Abstract
Lignocellulosic biomass is a potential source of renewable, low-carbon-footprint liquid fuels. Biomass recalcitrance and enzyme cost are key challenges associated with the large-scale production of cellulosic fuel. Kinetic modeling of enzymatic cellulose digestion has been complicated by the heterogeneous nature of the substrate and by the fact that a true steady state cannot be attained. We present a two-parameter kinetic model based on the Michaelis-Menten scheme (Michaelis L and Menten ML. (1913) Biochem Z 49:333–369), but with a time-dependent activity coefficient analogous to fractal-like kinetics formulated by Kopelman (Kopelman R. (1988) Science 241:1620–1626). We provide a mathematical derivation and experimental support to show that one of the parameters is a total activity coefficient and the other is an intrinsic constant that reflects the ability of the cellulases to overcome substrate recalcitrance. The model is applicable to individual cellulases and their mixtures at low-to-medium enzyme loads. Using biomass degrading enzymes from a cellulolytic bacterium Thermobifida fusca we show that the model can be used for mechanistic studies of enzymatic cellulose digestion. We also demonstrate that it applies to the crude supernatant of the widely studied cellulolytic fungus Trichoderma reesei and can thus be used to compare cellulases from different organisms. The two parameters may serve a similar role to Vmax, KM, and kcat in classical kinetics. A similar approach may be applicable to other enzymes with heterogeneous substrates and where a steady state is not achievable.
Lignocellulosic biomass is a potential source of sustainable, low-carbon-footprint transportation fuels. In order to enhance the positive attributes of cellulosic biofuels a number of challenges, including efficient enzymatic digestion of cellulose, have to be overcome. Cellulose digestion by most cellulolytic microorganisms is carried out by cellulases (EC 3.2.1.4) from multiple glycoside hydrolase (GH) families, many of which synergize with each other. In addition, a number of important non-cellulase auxiliary proteins have been recently discovered and studied (1, 2). The major function of these proteins appears to be disruption of the recalcitrant portions of cellulose, making them more accessible for digestion by cellulases. As modular proteins, many cellulases contain a catalytic domain (CD) attached to one or more carbohydrate binding modules (CBMs) via a flexible linker. The main function of the CBM is to increase the effective concentration of the CD on the cellulose surface (3, 4). There are two major classes of cellulases. Exocellulases have an active site inside a tunnel and attack cellulose chains from the ends. Most exocellulases appear to be processive and produce cellobiose (G2) as their major digestion product. Endocellulases have an active site inside an open cleft, which allows them to initiate hydrolysis anywhere along the cellulose chain. Most endocellulases seem to have low or no processivity, making random cleavages in accessible regions of the substrate. There is, however, a subclass of processive endocellulases, the most studied of which is Thermobifida fusca Cel9A (TfCel9A).
Digestion of insoluble cellulose involves a number of discrete steps. The enzyme must first bind to the substrate, usually via the CBM. It then must find an accessible site on the substrate and transfer an individual cellulose chain into the active site. Finally, the enzyme hydrolyzes the β-1,4 glycosidic bond, releases the product, and either translates along the chain or releases it. Exocellulases translate along the chain, cleaving off G2 units multiple times before releasing it. Once the cellulose chain is released from the active site, the CD may rebind to the same or a nearby chain or the enzyme may dissociate and rebind elsewhere. Given the fact that cellulase rates of hydrolysis of soluble substrates are much greater than those of insoluble substrates, it is generally believed that access to substrate is the rate limiting step in cellulose digestion. Recent studies have provided experimental support for this idea (4, 5).
Kinetic modeling of enzymatic cellulose digestion is complicated by the number of steps involved and by the heterogeneity of the substrate, which changes during the reaction. It appears that a constant rate of hydrolysis is never achieved in cellulose digestion and the rate begins to drop off rapidly after the initial digestion (6). Various kinetic models of cellulose digestion, some of which are based on the Michaelis-Menten scheme (7), have been proposed over the years (as reviewed in (8) and (9)). Some models include cellulase adsorption, most commonly expressed with the Langmuir isotherm. Incorporation of adsorption is complicated, however, by the fact that only a fraction of the cellulases is productively bound. This productively bound fraction is not directly measurable and most likely continuously changes over time. Nevertheless, despite their apparent limitations, many of the models are able to accurately fit experimental data and predict product formation under various conditions, sometimes up to high extents of digestion. The majority of the proposed models are of a predictive nature, which is particularly useful for the utilization of cellulases on an industrial scale. They typically rely on multiple mechanistic studies that allow for the determination of the relevant parameters included in the models.
Another goal of kinetic modeling is to better understand the mechanisms by which enzymes act on their substrates. The mechanisms and properties of enzymes acting on soluble substrates are commonly deduced using Michaelis-Menten kinetic parameters Vmax, KM, and kcat. The Michaelis-Menten kinetic scheme is based on a number of assumptions, which include steady state reaction conditions and a homogeneous substrate. Neither assumption is applicable to the hydrolysis of cellulose, however, which is a heterogeneous material that consists of highly crystalline, semi-crystalline, and amorphous regions (10–12). While the exact natures of cellulosic substrates have not been thoroughly characterized, due to their complexity and technological limitations, cellulose can be described as a collection of substrates with a spectrum of reactivities, such that some fractions are digested more easily than others. In this scenario one would expect preferential depletion of easily degradable regions early in the digestion followed by increasingly slower digestion of the recalcitrant fractions of the substrate. A decrease in substrate reactivity with the extent of substrate digestion has been demonstrated (13, 14), although the exact reasons for the increased recalcitrance are not easily determined. Some authors also have suggested that enzyme inactivation via irreversible non-productive binding is the primary cause of the continuously decreasing hydrolysis rate of cellulose (15, 16).
Proposed here is a simple two-parameter fit, which is based on the Michaelis-Menten scheme, but with a time dependent activity for the enzyme. This is analogous to the fractal and fractal-like kinetics approach formulated by Kopelman (17) for heterogeneous reactions limited by surface diffusion. The objective for this model was to develop a mathematical relationship, which yields an intrinsic constant that quantifies the different abilities of cellulases to overcome substrate recalcitrance. The two parameters are determined from time course data of cellulose hydrolysis and can provide insight into the mechanisms by which enzymes digest insoluble cellulose, alone and in mixtures. This is best illustrated by comparing the parameters of mutated versions of a cellulase, TfCel48A, with its native form. In addition, we demonstrate how addition of an auxiliary protein to an exocellulase affects its measured parameters, and thus confirms the presumed role of auxiliary disruptive proteins in cellulose hydrolysis. The model applies to low-to-medium enzyme-substrate ratios, and the two parameters can serve a similar role as Vmax, KM, and kcat in classical kinetics. In this study we mostly used enzymes from a model cellulolytic bacterium T. fusca to validate the proposed model and to demonstrate its utility, but we also provide data for the crude supernatant of a commonly studied cellulolytic fungus, Trichoderma reesei. We believe that the demonstrated modeling approach may prove useful for other enzymes that utilize a heterogeneous, continuously evolving substrate and do not reach steady state.
