation of the measured spatial frequencies and thus normally distributed with mean ρ0 = Ωs0 and covariance matrix Γ = ΩΦΩT. The measured py × px complex-valued image RC can be found by sequentially putting every px elements of the vector ρR + iρI into a matrix then taking the transpose.
An example of this methodology might be useful. Although explicit analytic expressions exist for the mean and covariance of the complex-valued transformed two-dimensional images measurements given the mean and covariance of the two-dimensional spatial frequency measurements, simulations were carried out to verify the analytic results in addition to determining those for magnitude-only image measurements where closed form analytic solutions do not exist due to the nonlinear and non one-to-one mapping. The simulations were performed under known conditions. These can be used to precisely characterize the signal and noise of the transformed measurements. All computations utilized Matlab (The Mathworks, Natick, MA, USA). Data was generated to mimic a two-dimensional magnetic resonance imaging experiment. Although this simulation is a mathematical ideal and possibly unrealistic, its results are useful in understanding the properties of the described methodology. A large number, L of random matrices of dimension py × px were generated for the pypx real spatial frequency measurements stacked upon the pypx imaginary spatial frequency measurements denoted by s1, . . ., sL that were generated from a normal distribution with mean s0 and covariance . Without loss of generality, s0 = 0 while Λ1, Λ2, and Λ3 are taken to be unit variance correlation matrices. The py × py correlation matrix Λ1 is taken to be an AR(1) correlation matrix with (i, j)th element where ϱ1 = 0.25, the 2 × 2 correlation matrix Λ2 is taken to have an off diagonal correlation ϱ = 0.5 while the px × px correlation matrix Λ3 is taken to be an AR(1) correlation matrix with (i, j)th element where ϱ3 = 0.5. The number of randomly generated vectors was selected to be L = 106.
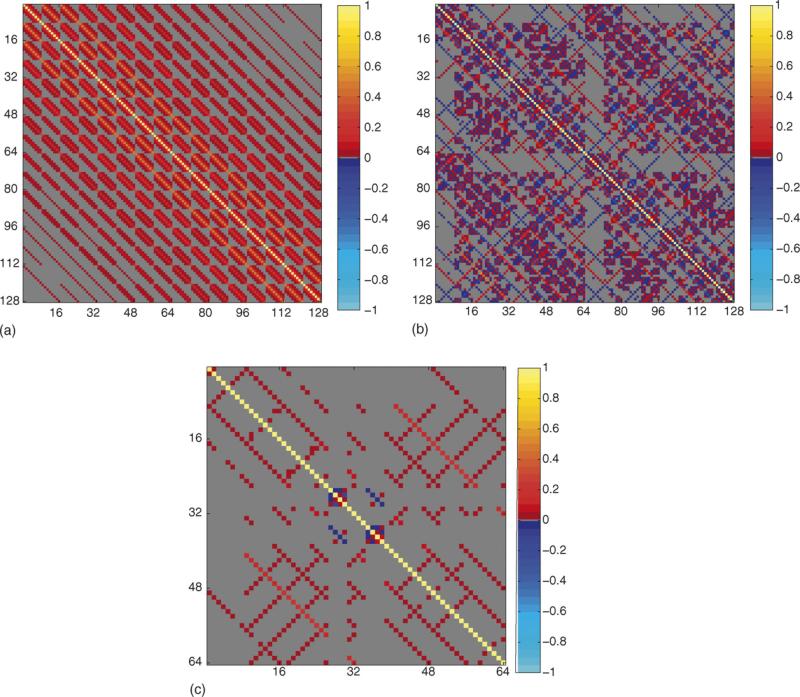
A value of py = px = 8 was chosen. Although the methodology equally applies to larger values, they are not shown to maintain the clarity of presentation. The corresponding sample correlation matrix from the L randomly generated noisy spatial frequency matrices in vector form was computed as displayed in Fig. 2a. Further, each random complex-valued spatial frequency matrix in vector form was pre-multiplied by Ω in Eq. (B.6) equilavent to pre- and post-multiplying in matrix form by the appropriate inverse Fourier transform matrices ΩCy and given in Eq. (B.1). The sample correlation matrix of the real-valued representation ρ of the complex-valued image measurements R was computed as displayed in Fig. 2b. The sample spatial frequency correlation matrix matched its theoretical population correlation matrix in Eq. (B.8) and the sample image correlation matrix matched the population value in Eq. (B.9) utilizing the previously described theory.
