Abstract
For a gene to be expressed, the functions of multiple molecular machines must be coordinated at the site of transcription. To understand the role of nuclear organization in transcription, it is necessary to visualize the dynamic interactions of regulatory factors with chromatin and RNA. It is currently possible to localize individual transcription sites in single living mammalian cells by engineering reporter gene constructs to include sequence elements which permit the visualization of nucleic acids in vivo. Upon stable integration, these transgenes form chromatinized arrays, which can be imaged during activation to obtain high-resolution quantitative information about transcriptional dynamics. Modeling can suggest new hypotheses about gene regulation, which can be tested both in the single-cell imaging system and at endogenous genes. This gene-specific imaging strategy has the potential to reveal regulatory mechanisms, which would be difficult to imagine outside of single living cells.
Keywords: Transcription, Chromatin, Single-cell imaging, Nuclear organization, Histone H3.3
Presentation
Genes are activated when transcription factors interact with specific DNA sequence elements to initiate a cascade of events, including chromatin remodeling, transcription and RNA processing (Coulon et al., 2013). Although these steps are often studied as distinct mechanisms, they are, in fact, highly interdependent (Hager et al., 2009). To achieve an integrated view of gene expression, it is necessary to understand how they are coordinated at transcription sites. By combining advances in live-cell imaging and auto-fluorescent protein technology with reporter gene engineering, it is possible to visualize gene activation at specific transcription sites in single cells. The information provided by this experimental approach about the timing and spatial organization of gene expression can lead to the discovery of novel regulatory mechanisms. This review describes the history and fundamental components of this gene imaging strategy, as well as its limitations and future applications.
Transcription and Chromatin: A Mutualistic Relationship
In cells, the genome is packaged into an assemblage of DNA and proteins called chromatin. Chromatin is the interface through which the transcription machinery gains access to the gene template. Therefore, chromatin is a critical regulator of transcription, and conversely, transcription drives changes in chromatin organization. Most conventional techniques for studying gene regulation (e.g. chromatin immunoprecipitation, RT-PCR, proteomics, etc.) require the disruption of cellular architecture rendering them unable to reveal how factors are temporally and spatially organized at genes during activation. Visualizing factor dynamics at individual transcription sites in single living cells, however, can provide insight into gene-specific regulatory mechanisms as well as genome organization and epigenetic inheritance because of the important role transcription plays in these processes (Denholtz and Plath, 2012; Misteli, 2010).
The Nucleus: A Dynamic Network of Functional Interactions
In addition to its effects on chromatin, transcription also significantly impacts nuclear organization (Bickmore and van Steensel, 2013; Ritland Politz et al., 2013). In the nucleus, factors with similar functions often co-localize in discreet membraneless structures called nuclear bodies (NBs) (Dundr and Misteli, 2010; Mao et al., 2011b) [e.g. PML nuclear bodies (PML-NBs), Cajal bodies, nuclear speckles, nucleoli, etc.]. Although the functions of most NBs are incompletely understood, alterations in their organization correlate with changes in gene expression in several diseases indicating that their structures are linked to functions. For example, the oncogenic fusion protein, PML/RARA (retinoic acid receptor α), which causes acute promyelocytic leukemia (APL), both represses genes required for myeloid differentiation and increases PML-NB numbers (de The et al., 2012). During herpes simplex virus-1 (HSV-1) infection, ICP0, the E3 ubiquitin ligase required to repress innate immunity and reverse viral latency, dissolves PML-NBs and alters host and viral gene expression (Boutell and Everett, 2013).