Model Development
In classical kinetics, the reaction rate is constant throughout the reaction. This is also true for the specific activity of enzymes acting on a soluble, homogeneous substrate, before other factors such as substrate depletion, thermal inactivation, and product inhibition become significant. In fractal and fractal-like systems, however, if the reaction is limited by surface diffusion of the reactants, the reaction rate is not constant, but is dependent on time, as demonstrated by Kopelman (17):
| (1) |
where t is time, K is the initial rate coefficient at t = 0, and h is the fractal factor, which can be determined experimentally and, in some cases, predicted mathematically.
Cellulose digestion can be represented as:
where S is insoluble cellulose and P any soluble oligosaccharide. In a classical kinetic system the rate of product formation at low enzyme (E) loads (E << Substrate) can be represented as:
| (2a) |
where k is the specific activity of the enzyme. Total generated product (Ptot) at any given time can be obtained by integrating Eqn. 2a with respect to time:
| (2b) |
It was determined experimentally that cellulose digestion shows the same time dependence of the specific activity coefficient as described for fractal systems. In addition, only some fraction of the added enzyme (Etot) is productively bound (EP), such that:
where EU is unable to generate a soluble product either because it is not bound or is bound unproductively. Most likely EP changes continuously during the reaction, but it is not directly measurable. Accounting for the time dependence of the specific activity of the cellulase and considering only the productively bound fraction gives:
| (3) |
It is also not possible to determine the exact value of K (i.e. the inherent specific activity) of any cellulase for its activity on insoluble substrate. Thus, for fitting purposes, the two variables, K and EP, are lumped into one term A, which represents the net activity of the total enzyme acting on the substrate, as determined by soluble products formed over time. While it has no effect on the utility of the model, it is important to note that for non-processive cellulases the value of A may be an underestimate of the actual number of cleavages carried out by these enzymes because in theory it should take multiple random cleavages to produce a soluble oligosaccharide product. However, pseudoprocessivity – a repeated non-processive attack in the same region, which produces soluble oligosaccharides – appears to be common among non-processive endocellulases, as evidenced by constant ratios of soluble and insoluble reducing ends generated by such enzymes over time. For the sake of simpler representation and to distinguish the present equation from fractal-like kinetics (discussed below), (1−h) is also simplified to a parameter b, which we call the “hydrolysis power factor”:
| (4) |
When b = 1, the specific activity of the enzyme is constant over time (extent of digestion) as would be the case in classical kinetics. It is useful to monitor cellulase activity with respect to substrate digestion. Knowing Ptot allows one to calculate directly the fraction of the digested substrate (X):
| (5) |
where Stot is the total substrate added to the reaction. Most of the data and empirically determined parameters in the manuscript are presented with respect to X, such that
| (6) |
where A contains Stot.
Materials and Methods
Substrates
Bacterial cellulose (BC) was a gift from Monsanto. BC cake was washed three times with deionized (DI) water by centrifugation and resuspended in DI water with 0.04% sodium azide (Sigma-Aldrich). Concentration was determined as dry weight per volume. Avicel® powder (PH-105; FMC Corporation, Philadelphia, PA, USA) was suspended in DI water with 0.04% sodium azide. Phosphoric acid swollen cellulose (PASC) was prepared from Avicel® powder using procedures described in (18) and was stored in DI water with 0.04% sodium azide.
Enzymes
All individual cloned T. fusca cellulases (except TfCel48A) and E7 were expressed in Escherichia coli BL21 or Streptomyces lividans strains, which secrete these enzymes during expression. The enzymes were purified from the culture supernatant as previously described (19, 20).
TfCel48A contains a 6-His tag on both the amino- and carboxyl termini and no signal peptide. WT and mutant TfCel48A were expressed inside E. coli BL21-CodonPlus® (DE3)-RIPL cells (Agilent Technologies) as follows. A starter culture was grown in Lysogeny Broth (LB) medium overnight at 37 °C. The starter culture was diluted 33 times in fresh LB and the cells were grown at 37 °C until optical density at 600 nm reached ~0.8 (about 2.5 hrs). The culture was transferred to 25 °C and Isopropyl β-D-1-thiogalactopyranoside was added to 0.8 mM. The culture was incubated with shaking for 16 hours. Cells were harvested by centrifugation and resuspended in 1/20 original culture volume of 20 mM sodium phosphate buffer, pH 8.0, 0.01 M Imidazole (Sigma-Aldrich), 0.5 M sodium chloride (Solution A). Cells were lysed with a French Press (at cell pressure of 20,000 PSI). Lysed cells were placed in a 50 °C water bath for 30 minutes (to precipitate out E. coli proteins) and centrifuged to remove the precipitate. The supernatant was diluted in 5 additional volumes of Solution A and loaded on a nickel Sepharose 6 Fast Flow (GE Healthcare) column by gravity. The protein was eluted by a gradient of 0.01 – 0.5 M Imidazole in 20 mM sodium phosphate buffer, pH 8.0, 0.5 M sodium chloride. Fractions containing TfCel48A, as determined by gel electrophoresis, were combined and diluted 10–15 times with DI water (to lower the conductivity of the solution to less than 0.5 mA). The protein was then loaded on a Q Sepharose (Sigma-Aldrich) column equilibrated with 0.1 M sodium chloride in 5 mM Bis-Tris buffer, pH 5.8, 10% glycerol (v/v). The protein was eluted by a gradient of 0.1–0.5 M sodium chloride in 5 mM Bis-Tris buffer, pH 5.8, 10% glycerol (v/v). Fractions containing TfCel48A, as determined by denaturing gel electrophoresis, were combined and concentrated using Millipore® centrifugal filter units with a 30 kDa cutoff membrane. Buffer exchange was carried out in the same filter units by washing the concentrated protein three times with 5 mM sodium acetate buffer, pH 5.5, 10% glycerol (v/v). Protein was filtered through a 0.22 µm filter using a syringe and stored at −20 °C.
All enzyme concentrations were determined by spectroscopy using NanoDrop® 1000 spectrophotometer (See Table S1 for relevant extinction coefficients and molecular weights). T. fusca crude supernatant was obtained by growing T. fusca ER1 strain on cellulose for 72 hours, as described in (21). T. reesei crude supernatant was obtained by growing T. reesei L27 strain on cellulose for 96 hrs, as described in (22). Cells were removed by centrifugation and the supernatant was stored at −70 °C. Crude supernatant protein concentration was determined by Bradford assay using Bio-Rad Protein Assay Dye Reagent Concentrate and bovine serum albumin for reference. TfCel48A mutants were generated using Agilent Quickchange® II XL Site Directed Mutagenesis Kit following the manufacturer’s instructions. All mutations and sequences were verified by Sanger sequencing. Mutant proteins were expressed and purified in the same way as WT.