Fig. 2.
Correlation maps, px = py = 8, L = 106. (a) Correlation between complex spatial frequency measurements, Corr(s, s). (b) Correlation between complex image voxel measurements, Corr(r, r). (c) Correlation between magnitude image voxel measurements, Corr(m, m).
Further, since a simple closed form analytic expression for the theoretical covariance matrix for magnitude-only image quantities cannot be found, the L matrices of dimension py = px containing real and imaginary measurements were converted to L matrices of dimension py × px containing magnitude-only image quantities. The sample correlation matrix of the magnitude-only image matrices was computed using the real-valued representation as displayed in Fig. 2c. Note that both complex-valued voxels and real-valued magnitude-only voxels are correlated.
3. Conclusions
This paper presented the resulting spatial correlation between voxels when Fourier reconstructing correlated spatial frequency measurements. However, the current methodology is applicable to any linear transformation. This includes non-Fourier reconstruction of Fourier encoded data (Cox and McCall, 2004) or non-Fourier reconstruction of non-Fourier encoded data (Panych et al., 1996). Additionally, the previously described Fourier Ω matrices can easily be adjusted to include phase terms as done when adjusting for magnetic field inhomogenieties with a field map (Jezzard and Balaban, 1995).
Spatially correlated voxels result from correlated spatial frequency measurements. These correlation results may have implications for functional magnetic resonance imaging. In particular, temporally autocorrelated spatial frequency measurements produce spatially correlated voxels. This may have specific implications for functional connectivity. The true voxel connectivity may be less than previously thought. This methodology could be utilized to characterize noise correlation in its original form and adjust for it. The baseline spatial correlation needs to be considered and accounted for when making statements regarding connectivity between voxels in fMRI. Although the normal distribution has been utilized in the present work, other statistical distributions could be used. Regardless of the chosen statistical distribution to model the noise, the mean and covariance results are still applicable. Additionally, some voxel correlation may be lost by the magnitude-only procedure. Making statistical inferences, interpreting analysis results, and drawing conclusions should be done in light of the current research.
Acknowledgments
This work was supported in part by NIH EB00215, AG020279, MH019992, and RR00058.
Appendix A. One-dimensional image
The px × 1 dimensional complex-valued spatial frequency measurements sC consisting of px × 1 dimensional true underlying noiseless complex-valued spatial frequencies s0C and px × 1 dimensional complex-valued measurement error ∈C can be represented as
| (A.1) |
where i is the imaginary unit while s0R, s0I, ∈R and ∈I are px × 1 dimensional real and imaginary vector valued parts of the true signal and measurement noise. Let ΩCx be a px × px complex-valued matrix such as a Fourier type matrix such that
| (A.2) |
where ΩRx and ΩIx are real and imaginary matrix valued parts. Then, the px × 1 dimensional complex-valued inverse Fourier transformation ρC of sC can be written (Strang, 1988) as the pre-multiplication by the complex-valued Fourier matrix as
where ρ0R, ρ0I, ηR, and ηI are real and imaginary vector valued parts of the Fourier transformed true signal (image) and transformed measurement noise. If ΩCx were a Fourier matrix, it is [ΩCx]jk = κ (ωJK) where κ = 1 and ω = exp[–i2π(j–1)(k – 1)/px] for the forward transformation while κ = 1/px and ω = exp[+i2π(j – 1)(k – 1)/px] for the inverse transformation, where j, k = 1, . . ., px.
This pre-multiplication of a complex-valued vector by a complex-valued matrix can be equivalently represented with the 2px dimensional real-valued representation:
| (A.3) |
As previously described, data collected from a scientific experiment is never precisely known and thus contains both true signal and measurement error. Scientific measurement error is quantified with statistical distributions and inferences drawn. In most instances, real-valued measurements are taken and real-valued statistical distributions utilized. However, in MRI complex-valued measurements are taken and thus a complex-valued statistical distribution needs to be utilized. The data can be represented using a real-valued representation and a multivariate normal distribution (Rowe, 2003). The real-valued representation used here is very general and within this framework contains the particular representation used to represent the complex-valued multivariate normal distribution (Anderson et al., 1995; Wooding, 1956). The transformation from complex-valued spatial frequency space to image space modifies properties of both the true noiseless signal and the measurement noise. The relationship between correlated complex-valued measurements made in spatial frequency space and the modified correlation between inverse Fourier transformed or reconstructed complex-valued measurements in image space is examined.