Initially, NBs were thought to be stably assembled structures because they appear to be highly organized in electron micrographs and immunofluorescence images (Rouquette et al., 2010). However, these techniques require fixation, and therefore, can only provide “snapshots” of cells at single time points. When auto-fluorescent proteins [e.g. green fluorescent protein (GFP)] were used to examine protein movement in living cells, they revealed that NBs are actually highly dynamic (Phair and Misteli, 2001). Although at steady state, their GFP-tagged components appear static, the rapid recovery of fluorescent signal after photobleaching suggests that they are continuously associating and dissociating (Misteli, 2008a). These studies indicate that the nucleus is simultaneously organized and dynamic, which is consistent with the ability of cells to rapidly respond to stimuli (e.g. growth factors, heat shock, viral infection, etc.) and efficiently execute regulatory events (e.g. DNA repair, transcription, DNA synthesis, etc.) (Misteli, 2008b).
Connecting NBs with Molecular Functions
Among NBs, the structure/function relationship is best defined for nucleoli, sites of RNA pol I transcription, pre-rRNA processing, and ribosome assembly (Hernandez-Verdun, 2006; Raska et al., 2006). The repetitiveness of ribosomal DNA (rDNA) provides the resolution needed to easily identify these RNA pol I transcription sites in single cells. The high concentration of substrate within nucleoli makes it possible to discern sub-domains, which likely function to accelerate enzymatic and assembly reactions (Boulon et al., 2010; Pederson, 2011). It is thought that nuclear factors self-organize around gene expression and genome regulatory events and that the structural integrity of NBs and their subdomains is defined by transient protein–protein and protein–nucleic acid interactions (Dundr, 2012; Dundr and Misteli, 2010).
In contrast to the multi-copy rDNA which comprises RNA pol I transcription sites, it is technically difficult at this time to visualize single endogenous RNA pol II genes in single living cells because of detection limitations. However, multi-copy arrays of transgenes containing sequence elements that allow nuclei acids to be visualized in vivo can be easily localized (Rafalska-Metcalf and Janicki, 2007; Yunger et al., 2013a). When RNA pol II transcription units are included in these transgenes, transcriptional dynamics can be directly visualized at these arrays. This approach has also been used to link NB biogenesis to transcriptional mechanisms (e.g. paraspeckles, histone locus bodies with associated Cajal bodies, and PML-NBs) (Mao et al., 2011a; Newhart et al., 2012; Shevtsov and Dundr, 2011). Therefore, in addition to transcriptional dynamics, this methodology can be used to study nuclear organization.
Assembling the “Visible Gene”
Synthetic reporter genes are routinely constructed to decipher the functions of genetic elements in transcription. These reporter gene assays are quantitative because measurable proteins (e.g. GFP, luciferase and β-galactosidase) are encoded in the transcription units. However, in its current usage, this methodology has a number of significant limitations. If cellular fixation and/or rupture are required for protein detection, it precludes analysis of the same sample over time resulting in limited and/or arbitrary time point selection. Assays which average effects in cell populations obscure transitory events and cell-to-cell variability. When reporter constructs are transiently expressed, transcription is measured from non-chromatinized templates. Additionally, protein-based gene activity assays are several degrees removed from events at the transcription site, and thus, provide only limited insight into cis-acting regulatory mechanisms.