Time course assays
All reactions were conducted in triplicate in Eppendorf 2 mL Protein LoBind plastic tubes. 1 mg substrate was combined with an indicated amount of enzymes in 0.6 mL 50 mM sodium acetate buffer, pH 5.5. In reactions involving E7, 1 mM glutathione (Sigma-Aldrich) was added to enhance activity of E7 (1). Only buffer and substrate were combined for negative controls. Upon mixing, BC and PASC reactions were immediately placed in a 50 °C (unless otherwise indicated) water bath. Avicel® reactions were carried out in a 50 °C incubator with rotation to prevent the substrate from settling. Samples (in triplicate) were removed at the given time points and placed on dry ice to stop the reaction. Frozen samples were later placed in a boiling bath for 10 minutes in order to denature the enzyme. It was verified experimentally that boiling does not alter soluble sugar profiles detected by High Performance Liquid Chromatography (HPLC). The remaining substrate was removed using Corning® Spin-X® Centrifuge tube filters and the soluble sugar concentrations were measured using a Shimadzu HPLC system fitted with a Bio-Rad Aminex® HPX-87P analytical column and a refractive index detector. The mobile phase was Milli-Q water at a flow rate of 0.6 ml/min. Sample injection (50 µl volume) was performed by an autosampler installed on the instrument.
Apparent KM assays
75 nM TfCel48A was combined with the indicated amount of BC in 0.6 ml of 50 mM sodium acetate, pH 5.5 and reactions were incubated in a 50 °C water bath for one hour. Samples were boiled for five minutes to denature the enzyme and were processed and analyzed by HPLC in the same manner as described for the time course assays.
Data analysis
HPLC data were processed with OriginPro 8. Product identities and concentrations were determined by Gaussian peak fitting, using standard solutions with known concentrations of soluble cellooligosaccharides for reference. Soluble sugar concentrations at time zero were subtracted from all of the subsequently obtained concentrations. Soluble sugar produced upon initial mixing of the enzyme and the substrate is primarily due to the burst activity, as described in (6, 23), whereas the model presented here is concerned with the digestion of the more recalcitrant portions of cellulose. Apparent KM and the A and b parameter values of time course profiles were determined using the nonlinear least squares fit of the Michaelis-Menten expression (Vmax [S]/(KM + [S])) and Eqn. 6 respectively. OriginPro 8 default settings (chi-square minimization) were used for all fits.
Results
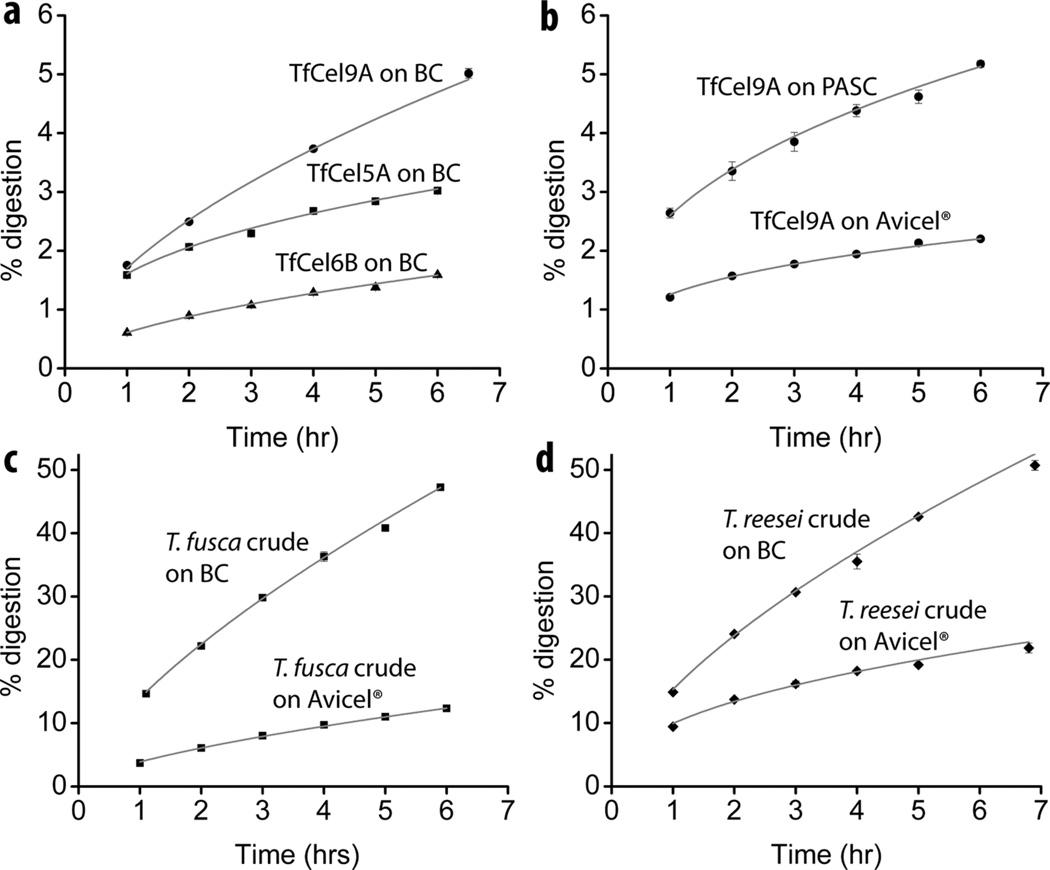
When the activity of different cellulases is monitored over time, their time course profiles are distinct (Fig. 1a). This is also true for the same cellulase acting on different substrates (Fig. 1b). It was empirically determined that initial time course profiles of all T. fusca cellulases follow Eqn. 6, a two parameter expression based on a pseudo zero order (with respect to the substrate) reaction with a time-dependent activity coefficient (Table 1). The same equation applies to mixtures of enzymes, as shown for T. fusca and T. reesei crude supernatants acting on bacterial cellulose (BC) and Avicel® (Fig. 1c and d).
Figure 1.
Fitted digestion curves for different enzymes on various substrates. (a) 33.3 nM T. fusca Cel9A, Cel5A, and Cel6B on BC; (b) T. fusca Cel9A on Avicel® (167 nM enzyme) and phosphoric acid swollen cellulose (PASC; 33.3 nM enzyme); (c) T. fusca crude supernatant on BC (95µg tot. protein/mg substrate) and Avicel® (159 µg tot. protein/mg substrate); (d) T. reesei crude supernatant on BC and Avicel®.
Table 1.