Using the real-valued representation in Eq. (A.3), let the 2px dimensional vector be multivariate normally distributed (Rowe, 2003) with mean and covariance matrix:
| (A.4) |
Complex multivariate normal structure occurs when Λ11 = Λ22 = Ψ, –Λ12 = Υ, and . That is, when the covariance matrix is of a skew-symmetric form as (Wooding, 1956; Anderson et al., 1995). The current representation is more general and less restrictive than multivariate complex normal structure. By carrying out a multivariate transformation of variable with the real-valued representation from s to ρ through ρ = Ωxs, the statistical distribution of ρ is also multivariate normally distributed but with mean ρ0 given by
and covariance matrix , given by
| (A.5) |
where Ωx is of full rank if it is a Fourier matrix. Again, this representation is more general and less restrictive than multivariate complex normal structure (Anderson et al., 1995; Wooding, 1956). In the multivariate complex normal case (Anderson et al., 1995; Wooding, 1956) where Λ11 = Λ22 = Ψ, –Λ12 = Υ , and , the covariance matrix Δ is
| (A.6) |
where Υ is a skew symmetric matrix, ΥT = –Υ .
It can readily be seen that if the measurement process that generates the data produces uncorrelated real and imaginary channels, that is, but correlated within the real imaginary channels, then after transformation the real and imaginary channels are correlated both between and within. It should be noted that if Υ = 0 and Ψ = ψ2Ipx, then where δ = ψ2/px for the inverse transformation and δ = ψ2px for the forward transformation. The Kronecker product ⊗ was utilized which multiplies every element of its first matrix argument by its entire second matrix argument.
The above specific multivariate complex normal structure could alternatively be developed utilizing the complex multivariate normal distribution (Anderson et al., 1995; Wooding, 1956). A property of the complex multivariate normal distribution is that if sC ~ NC (s0C, ΛC), then ρC = ΩCxsC is also complex normal distributed, ρC ~ NC (ΩCxs0C, ) where ΛC = Ψ + iΥ and “H” denotes the Hermitian or complex conjugate transpose.
After image reconstruction, the usual procedure is to convert from real and imaginary images to magnitude and phase images. The phase is generally discarded in fMRI and magnitude-only time course data are analyzed. The conversion from real and imaginary images to magnitude and phase images is a nonlinear transformation and thus the joint distribution of the magnitude image measurements is not straightforward. On an individual basis, the measured magnitude quantity in voxel j in each magnitude image is
where ρ0Rj and ρ0Ij are the means in the real and imaginary parts while ηRj and ηIj are the zero mean real and imaginary Gaussian error terms with variances Δjj and Δpx+j, px+j, j = 1, . . ., px, generally assumed to be the same. It is well known (Gudbjartsson and Patz, 1995; Rice, 1954; Rowe and Logan, 2004) that the measured magnitude voxel intensity mj is Ricean distributed with parameters , being the pixel magnitude intensity in the absence of noise, and Δj = Δjj = Δpx+j,px+j, being the equal variances of the real and imaginary parts. The population correlation between Ricean distributed magnitude image measurements will be examined through simulation.
Appendix B. Two-dimensional image
The py × px dimensional complex-valued spatial frequency measurements SC consisting of py × px dimensional true under lying noiseless complex-valued spatial frequencies S0C and py × px dimensional complex-valued measurement error EC can be represented as
where i is the imaginary unit while S0R, S0I, ER and EI are real and imaginary matrix valued parts of the true spatial frequencies signal and measurement noise. Let ΩCx and ΩCy be px × px and py × py Fourier matrices such that
| (B.1) |
where ΩRy and ΩRx are real while ΩIy and ΩIx are imaginary matrix valued parts.
Then, the py × px complex-valued inverse Fourier transformation RC of SC can be written as
where
are real and imaginary matrix valued parts of the inverse Fourier transformed true signal (image) and measurement noise. Each row in the curled bracket part of the expression for RC is a one-dimensional complex-valued transformation:
| (B.2) |
as in the previous one-dimensional case where represents the jth row in SC that is px dimensional, j = 1, . . ., py. The complex matrices ΩCy and ΩCx can be Fourier matrices. This preand post-multiplication of a complex-valued matrix by complex-valued matrices could be equivalently represented with a similar real-valued representation. This could be accomplished by forming a py × 2px dimensional matrix where a given row j, is () a real-valued representation of the rows of SC. This real-valued matrix is then post-multiplied by . The resultant real-valued matrix is then reformed into a complex-valued matrix and another real-valued representation made from the columns to form a 2py × px matrix. This new real-valued representation is then pre-multiplied by Ωy and the resultant real-valued matrix is then reformed into a complex-valued matrix being the measured two-dimensional image. In the procedure just described, it is difficult to keep track of individual measurements and the correlations between other measurements within the array.
