Abstract
Common variable immunodeficiency (CVID) has been associated recently with a dramatic increase in total copy number variation burden, the cause of which is unclear. In order to explore further the origin and clinical relevance of this finding, we quantified the total genomic copy number variation (CNV) burden in affected patients and evaluated clinical details in relationship to total CNV burden. No correlation was found between total CNV burden and either patient age or time elapsed since symptom onset, and higher total burden did not correlate with incidence of malignancy or other subphenotypes. These findings suggest that the increased CNV burden is static and intrinsic to CVID as a disease.
Keywords: CVID, copy number variation, immunodeficiency, immunoglobulin
Introduction
Among forms of primary immunodeficiency, common variable immunodeficiency disorder (CVID) is one of the most frequently diagnosed and treated entities. To date, 10 genes have been identified as monogenic associations and/or causes of CVID 1,2. However, the cause of greater than 80% of CVID cases remains unclear. Given the wide phenotypical variety and variable onset of CVIDs, a multi-centre consortium sought to explore the genetics using a genome-wide single nucleotide polymorphism (SNP)-array 3.
This study unearthed associations with both CVID and CVID subphenotypes and many rare copy number variants and SNPs. An unexpected result of this study was a significantly increased overall burden of copy number variation (CNV) in patients versus controls. Although several of the individual variants are currently under investigation, the reason behind the high overall CNV burden was unclear. Further, as the study was performed using DNA from peripheral leucocytes, it was not clear if these results represented true germline or accumulated somatic CNV.
One of the clinical features of CVID includes an increased incidence of cancer, primarily lymphoma 4. Additionally, an association between CVID and mutations in DNA mismatch-repair genes (Lynch syndrome) has been described previously 5. Furthermore, several published reports have demonstrated radiosensitivity in peripheral leucocytes from selected CVID patients 6,7. Because the acquisition of CNV is well described in cancer, we hypothesized that genomic instability may exist in a subset of CVID patients, leading to a gradual acquisition of a CNV burden over time. If this were the case, then a higher total CNV burden may correlate with complex clinical phenotypes, such as malignancy. In order to address this question, we sought to quantify the genomic burden of CNV for each patient and determine if a correlation existed between this quantity and either patient age or years elapsed since symptomatic onset.
Material and methods
Data were acquired as described previously 3. Briefly, genotyping was performed on the Infinium Humanhap610 BeadChip array, and additionally revalidated with the Affymetrics Cytogenetics Whole-Genome 2·7 M array. Subjects from the study were included in analyses if their birth year and age at symptom onset was recorded. In order to determine genomic CNV burden, the sum of the base pairs impacted by deletions or duplications was calculated. The approximate subject ages were calculated based on their birth year and the sampling date ranges, which produced a standard deviation of approximately ± 1 year. Chart review was performed to determine relevant clinical patient characteristics. Linear regression was performed in Microsoft Excel, and a two-tailed Student's t-test was used for comparison of CNV between clinical groups.
Results
Patient characteristics are listed in Table 1. Linear regression between total CNV burden and either patient age (Fig. 1a) or years of symptoms (Fig. 1b) did not show any correlation. Most patients with total CNV >5 MBp had individual CNVs greater than 1 MBp in size (Fig. 1c). Of these 11 patients, no clear phenotypical similarities were noted in terms of autoimmunity, lung disease, enteropathy or malignancy (data not shown). A comparison between the 21 patients with malignancies and the remainder of the group showed no significant difference in total CNV burden (Fig. 1d, P = 0·35), nor did they correlate with having > 1 MBp individual CNVs.
Table 1.
Patient demographics and genomic data.
| Patient characteristics | Value |
|---|---|
| Median age* | 46 (range 6–88 years) |
| Median years symptoms (n = 130) | 19 (range 2–71 years) |
| Median age at diagnosis (n = 148) | 35 (range 1–79 years) |
| Mean CNV (MBp)* | 2·04 (s.d.: ± 1·72) |
| Patients with single CNV >1 MBp* | 11 |
| Patients with malignancies* | 21 |
n = 230 except where noted; CNV = copy number variation; s.d. = standard deviation.
Fig. 1.