A and b values of all T. fusca cellulases digestion time course profiles on BC.
| Enzyme* | Endo/ Exo |
A# (% digestion hr−1) |
b# | Processivity% |
|---|---|---|---|---|
| TfCel9A | Endo | 1.68 ± 0.04 | 0.58 ± 0.02 | 6.9 |
| TfCel9A-CD | Endo | 0.90 ± 0.01 | 0.58 ± 0.01 | 2.5–3.5 |
| TfCel5A | Endo | 1.61 ± 0.02 | 0.33 ± 0.02 | N/A |
| TfCel9B | Endo | 1.11 ± 0.03 | 0.31 ± 0.01 | N/A |
| TfCel6A | Endo | 1.18 ± 0.07 | 0.38 ± 0.01 | N/A |
| TfCel6B | Exo | 0.48 ± 0.01 | 0.50 ± 0.01 | 20 |
| TfCel48A | Exo | 0.52 ± 0.01 | 0.33 ± 0.01 | 15 |
| TfCel48A-CD | Exo | 0.24 ± 0.01 | 0.33 ± 0.01 | 15 |
| T. fusca crude supernatant | N/A | 13.97 ± 0.23 | 0.69 ± 0.01 | N/A |
| T. reesei crude supernatant | N/A | 15.32 ± 0.22 | 0.64 ± 0.01 | N/A |
Individual cellulase concentrations, except TfCel48A, were 33.3 nM; TfCel48A and TfCel48A-CD concentrations were 100 nM; T. fusca and T. reesei crude supernatant total protein added per reaction was 95 µg and 8 µg respectively.
N = 3 for all data points. Averaged values with standard deviations were fit to Equation 6 to obtain A and b values. Standard error is provided for all values. All R2 > 0.97.
Values for TfCel9A are the ratio of soluble and insoluble reducing ends after digestion of filter paper and were previously published (20, 42). Values for TfCel6B and TfCel48A are the ratio of cellobiose to cellotriose concentrations (33) after digestion of BC.
N/A = not applicable.
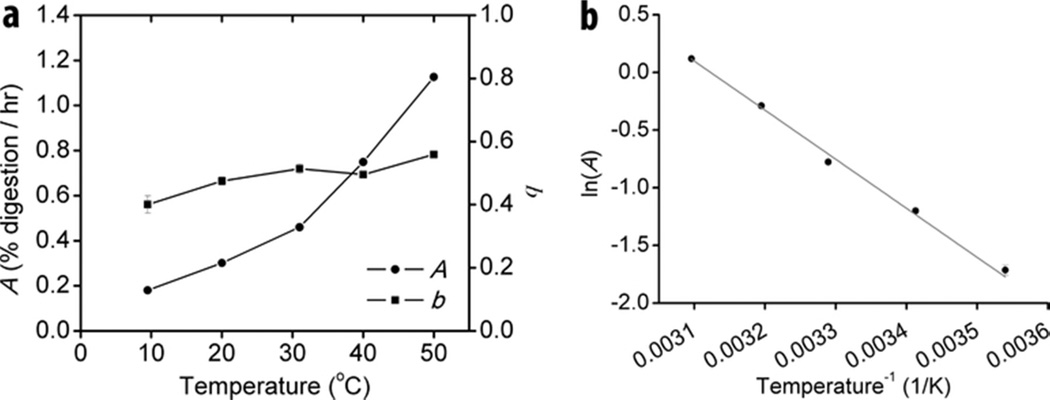
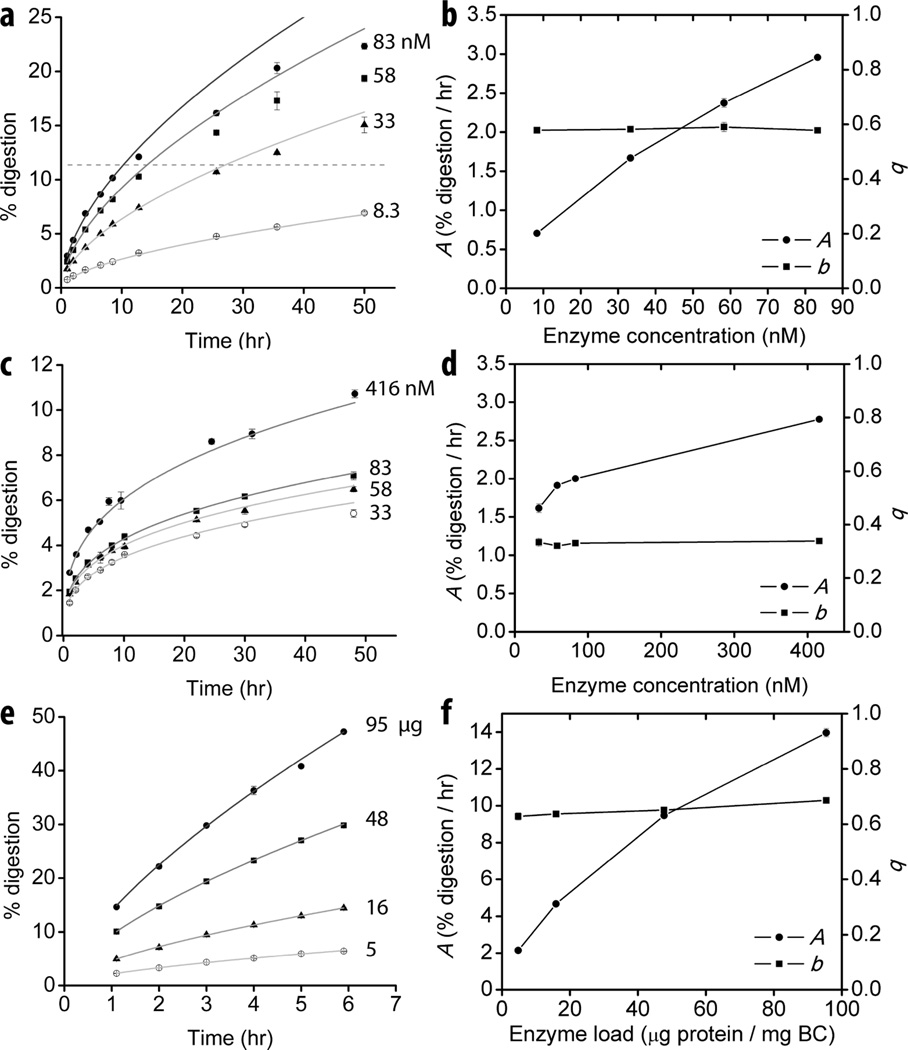
Based on the derivation of Eqn. 4, the two parameters quantify the net activity of the added cellulase (A) and the intrinsic ability of the cellulase to overcome substrate recalcitrance (b, the curvature of the time course profile). In order to verify the mathematically implied significance of the parameters experimentally the effect of reaction temperature (Fig. 2) and enzyme concentration (Fig. 3) on their values was tested. A and b values were determined for a processive endocellulase TfCel9A on BC at temperatures between 10 and 50 °C. An increase in the reaction temperature had a strong effect on the measured value of A. The relationship between A and temperature follows the Arrhenius equation (Fig. 2b), which holds true for most reaction rate constants. On the other hand, the value of b increased only slightly with temperature, indicating that temperature plays a minor role in the enzyme’s ability to overcome substrate recalcitrance. The effect of enzyme concentration on the A and b values was determined for a processive endocellulase TfCel9A, a non-processive endocellulase TfCel5A, and T. fusca crude supernatant on BC (Fig. 3). For both individual cellulases, A values increased with increasing enzyme concentration. On the other hand, the b value was constant, consistent with it being an intrinsic constant. For crude supernatant, the results are similar to those obtained for individual cellulases, but there is a slight increase in the value of the b parameter from 0.63 ± 0.01 to 0.69 ± 0.01 with increased enzyme load. This may be due to an improved synergistic effect caused by more densely crowded cellulases on the cellulose surface (24).