A simple representation from matrix algebra can be utilized to assist with this endeavor. It is known (Harville, 1999) that the vectorization of the triple product of conformable matrices A, B, and C can be written as
which translates to our application as
where vec is the vectorization operator that stacks the columns of its matrix argument and .
As previously noted, the matrix of spatial frequency measurements can be described with a real-valued representation. The large Kronecker product can be represented as
| (B.3) |
| (B.4) |
| (B.5) |
| (B.6) |
The complex-valued image vector can be represented as ρC = ΩCsC. We can pre-multiply the complex-valued spatial frequency vector by this complex-valued matrix as in the one dimensional image case, or equivalently with a similar real-valued representation:
| (B.7) |
where the real-valued vector of spatial frequencies is formed by
while Re( ) and Im( ) denote the operators that return the real and imaginary parts of their arguments and vec( ) denotes the vectorization operator that stacks the columns of its matrix argument.
The Fourier image reconstruction process to generate a complex-valued measured image RC consists of pre-multiplying the measured spatial frequencies SC by the Fourier matrix ΩCy in Eq. (B.1) and post-multiplying it by ΩTCx in Eq. (B.1). As shown above, this is equivalently represented as the pre-multiplication of the real-valued vector of measured spatial frequencies s by the real-valued matrix Ω in Eq. (B.6) to arrive at the real-valued representation of the measured image ρ. The vector s is assumed to be characterized as having a multivariate normal distribution with mean s0 and covariance matrix Φ denoted as
| (B.8) |
The real-valued representation of the measured image ρ is a linear transformation of the real-valued representation of the measured spatial frequencies and thus normally distributed with mean ρ0 = Ωs0 and covariance matrix Γ = ΩΦΩT denoted as
| (B.9) |
The measured py × px complex-valued image RC can be found by sequentially putting every px elements of the vector ρR + iρI into a matrix then taking the transpose.
In terms of complex-valued matrices, the mean of the transformed variables can be written as
as previously defined but the covariance of the transformed measurements is not easily represented with complex numbers and requires the larger real-valued representation.
Again, after image reconstruction, the usual procedure is to convert from real and imaginary images to magnitude and phase images. The phase is generally discarded in fMRI and magnitude-only time course data is analyzed. The conversion from real and imaginary images to magnitude and phase images is a nonlinear not one-to-one transformation and thus the joint distribution of the magnitude-only image quantities is not straightforward. However an approximation to the correlation of the square of the magnitudes, or in general any quadratic form exists. Further, the correlation of squared magnitudes is a good approximation to the correlation of magnitudes. If any pair of random variables is transformed by the same function (here the square root), their correlation remains roughly the same, by a Taylor series expansion argument.
On an individual basis, the measured magnitude quantity in voxel (i, j) in each magnitude image is
where R0Rjk and R0Ijk are the means in the real and imaginary parts, NRjK and NIjk are the zero mean real and imaginary Gaussian error terms with variances Γjpx+k,jpx+k and Γpxpy+jpx+k,pxpy+jpx+k, j = 1, . . ., px = 1, . . ., py, generally assumed to be the same.
It is well known (Gudbjartsson and Patz, 1995; Rice, 1954; Rowe and Logan, 2004) that the measured magnitude voxel intensity mj is Ricean distributed with parameters , being the pixel magnitude intensity in the absence of noise, and Γpxpy+jpx+k,pxpy+jpx+k, being the equal variances of the real and imaginary parts. The population correlation between Ricean distributed magnitude image measurements will be examined through simulation.