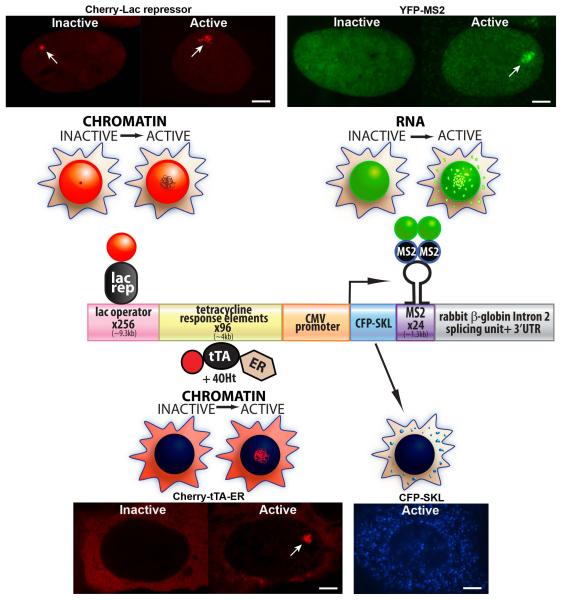
A pivotal advance in our ability to directly study transcription in single cells came from the use of protein/nucleic acid interaction units from bacteria and bacteriophage to visualize DNA (e.g. the lac repressor/operator) and RNA (e.g. MS2 coat protein and Pseudomonas aeruginosa PP7 bacteriophage/hairpin sequences) in living cells (Darzacq et al., 2009; Hocine et al., 2013; Rafalska-Metcalf and Janicki, 2007; Wu et al., 2011). When synthetic reporter genes, containing repeats of these elements, are stably expressed, they integrate into single genomic sites as chromatinized multi-copy arrays (Rafalska-Metcalf and Janicki, 2013). Figure 1 shows the linear structure of this type of transgene and the cellular localization patterns of the proteins used to track gene expression. Lac repressor, fused to an auto-fluorescent protein, makes it possible to visualize the “transcription site” chromatin in both the inactive (condensed) and activated (decondensed) states (Figure 1). Similarly, the MS2 coat protein, which interacts with the stem loop structure of the translational operator as a dimer, permits visualization of the transcribed RNA at both the transcription site and throughout the nucleus (Figure 1). The mRNA encodes Cyan Fluorescent Protein (CFP) fused to a peroxisomal targeting signal (SKL). Its expression provides real-time confirmation that all of the steps required for gene expression have been completed (Janicki et al., 2004; Tsukamoto et al., 2000) (Figure 1). The concentration of CFPSKL in the peroxisomes both improves the signal-to-noise ratio during imaging and segregates it from the nucleus so that CFP can also be used to tag transcriptional regulators for simultaneous multi-color imaging.
Figure 1.
Molecular and Cellular Characterization of the Single-Cell Imaging System The diagram of the single-cell imaging transgene shows the modular organization of its sequence components. The proteins, which permit the analysis of DNA and RNA in single living cells, are shown in association with the sequence elements to which they bind. The cell diagrams above each factor depict their localization patterns in inactive and activated cells. The cell images are of the U2OS cell line (2-6-3), which contains a multi-copy array of the CMV promoter regulated transgene. Cherry-lac repressor allows the transgene to be visualized in both the inactive (condensed) and active (deccondensed) states. Transcription is induced from the minimal CMV promoter by the activator, Cherry-tTA-ER, which is composed of the tetracycline transactivator (tTA) fused to Cherry and the estrogen receptor (ER) hormone-binding domain. Cherry-tTA-ER is retained in the cytoplasm until 4-hydroxytamoxifen (4-OHT) induces its entry into the nucleus where it accumulates at the transgene array and activates transcription. The transcription unit encodes Cyan Fluorescent Protein (CFP) fused to a peroxisomal targeting signal (SKL), MS2 bacteriophage translational operator repeats, and the intron 2 splicing unit of the rabbit β-globin gene. In inactive cells, YFP-MS2 is diffusely distributed in the nucleus. Upon activation, YFP-MS2 binds to the MS2 stem loop structure in the transcribe RNA both at the transcription site and throughout the nucleus. The enrichment of the protein product, CFP-SKL, in the peroxisomes provides real-time confirmation that all of the steps required for gene expression have been successfully completed. Scale bars =5 9m.
Transcriptional Activators: Modular Tools for Probing Chromatin Organization
The transcriptional activator used in this system [tetracycline transactivator (tTA)] consists of a fusion between the bacterial DNA-binding protein, tetracycline repressor (TetR), and the HSV-1 VP16 transactivation domain (TAD)(Gossen and Bujard, 1992) (Figure 1 and Figure 2B). In the absence of tetracycline and its derivatives (e.g. doxycycline), tTA binds constitutively to the tetracycline response elements (TREs) in the transgene and initiates transcription from the minimal CMV promoter. When the activator is fused to an auto-fluorescent protein, such as Cherry, it permits the specific visualization of transcriptionally active chromatin (Figures 1). Tighter transcriptional control can be achieved by fusing tTA to the 4-hydroxytamoxifen (4-OHT)-inducible hormone-binding domain of the estrogen receptor (ER), which sequesters it in the cytoplasm prior to activation (Figure 1). In the absence of 4-OHT, ER associates with heat shock protein 90 (Hsp90) (Eilers et al., 1991). When 4-OHT is added to the media, ER sheds Hsp90 and enters the nucleus.