Figure 2.
Dependence of A and b on temperature. (a) Experimentally determined A and b values for TfCel9A digestion of BC at different temperatures; (b) Arrhenius plot of A. A time course profile was obtained for each temperature (N=3) and the data were fit with Eqn. 6 to obtain A and b values. Error bars represent standard error of the fit.
Figure 3.
Dependence of A and b on enzyme concentration. Different concentrations of TfCel9A ((a) and (b)) TfCel5A ((c) and (d)), and T. fusca crude supernatant ((e) and (f)) on BC. All time course data were obtained in triplicate. Error bars in (a), (c), and (e) represent standard deviation. Time course data were fit with Eqn. 6 and the obtained A and b values are plotted in (b), (d), and (f); error bars represent standard error. Dashed line in (b) indicates the approximate “drop-off” value, above which digestion rate decreases more rapidly than predicted by the initial fit.
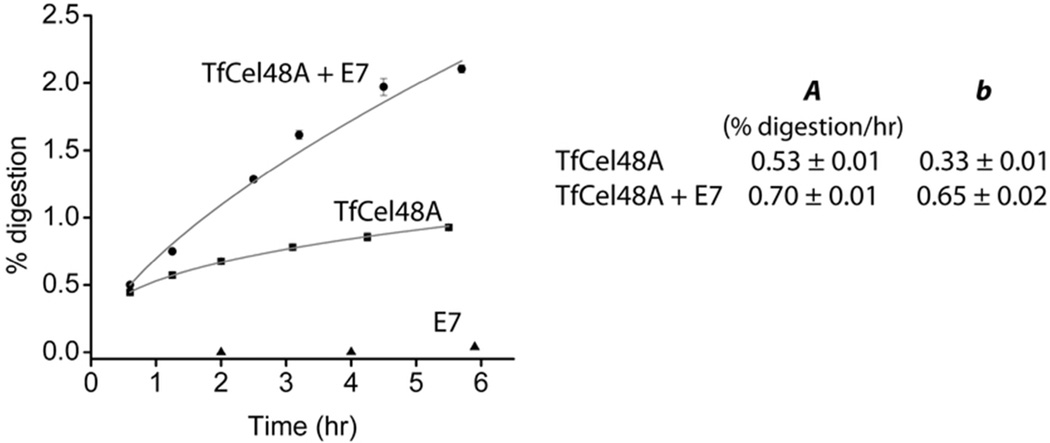
In recent years, a number of disruptive enzymes, which are important for enzymatic biomass digestion, have been discovered (1, 2). These enzymes belong to auxiliary activity families AA10 (formerly CBM33) in bacteria and AA9 (formerly GH61) in fungi, and use an oxidoreductive mechanism rather than general acid/base hydrolysis to cleave cellulose chains at random locations on the bulk substrate (1, 25, 26). Their activity is believed to disrupt the crystalline regions of cellulose and thus make it more accessible to cellulases. Addition of these enzymes to cellulases acting on cellulose can significantly increase the rate of cellulose digestion even though they produce very few soluble products by themselves. T. fusca secretes two known AA10 enzymes – E7 and E8 – which have been shown previously to stimulate the activity of other T. fusca cellulases on BC (27). In order to determine the effect of E7 addition on the b parameter of an individual cellulase, time course experiments were carried out on BC using T. fusca exocellulase TfCel48A in combination with E7 (Fig. 4). TfCel48A has the lowest activity of all T. fusca cellulases on insoluble cellulose (Table 1). Due to E7’s extremely low generation of soluble products and different mechanism of hydrolysis, it is not possible to fit its time course data with the proposed model. When E7 is added to TfCel48A, the b parameter of the digestion curve is increased from 0.34 ± 0.01 for TfCel48A alone to 0.65 ± 0.01 for the mixture of the two enzymes. The A parameter is increased from 0.53 ± 0.02 to 0.70 ± 0.01.
Figure 4.
Effect of the auxiliary protein T. fusca E7 (AA10) on the digestion of BC by a weak T. fusca exocellulase TfCel48A. Time course data were collected in triplicate and were fit to Eqn. 6. The calculated parameters are presented in the table inset. E7 alone produces only trace amounts of soluble sugars and its time course profile does not follow Eqn. 6.