References
- Anderson HH, Højbjerre M, Sørensen D, Eriksen PS. Linear and graphical models for the multivariate complex normal distribution. Springer-Verlag; New York, NY, USA: 1995. [Google Scholar]
- Bandettini PM, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional MRI of the human brain. Magn Reson Med. 1993;30(2):161–73. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Buchel C, Coull JT, Friston KJ. The predictive value of changes in effective connectivity for human learning. Science. 1999;283(5407):1538–41. doi: 10.1126/science.283.5407.1538. [DOI] [PubMed] [Google Scholar]
- Cox RW, McCall LA. Non-Fourier reconstruction can prevent image dropouts. Proc Soc Magn Reson Med. 2004;12:2131. [Google Scholar]
- Friston KJ, Jezzard P, Turner RS. Analysis of functional MRI time-series. Hum Brain Mapp. 1994;1(8):153–71. [Google Scholar]
- Gudbjartsson H, Patz S. The Rician distribution of noisy data. Magn Reson Med. 1995;34(6):910–4. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haacke EM, Brown R, Thompson M, Venkatesan R. Magnetic resonance imaging: principles and sequence design. John Wiley and Sons; New York: 1999. [Google Scholar]
- Harville DA. Matrix algebra from a statistician's perspective. Springer-Verlag; New York: 1999. [Google Scholar]
- Jezzard P, Balaban RS. Correction for geometric distorition in echo planar images from B0 field variations. Magn Reson Med. 1995;34(1):65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- Logan BR, Rowe DB. An evaluation of thresholding techniques in fMRI analysis. Neuroimage. 2004;22(1):95–108. doi: 10.1016/j.neuroimage.2003.12.047. [DOI] [PubMed] [Google Scholar]
- Menon RS. Postacquisition suppression of large-vessel BOLD signals in high-resolution fMRI. Magn Reson Med. 2002;47(1):1–9. doi: 10.1002/mrm.10041. [DOI] [PubMed] [Google Scholar]
- Nan FY, Nowak RD. Generalized likelihood ratio detection for fMRI using complex data. IEEE Trans Med Imag. 1999;18(4):320–9. doi: 10.1109/42.768841. [DOI] [PubMed] [Google Scholar]
- Nencka AS, Rowe DB. Complex constant phase method removes venous BOLD component in fMRI. Proc Soc Magn Reson Med. 2005;13:495. [Google Scholar]
- Nencka AS, Rowe DB. Theoretical results demonstrate fundamental differences in venous BOLD reducing activation methods. Proc Soc Magn Reson Med. 2006;14:3269. [Google Scholar]
- Panych LP, Oesterle C, Zientara GP, Hening J. Implementation of a fast gradient-echo SVD encoding technique for dynamic imaging. Magn Reson Med. 1996;35(4):554–62. doi: 10.1002/mrm.1910350415. [DOI] [PubMed] [Google Scholar]
- Rice SO. Mathematical analysis of random noise. Bell Syst Technol J. 1944;23:282, 1944. Reprinted by Wax N. Selected papers on noise and stochastic process. Dover Publication; 1954. QA273W3.
- Rowe DB. Multivariate Bayesian statistics. CRC Press; Boca Raton, FL, USA: 2003. [Google Scholar]
- Rowe DB. Modeling both the magnitude and phase of complex-valued fMRI data. Neuroimage. 2005a;25(4):1310–24. doi: 10.1016/j.neuroimage.2005.01.034. [DOI] [PubMed] [Google Scholar]
- Rowe DB. Parameter estimation in the magnitude-only and complex-valued fMRI data models. Neuroimage. 2005b;25(4):1124–32. doi: 10.1016/j.neuroimage.2004.12.048. [DOI] [PubMed] [Google Scholar]
- Rowe DB. Complex activation is more focal and concentrated to parenchymal tissue. Proc Soc Magn Reson Med. 2005c;13:1575. [Google Scholar]
- Rowe DB, Logan BR. A complex way to compute fMRI activation. Neuroimage. 2004;23(3):1078–92. doi: 10.1016/j.neuroimage.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Rowe DB, Logan BR. Complex fMRI analysis with unrestricted phase is equivalent to a magnitude-only model. Neuroimage. 2005;24(2):603–6. doi: 10.1016/j.neuroimage.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Rowe DB, Nencka AS. Complex activation suppresses venous BOLD in GE-EPI fMRI data. Proc Soc Magn Reson Med. 2006;14:2834. [Google Scholar]
- Strang G. Linear algebra and its applications. 3rd ed. Harcourt Brace Jovanovich; Orlando, Florida: 1988. [Google Scholar]
- Wooding RA. The multivariate distribution of complex normal variables. Biometrika. 1956;43:212–5. [Google Scholar]