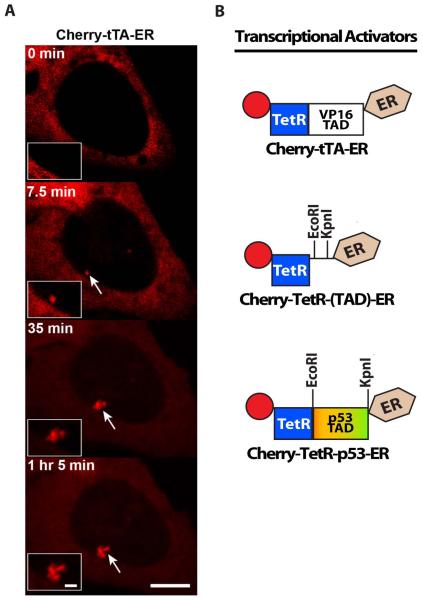
Figure 2.
- Images from a time series of the recruitment of the activator, Cherry-tTA-ER, to the CMV-promoter regulated transgene array in the U2OS cell line (2-6-3). Arrows indicate the location of the array. Scale bar =5 ,m. Scale bar in enlarged inset =1 ,m.
- (B) Schematic diagram of the synthetic transcriptional activator constructs, which can be used to analyze the effects of different transcriptional activation domains (TADs) on transcription and chromatin organization in cell lines with arrays that include the tetracycline response element repeats (Figure 1). Cherry-tTA-ER consists of the auto-fluorescent protein, Cherry, the tetracycline transcriptional activator (tTA), and the estrogen receptor hormone-binding domain (ER). tTA, itself, is a fusion between the tetracycline repressor (TetR) and the VP16 TAD. Cherry-TetR-(TAD)-ER was constructed for easy introduction of dirent TADs by directional cloning using EcoRI and KpnI. Cherry-TetR-p53 was constructed by introducing the p53 TAD into Cherry-TetR-(TAD)-ER.
Due to its specific interaction with active chromatin, the Cherry-tTA-ER activator can be used to determine the timing of transcriptional events. Cherry-tTA-ER is first detected at the CMV-promoter-regulated transgene array in the U2OS cell line (2-6-3) ~7 min after the addition of 4-OHT to the media (Figure 2A). The simultaneous detection of the RNA, marked by YFP-MS2, with Cherry-tTA-ER indicates that, in U2OS cells, transcription is rapidly induced (Rafalska-Metcalf et al., 2010). Therefore, the activation time includes the time required for Cherry-tTAER to enter the nucleus and accumulate to detectable levels at the array. Simultaneous imaging of Cherry-tTA-ER with YFP-tagged regulators (e.g. Brd4, GCN5, and histone H3.3) can be done to determine the timing of their recruitment. Modeling these types of data can suggest novel mechanisms of gene regulation (Darzacq et al., 2007; Newhart et al., 2013b; Rafalska-Metcalf et al., 2010; Zhao et al., 2011).
The modular structure of the transcriptional activator lends itself to development as a tool for comparing the chromatin decondensation and transcriptional activation potentials of different cellular and viral TADs. The Cherry-TetR-(TAD)-ER construct was designed to construct new activators by directional cloning (Figure 2B) (Rafalska-Metcalf et al., 2010). Comparison of the strong VP16 and weak p53 TADs in the U2OS cell line (2-6-3) demonstrated that their differential effects on transcription and chromatin organization are inherent to their amino acid sequence, which likely regulates the levels and types of co-activators recruited (Rafalska-Metcalf et al., 2010).