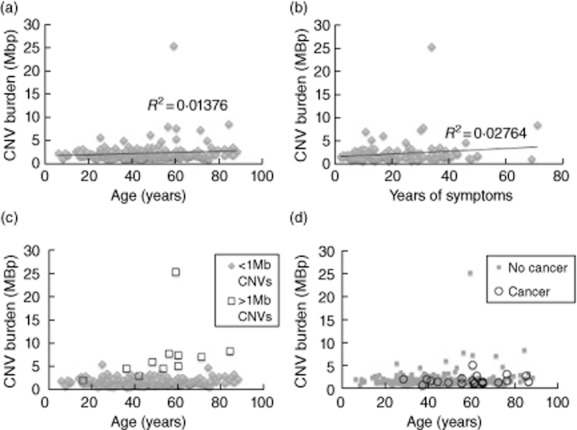
Copy number variation (CNV) burden showed no correlation with patient age (a) or years elapsed since symptom onset (b). Correlation coefficients (R2) are displayed. A subset of patients had large individual CNVs of greater than 1 MBp (c, n = 11). Phenotypical details including malignancies (d, n = 21) did not correlate with CNV burden.
Discussion
These data suggest that there is no increase in CNV burden over time in patients with CVID, nor does overall CNV quantity seem to correlate with clinical phenotype. Rather, specific CNVs, as noted in the previous study, are probably of much greater phenotypical and mechanistic consequence. Although these results do not rule out definitively the possibility of somatic CNV, they suggest that the CNV burden is fairly static over a wide age range, and thereby likely to be germline. Although specific details regarding infections were not collected, due to the static nature of the burden, we would also theorize that it is not caused by accrued damage due to infections.
Limitations of this study include the range of sampling dates, which means that patient ages could be miscalculated by 1–2 years, as well as the possible recall bias of the patients regarding the age at symptom onset. However, the complete lack of correlation suggests that the effect of this error is not consequential.
In summary, the increased CNV burden in CVID appears to be static, and the reason for this high degree of CNV remains unclear. In concert with further studies on specific rare CNV, familial studies in CVID may be valuable to determine the patterns of CNV inheritance and to evaluate for possible anticipation.
Acknowledgments
We thank Rosetta Chiavacci and Cecilia Kim for their support of this work. Support was from the Children's Hospital of Philadelphia Institutional Development Award to the Center for Applied Genomics, which funded all genotyping (to H. H.); a Research Development Award from the Cotswold Foundation (to H. H.); the Jeffrey Modell foundation (to J. S. O.); and National Institutes of Health (NIH) grant AI-079731 (to J. S. O.). This study was performed with approval from the institutional review boards at the host institutes.
Disclosures
The authors report no conflicts of interest relevant to this manuscript.
Author contributions
M. D. K., J. S. O. and H. H. wrote the manuscript. M. D. K., J. G., C. C. R., J. S. O. and and H. H. designed the study. M. D. K., J. G. and E. R. collected and analysed the data. E. R., E. P., H. C., M. L., K. S., C. C. R., J. S. O. and H. H. provided data and edited the manuscript.
References
- 1.Yong PF, Thaventhiran JE, Grimbacher B. ‘A rose is a rose is a rose’, but CVID is not CVID common variable immune deficiency (CVID), what do we know in 2011? Adv Immunol. 2011;111:47–107. doi: 10.1016/B978-0-12-385991-4.00002-7. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Herrera G, Tampella G, Pan-Hammarstrom Q, et al. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet. 2012;90:986–1001. doi: 10.1016/j.ajhg.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orange JS, Glessner JT, Resnick E, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127:1360–7 e1366. doi: 10.1016/j.jaci.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119:1650–1657. doi: 10.1182/blood-2011-09-377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Offer SM, Pan-Hammarstrom Q, Hammarstrom L, Harris RS. Unique DNA repair gene variations and potential associations with the primary antibody deficiency syndromes IgAD and CVID. PLOS One. 2010;5:e12260. doi: 10.1371/journal.pone.0012260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aghamohammadi A, Moin M, Kouhi A, et al. Chromosomal radiosensitivity in patients with common variable immunodeficiency. Immunobiology. 2008;213:447–454. doi: 10.1016/j.imbio.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Palanduz S, Palanduz A, Yalcin I, et al. In vitro chromosomal radiosensitivity in common variable immune deficiency. Clin Immunol Immunopathol. 1998;86:180–182. doi: 10.1006/clin.1997.4478. [DOI] [PubMed] [Google Scholar]