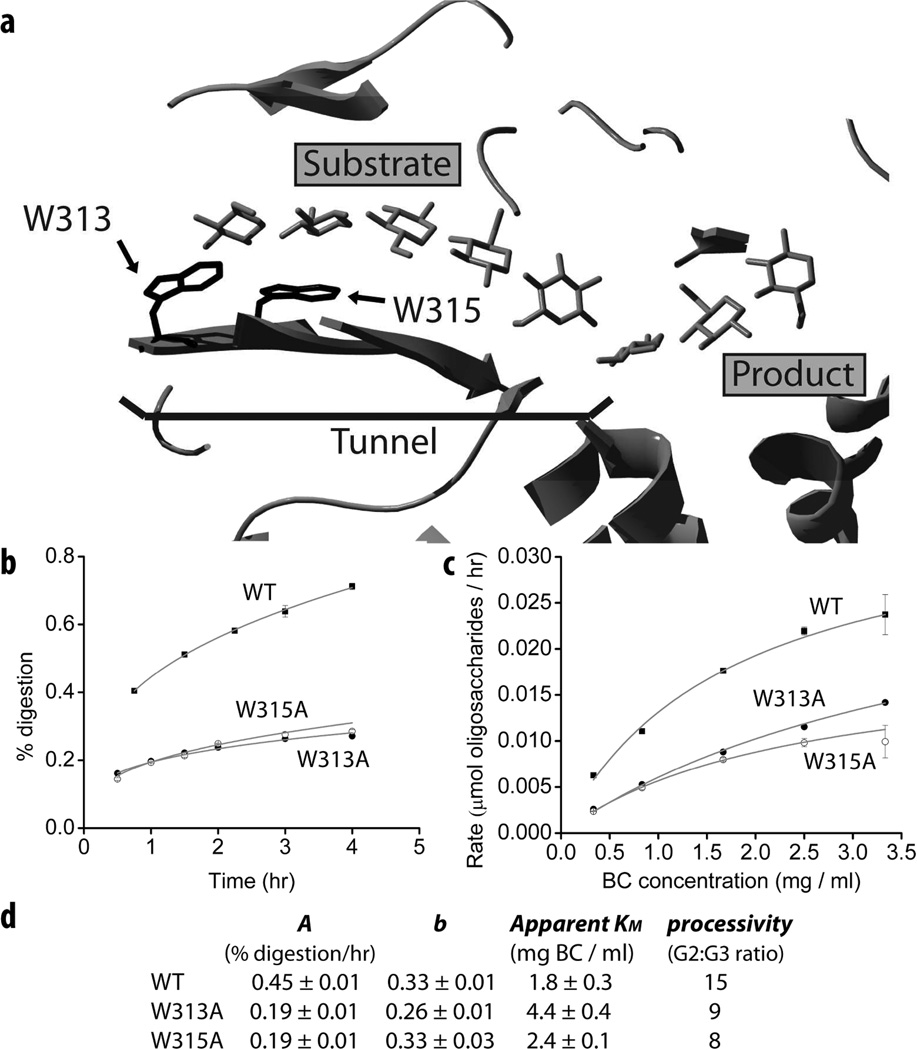
Access to substrate is widely accepted to be the rate limiting step of enzymatic crystalline cellulose digestion (5, 28) and it is likely that cellulases have evolved residues that play a direct role in the separation of an individual cellulose chain from the bulk substrate. The mutation of such residues should decrease the activity of a cellulase on crystalline cellulose. As for all exocellulases, the active site of TfCel48A is located inside a tunnel (29), which is presumed to be important for the processivity of these enzymes. Our group is in the process of studying residues around the tunnel entrance of TfCel48A in regard to their role in crystalline cellulose digestion. As part of this study, the effect of these residues on the b parameter value of the enzyme was determined. Fig. 5b shows time course data for two TfCel48A mutants, W313A, and W315A on BC. Both residues are conserved in the GH48 family and aromatic residues of various cellulases have been shown to play an important role in substrate binding and cellulase processivity (30–33). W313 and W315 are located at the mouth and inside the tunnel respectively (Fig. 5a). Both W313A and W315A mutants have strongly reduced activity on BC, as shown by their time course profiles. Both mutants have lower than WT processivity (Fig. 5d), which is determined by their cellobiose to cellotriose ratio (33), and a similarly reduced A parameter value. However, only the b parameter of W313A is significantly reduced in comparison to WT. A similar result is observed for apparent KM of the mutants on BC (Fig. 5c). The mutation of W315 results in a slight increase in apparent KM of the enzyme from 1.8 ± 0.3 to 2.4 ± 0.1 mg/ml BC, but the mutation of W313 results in a much greater increase of apparent KM to 4.4 ± 0.4 mg/ml BC. These results are consistent with the relative position of the two residues: one at the mouth and one inside the tunnel, as only residues at or near the surface of the protein should directly affect the enzyme’s interaction with the bulk substrate.
Figure 5.
Role of W313 and W315 in the digestion of BC by TfCel48A. (a) The tunnel and active site of TfCel48A. W313 and W315 residues are located at the mouth and inside the tunnel respectively; (b) time course profiles of WT, W313A, and W315A on BC (N=3), fitted with Eqn. 6; (c) apparent KM experiments with WT, W313A, and W315A on BC; (d) Eqn. 6 parameter values, processivity, and apparent KM of WT and mutated TfCel48A. Processivity is calculated as a ratio of cellobiose (G2) to cellotriose (G3) (33).
Discussion
The distinct time course profiles of different cellulases acting on the same substrates or the same cellulase acting on different substrates (Fig. 1) indicate that the detailed change in substrate during digestion is not universal, but is specific to each cellulase. Hence, the time course profile is a reflection of the ability of a cellulase to digest various forms of cellulose. Enzymatic cellulose digestion is a multistep process, which is further complicated by the continuously changing substrate. Most models of cellulose hydrolysis thus utilize multiple parameters that quantify the different factors involved in digestion (e.g. enzyme binding, change in the average degree of polymerization, change in cellulose crystallinity, etc.).
Proposed here is an alternative approach, which treats cellulose digestion similarly to classical kinetics, but replaces the specific activity constant with a coefficient that is dependent on time (extent of substrate digestion). The results indicate that cellulases have different time-dependencies of their activity coefficients when acting on insoluble substrates, as quantified by parameter b (curvature of the time course profile) of Eqns. 4 and 6. This parameter is not dependent on the enzyme concentration within the tested range (Fig. 3) and appears to be only slightly dependent on temperature (Fig. 2). These observations support the idea that the value of b quantifies the intrinsic ability of the cellulase to digest bulk cellulose. Based on the derivation of Eqn. 4, the value of b will fall between zero, when no products are produced over time, and one, when the specific activity is constant, as is in classical kinetics. A relatively robust cellulase, which can effectively hydrolyze recalcitrant portions of cellulose, will have a higher b value than a cellulase that is only effective on the easily accessible fractions of the substrate. This holds true for T. fusca cellulases acting on BC (Table I). In particular, TfCel9A, which is the most effective T. fusca cellulase on crystalline substrates (19) has the highest b value of 0.58 ± 0.02. Other cellulases, which are variably effective in crystalline cellulose digestion, have b values in the range from 0.31 to 0.50. The b value does not change when the CBM is removed from the cellulase (Table I), which leads to a reduced binding efficiency of the cellulase (20, 34). This suggests that the CBM of the tested enzymes (TfCel9A and TfCel48A) does not directly assist the CD in substrate digestion, as has been suggested for some cellulases in the literature (35–37). The constant b parameter value with and without the CBM attached to the CD also indicates that the time dependence of the cellulase’s specific activity is not related to surface diffusion limitations, which form the basis of fractal-like kinetic modeling (17). Rather, in the case of cellulose digestion, the continuously decreasing specific activity is most likely due to increasing substrate recalcitrance as easier to digest fractions are preferentially hydrolyzed by the enzyme(s).
The value of A follows the Arrhenius relationship as a function of temperature (Fig. 2), which is consistent with its proposed significance. Further support is provided by the data presented in Table I. When the CBM is removed from TfCel9A and TfCel48A, the A value is significantly reduced, presumably due to the lower binding efficiency of the CD alone. Both exocellulases (TfCel48A and TfCel6b) have much lower A parameters than the endocellulases, which probably reflects the fewer productive binding sites (chain ends) available to these enzymes. Finally, the A value is strongly dependent on enzyme concentration (Fig. 3), as would be expected for the productively bound concentration at low enzyme-substrate ratios. The fact that no two T. fusca cellulases have the same combination of A and b values suggests that no two cellulases act in the same way. It would be of particular interest to identify the unique features of the different endocellulases (Cel5A, Cel6B, and Cel9B in T. fusca), as they may have unique roles in the synergistic mixtures during substrate digestion.