Exploring the Interface Between Transcription and Nuclear Organization
Numerous multi-subunit transcriptional regulators have been identified through biochemical and genetic analyses. The single-cell imaging strategy described in this review provides the opportunity to directly relate factor recruitment to changes in chromatin organization and RNA accumulation at molecularly defined transcription sites (Darzacq et al., 2007). Knockdown analyses can be combined with conventional techniques to validate the visual data. For example, strand-specific quantitative RT-PCR, high-throughput sequencing, and RNA FISH have been used to analyze RNA expression (Newhart et al., 2013b). Chromatin immunoprecipitation has been used to monitor factor recruitment and histone post-translational modifications (PTMs) as well as to identify sequence specific enrichment (Newhart et al., 2013a; Newhart et al., 2012; Shanbhag et al., 2010; Tang et al., 2013; Zhao et al., 2011). Validated hypotheses about gene regulatory mechanisms can be tested at endogenous genes.
De Novo Transcription Sites: Giving Genes a “Fresh Start”
In addition to providing a previously unavailable view of transcriptional dynamics, the ability to create de novo transcription sites has several other significant advantages. Since the CMV promoter-regulated transgene is inducible and its expression is not required for viability, it can be propagated in cells in an inactive state until the synthetic activator, Cherry-tTA-ER, is expressed (Figure 1). In this way, transcriptional activation can be tracked from a naive inactive chromatin state. Using the MS2 tag to visualize the expressed RNA also makes it possible to define the window of linear transcriptional increase. Examining the recruitment of factors to chromatin as it transitions to the active state has the potential to illuminate regulatory mechanisms.
The modularity of single-cell imaging transgenes, such as the one described in Figure 1, make it possible to construct new versions in order to directly examine the effects wild-type and mutated genetic elements have on transcription and chromatin organization. By inserting different promoters, it is possible to study the effects of cell cycle, developmental, and environmental stimuli (Hocine et al., 2013; Larson et al., 2011; Maiuri et al., 2011; Yunger et al., 2010). Gene-specific RNA processing events, including mRNA splicing, polyadenylation, RNA degradation and mRNA export, can be studied by introducing different 3′ sequence elements (Ben-Ari et al., 2010; Brody et al., 2011; Katz et al., 2012; Martins et al., 2011). Using this approach to analyze viral genetic elements could provide insight into intrinsic immunity and viral latency.
Oh What a Difference Genetic Background Makes
Transgene arrays can be integrated into different cell lines to investigate the effects of genetic background on transcription. For example, the CMV-promoter regulated transgene (Figure 1) is refractory to activation in HeLa cells, which express both the histone H3.3 chaperone, Daxx, and the SNF2-type ATP-dependent chromatin-remodeling factor, ATRX (Newhart et al., 2012). Daxx and ATRX regulate the replication independent (RI) incorporation of H3.3 into telomeres and centromeres (Drane et al., 2010; Goldberg et al., 2010). In contrast to HeLa cells, transcription begins ~7 min after activation in the ATRX-null U2OS cell line (2-6-3) (Figure 2A), which demonstrates the important role RI H3.3 chromatin assembly plays in regulating transcriptional repression and heterochromatin organization (Rafalska-Metcalf et al., 2010). Prior to being identified as RI H3.3 chromatin assembly factors, Daxx and ATRX were known to be PML-NB components, viral targets for disorganization and/or degradation, and regulators of gene silencing (Boutell and Everett, 2013; Lallemand-Breitenbach and de The, 2010). The differential expression patterns of the CMV-promoter transgene in HeLa compared to U2OS cells are consistent with previous reports on Daxx and ATRX function.