A potentially useful aspect of the model is the variable range of the extent of digestion over which the model is applicable. At least some individual cellulases appear to reach a “drop-off” value, above which the rate of digestion decreases more rapidly than would be predicted by the b parameter. For TfCel9A (Fig. 3a) and TfCel48A (Fig. S1) acting on BC the “drop-off” values are approximately at 11% and 2% digestion respectively. On the other hand, the model fits T. fusca and T. reesei crude supernatant digestion of BC up to at least 50% (Fig. 1 c and d). This observation suggests that as different biomass degrading enzymes are combined in synergistic mixtures, their ability to maintain initial-like conditions are enhanced. It is likely that during cellulose digestion by individual cellulases or incomplete synergistic mixtures, factors other than increased substrate recalcitrance contribute to the drop in the digestion rate earlier, which results in a more significant rate decline than would be predicted from the initial data. Such factors may be attributed, for example, to an increasing fraction of unproductively bound enzymes (15, 16) or the formation of obstacles on the eroded surface of the substrate (38). A constant b value over a large extent of digestion by the crude extract of T. fusca and T. reesei suggests that relief of such factors plays a significant role in the synergism between cellulases with complementary properties (38, 39).
Eqns. 4 and 6 apply to both individual cellulases and their mixtures (Fig. 1). This is an important feature of the model, as it makes it possible to design experiments aimed at understanding synergistic interactions between cellulases and other important proteins. In comparison to the individual cellulases, the T. fusca crude supernatant time course profile on BC has the highest b value, which suggests that the synergistic interactions between proteins in the crude supernatant enhance the ability of the mixture to digest a crystalline substrate more effectively (not only faster) than any cellulase alone. Cellulases can be combined in mixtures of two or more and the changes in the parameter values can help determine the mechanisms of synergistic interactions between those enzymes. The data presented for BC digestion by TfCel48A (Fig. 4) with and without the disruptive T. fusca AA10 protein E7 shows that E7 increases the b and A parameter values of Cel48A by 97% and 32% respectively. This is consistent with other studies, which show that AA10 (CBM33) proteins are likely to disrupt the recalcitrant portions of crystalline cellulose with minimal formation of soluble products (27, 40). The relatively minor increase in the A parameter indicates that the specific activity of TfCel48A acting on BC probably is not increased in the presence of E7. Rather, E7 seems to increase the accessible fraction of the substrate, which results in a higher fraction of productively bound enzyme. Our group is also carrying out similar time course experiments as part of a study aimed to understand the synergistic interactions between TfCel9A and TfCel48A.
It is important to verify that Eqn. 6 applies to biomass-degrading enzymes from other organisms. For this reason the crude supernatant of T. reesei was tested on BC and Avicel®. As Fig. 1d shows, Eqn. 6 fits well T. reesei crude supernatant digestion of BC and Avicel® up to at least 50% and 20% digestion respectively. While comparison of the T. fusca and T. reesei crude supernatant time course profiles may provide interesting information about the possible differences between the bacterial and fungal biomass digestion mechanisms, such a study would require additional rigorous experiments. For this reason, no conclusions are made based on the experiments described here, but the results demonstrate that the kinetic model described by Eqns. 4 and 6 is applicable to both bacterial and fungal systems.
Dependence of the cellulose hydrolysis rate coefficient on time (extent of digestion) had been previously shown by Valjamae and co-workers (24). Similarly, Ohmine and co-workers demonstrated that the time course profiles of cellulose digestion can be defined by two empirically determined parameters that quantify the interaction between a cellulase mixture and its substrate (41). In both studies, however, only mixtures of cellulases were tested under pseudo-first order (with respect to the substrate) conditions (i.e. much higher enzyme to substrate ratios). This leads to important differences between the significance of our model and those published in the above studies. All three models contain a parameter that quantifies the shape of the time course profile (b parameter in the proposed model). In both of the above studies this parameter is dependent on enzyme-substrate ratio, while the b parameter in this model is constant within the tested range for individual cellulases. On the other hand, a slight dependence of the b value on enzyme load was observed for T. fusca crude digestion of BC. This is consistent with the explanation offered by Valjamae et al. (24) that at increasing enzyme loads the distances between the synergistically acting enzymes decrease, leading to a more efficient digestion of the substrate. Since there is no auto-synergism by individual cellulases, an increase in enzyme to substrate ratio, should not alter the shape of the digestion profile for a single cellulase. In addition, by working at much lower enzyme to substrate ratios than those used in the above studies, we avoid the contribution of substrate depletion to the multiple factors responsible for the continuous drop in the digestion rate of cellulose.
In comparison to the more complex kinetic models of enzymatic cellulose digestion developed to date, our model has unique advantages for the studies of mechanisms by which cellulases degrade insoluble cellulose. Only small amounts of enzyme are required for initial studies. No preliminary information about the cellulase is required; the empirically obtained A and b parameters are themselves meaningful measures of the cellulase properties and the b parameter in particular can be used to compare cellulases across different studies. Correlation between induced modifications to the cellulases (e.g. TfCel48A mutations, Fig. 5) or to their substrates (e.g. PASC derived from Avicel®) and the corresponding changes in parameter values can provide information about the effects of the induced modifications. Because the model applies both to individual cellulases and their mixtures, one can design synergistic experiments that lead to a better understanding of the specific interactions between the enzymes of interest (e.g. TfCel48a in combination with E7, Fig. 4). Hence, the presented model can be useful for basic studies of enzymatic digestion of cellulose, as well as for the design of improved synergistic biomass-degrading cocktails and pretreatment methods of lignocellulosic biomass.