Multi-Copy Transgene Arrays: Regulating Repetition
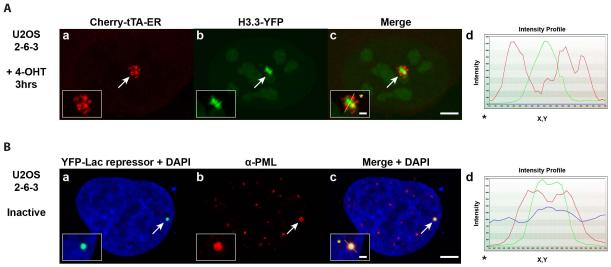
An important feature of the single-cell imaging system is that the transgenes integrate into cellular genomes as multi-copy arrays. Not only does this provide the DNA volume needed to visualize changes in chromatin organization during transcriptional activation (Janicki et al., 2004; Rafalska-Metcalf et al., 2010), but it also makes it possible to discern patterns of regulatory factor organization at the site. For example, H3.3 is strongly recruited to the activated transgene array in the ATRX-null U2OS cell line (2-6-3), despite the fact that chromatin assembly is blocked (Newhart et al., 2012). Importantly, H3.3 accumulates in regions that are distinct from the chromatin, marked by DNA-binding proteins including the activator, Cherry-tTA-ER (Figure 3A, panels a-d). The intensity profile (Figure 3, panel d), which graphs pixel measurements along the line drawn through the array (note the yellow line in enlarged inset in the merged image, panel c), shows that the H3.3-YFP and Cherry-tTA-ER peaks do not overlap. This suggests that when chromatin assembly is blocked, H3.3-YFP accumulates with factors that function upstream of nucleosomal deposition.
Figure 3.
Images of Regulatory Factor Organization at the Transgene Array.
(A) Histone H3.3-YFP (a) is strongly recruited to the activated transgene array in the ATRX-null U2OS cell line (2-6-3) in which replication independent histone H3.3 chromatin assembly is blocked. H3.3-YFP does not co-localize with the chromatin of the activated array marked by Cherry-tTA-ER (b). Arrows mark the location of the transgene array. Yellow lines in enlarged merge insets (c) show the path through which the signal intensities were measured in the intensity profiles (d). Asterisks mark the start of the measured line. Scale bar =5 0m. Scale bar in enlarged inset =1 0m.
(B) PML protein (b) is enriched at the inactive CMV-promoter regulated transgene array in the U2OS cell line (2-6-3), marked by YFP-lac repressor (a). DAPI DNA staining is shown in the merged images (a and c). (b)PML immunofluoresence staining was done with antibody PG-M3 (Santa Cruz Biotechnology).
Analyses of the H3.3 localization pattern revealed that it accumulates with sense and antisense RNA at the active transcription site, which suggests that it is recruited to its incorporation sites through an RNA-mediated mechanism (Newhart et al., 2013b). RI H3.3 chromatin assembly is a highly conserved but incompletely understood genome regulatory mechanism. Daxx and ATRX are mutated in several neurological cancers and gain-of-function mutations in H3.3 have been identified in pediatric glioblastoma (Jiao et al., 2011; Molenaar et al., 2012; Schwartzentruber et al., 2012; Wu et al., 2012). Therefore, the ability to visualize the dynamic interaction of H3.3 with a transcription site at which RI chromatin assembly is blocked could provide new insight into both genome regulation and disease pathogenesis.
A large body of literature indicates that multi-copy transgene arrays are silenced as a result of transcription through inverted repeats (Calero-Nieto et al., 2010; Davis and MacDonald, 1988; Dorer and Henikoff, 1994; Garrick et al., 1998; Henikoff, 1998). As the methods used to isolate these array-containing cell lines do not control for orientation, it is likely that they all contain inverted repeats. Although transcription is rapidly induced when Daxx and ATRX-regulated RI H3.3 chromatin assembly is disrupted (Figure 2A), the effects of transgene repetitiveness and inversion are not yet fully understood. In ATRX-null U2OS cells, the inactive CMV-promoter-regulated array is highly condensed, which suggests that additional mechanisms may function to compact the chromatin in order to preserve genomic integrity (Figure 1; Cherry-lac repressor image). Indeed the condensed structure of the inactive array is itself an impediment to transcription because the weak p53 TAD construct cannot induce decondensation to the same extent in the U2OS cell line as Cherry-tTA-ER, which contains the strong VP16 TAD (Rafalska-Metcalf et al., 2010). Therefore, this system provides an important opportunity to study the mechanisms which regulate repetitive DNA in mammalian cells.
Deciphering the Connection Between PML-NBs and Transcription
Overall, our analyses indicate that trans-acting factors (i.e. Daxx and ATRX) have a dominant effect on transcription compared to cis-acting elements (i.e. the genomic site of the array integration and the transgene repeats). For example, all of the CMV-promoter regulated transgene arrays independently integrated into ATRX-null U2OS cells (e.g. 2-6-3 and 60-1) (Newhart et al., 2012) (smj unpublished data) have been rapidly activated. In contrast, the array is refractory to activation in HeLa cells (cell line HI 1-1) unless ICP0, which depletes Daxx, ATRX and PML, is co-expressed (Newhart et al., 2012). PML-NBs are defined by the presence of PML protein (Lallemand-Breitenbach and de The, 2010). Therefore, the enrichment of PML at the transgene array (Figure 3B), suggests that it is a PML-NB and that PML-NBs are transcription sites. ChIP analyses indicate that Daxx, ATRX, PML and Sp100 are all specifically enriched at the CMV promoter (Newhart et al., 2013a; Newhart et al., 2012). This suggests that promoter sequences seed their assembly at the site. What is not known, however, is whether PML-NB proteins are recruited to specific promoters (e.g. viral promoters such as CMV) or all promoters in repetitive arrays. Overall, these results demonstrate the power of this imaging strategy for deciphering mechanisms of transcription, nuclear organization, and innate immunity.
Future Directions
The inherent features of this experimental system lend it to continual development as a discovery tool. Below is a discussion of the ways in which it can be expanded and refined.
Biochemical Purification
The protein/nucleic acid interaction units used for DNA and RNA visualization (Figure 1) can be adapted (e.g. immunological or streptavidin-based tagging systems) in order to purify the transgene chromatin and expressed RNA for comprehensive analysis of associated factors, PTMs, and RNA.
Targeted Integration
Genomic location is extremely important for its function. Additionally, it is desirable to visualize regulatory events at single-copy genes because the effects of the arrays on transcription are not fully understood. Therefore, site-specific homologous recombination can be used to create cell lines with single copies of these transgenes (Yunger et al., 2010; Yunger et al., 2013b). As detection sensitivity increases, it will also be possible to use high-resolution imaging methods to visualize regulatory factors dynamics at single-copy sites (Cisse et al., 2013; Gebhardt et al., 2013).
Using Transgene Engineering to Study Transcriptional Mechanisms
As mentioned above, the modularity of this transgene makes it possible to introduce different cellular and viral wild-type and mutated genetic elements into it in order to study their effects on transcription and chromatin organization. The development of reporter constructs with RNA pol I and RNA pol III regulated transcription units could also bring new insight into the dynamics of these transcriptional machineries. Boundary elements could also be included in order to understand their role in regulating chromatin and nuclear organization.
Summary
The limits to our understanding of cellular regulation are directly related to the limitations of our experimental techniques. Insights gained from using new technologies must always be rigorously evaluated in order to effectively integrate them into existing regulatory paradigms. The ability to visualize molecularly defined transcription sites in single living mammalian cells now provides unprecedented access to information about the timing and spatial organization of transcription. Only by directly examining the dynamic interactions of factors with specific sequence elements will it be possible to fully understand how cells “translate” the information encoded in DNA into heritable patterns of gene expression.
Acknowledgements
We would like to thank Michael Showe and Dario Altieri for critical comments on the manuscript, Sylvie Schaffer for her contributions to the artwork, and the Wistar Imaging Facility for the confocal images.
Contract grant numbers: R01 GM093000-02 and P30 CA10815).
References
- Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. The life of an mRNA in space and time. J Cell Sci. 2010;123(Pt 10):1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40(2):216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94(Pt 3):465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co transcriptional splicing. PLoS Biol. 2011;9(1):e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Nieto FJ, Bert AG, Cockerill PN. Transcription-dependent silencing of inducible convergent transgenes in transgenic mice. Epigenetics Chromatin. 2010;3(1):3. doi: 10.1186/1756-8935-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science. 2013 doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013 doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14(9):796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, Tjian R, Singer RH. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BP, MacDonald RJ. Limited transcription of rat elastase I transgene repeats in transgenic mice. Genes Dev. 1988;2(1):13–22. doi: 10.1101/gad.2.1.13. [DOI] [PubMed] [Google Scholar]
- de The H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198(1):11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Plath K. Pluripotency in 3D: genome organization in pluripotent cells. Curr Opin Cell Biol. 2012;24(6):793–801. doi: 10.1016/j.ceb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77(7):993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24(3):415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(12):a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Schirm S, Bishop JM. The MYC protein activates transcription of the alpha-prothymosin gene. Embo J. 1991;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18(1):56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10(5):421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35(6):741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20(7):532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. The nucleolus: a model for the organization of nuclear functions. Histochem Cell Biol. 2006;126(2):135–148. doi: 10.1007/s00418-006-0212-3. [DOI] [PubMed] [Google Scholar]
- Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10(2):119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116(5):683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr., Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ZB, Wells AL, Park HY, Wu B, Shenoy SM, Singer RH. beta-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26(17):1885–1890. doi: 10.1101/gad.190413.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011;12(12):1280–1285. doi: 10.1038/embor.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011a;13(1):95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011b;27(8):295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol. 2011;18(10):1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell biology: Nuclear order out of chaos. Nature. 2008a;456(7220):333–334. doi: 10.1038/456333a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol. 2008b;129(1):5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2(8):a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM, Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Newhart A, Negorev DG, Rafalska-Metcalf IU, Yang T, Maul GG, Janicki SM. Sp100A promotes chromatin decondensation at a CMV-promoter regulated transcription site. Mol Biol Cell. 2013a doi: 10.1091/mbc.E12-09-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Rafalska-Metcalf IU, Yang T, Joo LM, Powers SL, Kossenkov AV, Lopez-Jones M, Singer RH, Showe LC, Skordalakes E, Janicki SM. Single-cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site. J Biol Chem. 2013b doi: 10.1074/jbc.M113.473181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. Single cell analysis of Daxx and ATRX-dependent transcriptional repression. J Cell Sci. 2012 doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3(3) doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2(12):898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Janicki SM. Show and tell: visualizing gene expression in living cells. J Cell Sci. 2007;120(Pt 14):2301–2307. doi: 10.1242/jcs.008664. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Janicki SM. Preparation of cell lines for single-cell analysis of transcriptional activation dynamics. Methods Mol Biol. 2013;977:249–258. doi: 10.1007/978-1-62703-284-1_20. [DOI] [PubMed] [Google Scholar]
- Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One. 2010;5(4):e10272. doi: 10.1371/journal.pone.0010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18(3):325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Ritland Politz JC, Scalzo D, Groudine M. Something Silent This Way Forms: The Functional Organization of the Nuclear Repressive Compartment. Annu Rev Cell Dev Biol. 2013 doi: 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Cremer C, Cremer T, Fakan S. Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol. 2010;282:1–90. doi: 10.1016/S1937-6448(10)82001-5. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012 doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141(6):970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13(2):167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20(3):317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2(12):871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Curr Opin Cell Biol. 2011;23(3):310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012 doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunger S, Kalo A, Kafri P, Sheinberger J, Lavi E, Neufeld N, Shav-Tal Y. Zooming in on single active genes in living mammalian cells. Histochem Cell Biol. 2013a;140(1):71–79. doi: 10.1007/s00418-013-1100-2. [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7(8):631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Quantifying the transcriptional output of single alleles in single living mammalian cells. Nat Protoc. 2013b;8(2):393–408. doi: 10.1038/nprot.2013.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13(11):1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]