Acknowledgments
Funding statement:
This work was supported by the BioEnergy Science Center, a U.S. Department of Energy (DOE) center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Abbreviations
- GH
glycoside hydrolase
- CD
catalytic domain
- G2
cellobiose
- G3
cellotriose
- CBM
carbohydrate binding module
- BC
bacterial cellulose
- PASC
phosphoric acid swollen cellulose
Footnotes
Supporting Information Available
Supporting information contains an additional figure and a table of extinction coefficients and molecular weights of the proteins used. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VG. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 2.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 3.Stahlberg J, Johansson G, Pettersson G. A New Model for Enzymatic-Hydrolysis of Cellulose Based on the 2-Domain Structure of Cellobiohydrolase-I. Bio-Technol. 1991;9:286–290. [Google Scholar]
- 4.Kostylev M, Moran-Mirabal JM, Walker LP, Wilson DB. Determination of the molecular states of the processive endocellulase Thermobifida fusca Cel9A during crystalline cellulose depolymerization. Biotechnol Bioeng. 2012;109:295–299. doi: 10.1002/bit.23299. [DOI] [PubMed] [Google Scholar]
- 5.Fox JM, Levine SE, Clark DS, Blanch HW. Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry. 2012;51:442–452. doi: 10.1021/bi2011543. [DOI] [PubMed] [Google Scholar]
- 6.Cruys-Bagger N, Elmerdahl J, Praestgaard E, Tatsumi H, Spodsberg N, Borch K, Westh P. Pre-steady-state kinetics for hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J Biol Chem. 2012;287:18451–18458. doi: 10.1074/jbc.M111.334946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaelis L, Menten ML. The kenetics of the inversion effect. Biochem Z. 1913;49:333–369. [Google Scholar]
- 8.Zhang YH, Lynd LR. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol Bioeng. 2004;88:797–824. doi: 10.1002/bit.20282. [DOI] [PubMed] [Google Scholar]
- 9.Bansal P, Hall M, Realff MJ, Lee JH, Bommarius AS. Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol Adv. 2009;27:833–848. doi: 10.1016/j.biotechadv.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Kostylev M, Wilson D. Synergistic interactions in cellulose hydrolysis. Biofuels. 2012;3:61–70. [Google Scholar]
- 11.Park S, Baker JO, Himmel ME, Parilla PA, Johnson DK. Cellulose crystallinity index: measurement techniques and their impact on interpreting cellulase performance. Biotechnol Biofuels. 2010;3:10. doi: 10.1186/1754-6834-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsson P, Wickholm K, Iversen T. A CP/MAS13C NMR investigation of molecular ordering in celluloses. Carbohydr Res. 1997;302:19–25. [Google Scholar]
- 13.Zhang S, Wolfgang DE, Wilson DB. Substrate heterogeneity causes the nonlinear kinetics of insoluble cellulose hydrolysis. Biotechnol Bioeng. 1999;66:35–41. doi: 10.1002/(sici)1097-0290(1999)66:1<35::aid-bit3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 14.Nidetzky B, Steiner W. A new approach for modeling cellulase-cellulose adsorption and the kinetics of the enzymatic hydrolysis of microcrystalline cellulose. Biotechnol Bioeng. 1993;42:469–479. doi: 10.1002/bit.260420410. [DOI] [PubMed] [Google Scholar]
- 15.Yang B, Willies DM, Wyman CE. Changes in the enzymatic hydrolysis rate of Avicel cellulose with conversion. Biotechnol Bioeng. 2006;94:1122–1128. doi: 10.1002/bit.20942. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson T, Karlsson J, Tjerneld F. A Model Explaining Declining Rate in Hydrolysis of Lignocellulose Substrates with Cellobiohydrolase I (Cel7A) and Endoglucanase I (Cel7B) of Trichoderma reesei. Appl Biochem Biotechnol. 2002;101:41–60. doi: 10.1385/abab:101:1:41. [DOI] [PubMed] [Google Scholar]
- 17.Kopelman R. Fractal Reaction-Kinetics. Science. 1988;241:1620–1626. doi: 10.1126/science.241.4873.1620. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YH, Cui J, Lynd LR, Kuang LR. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules. 2006;7:644–648. doi: 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 19.Irwin DC, Spezio M, Walker LP, Wilson DB. Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 20.Irwin D, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J Bacteriol. 1998;180:1709–1714. doi: 10.1128/jb.180.7.1709-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irwin D, Leathers TD, Greene RV, Wilson DB. Corn fiber hydrolysis by Thermobifida fusca extracellular enzymes. Appl Microbiol Biotechnol. 2003;61:352–358. doi: 10.1007/s00253-002-1210-6. [DOI] [PubMed] [Google Scholar]
- 22.Walker LP, Wilson DB, Irwin DC, Mcquire C, Price M. Fragmentation of Cellulose by the Major Thermomonospora Fusca Cellulases, Trichoderma Rees ei Cbhi, and Their Mixtures. Biotechnol Bioeng. 1992;40:1019–1026. doi: 10.1002/bit.260400905. [DOI] [PubMed] [Google Scholar]
- 23.Praestgaard E, Elmerdahl J, Murphy L, Nymand S, McFarland KC, Borch K, Westh P. A kinetic model for the burst phase of processive cellulases. Febs J. 2011;278:1547–1560. doi: 10.1111/j.1742-4658.2011.08078.x. [DOI] [PubMed] [Google Scholar]
- 24.Valjamae P, Kipper K, Pettersson G, Johansson G. Synergistic cellulose hydrolysis can be described in terms of fractal-like kinetics. Biotechnol Bioeng. 2003;84:254–257. doi: 10.1002/bit.10775. [DOI] [PubMed] [Google Scholar]
- 25.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose Dehydrogenase and a Copper-Dependent Polysaccharide Monooxygenase Potentiate Cellulose Degradation by Neurospora crassa. ACS Chem Biol. 2011 doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 26.Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser F, Irwin D, Chen S, Wilson DB. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol Bioeng. 2008;100:1066–1077. doi: 10.1002/bit.21856. [DOI] [PubMed] [Google Scholar]
- 28.Kostylev M, Moran-Mirabal JM, Walker LP, Wilson DB. Determination of the molecular states of the processive endocellulase Thermobifida fusca Cel9A during crystalline cellulose depolymerization. Biotechnol Bioeng. 2011 doi: 10.1002/bit.23299. [DOI] [PubMed] [Google Scholar]
- 29.Reverbel-Leroy C, Parsiegla G, Moreau V, Juy M, Tardif C, Driguez H, Belaich JP, Haser R. Crystallization of the catalytic domain of Clostridium cellulolyticum CeLF cellulase in the presence of a newly synthesized cellulase inhibitor. Acta Crystallogr D Biol Crystallogr. 1998;54:114–118. doi: 10.1107/s090744499700797x. [DOI] [PubMed] [Google Scholar]
- 30.Koivula A, Kinnari T, Harjunpaa V, Ruohonen L, Teleman A, Drakenberg T, Rouvinen J, Jones TA, Teeri TT. Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 1998;429:341–346. doi: 10.1016/s0014-5793(98)00596-1. [DOI] [PubMed] [Google Scholar]
- 31.Horn SJ, Sikorski P, Cederkvist JB, Vaaje-Kolstad G, Sorlie M, Synstad B, Vriend G, Varum KM, Eijsink VG. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc Natl Acad Sci U S A. 2006;103:18089–18094. doi: 10.1073/pnas.0608909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Wilson DB. Surface residue mutations which change the substrate specificity of Thermomonospora fusca endoglucanase E2. J Biotechnol. 1997;57:101–113. doi: 10.1016/s0168-1656(97)00093-x. [DOI] [PubMed] [Google Scholar]
- 33.Vuong TV, Wilson DB. Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl Environ Microbiol. 2009;75:6655–6661. doi: 10.1128/AEM.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irwin DC, Zhang S, Wilson DB. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur J Biochem. 2000;267:4988–4997. doi: 10.1046/j.1432-1327.2000.01546.x. [DOI] [PubMed] [Google Scholar]
- 35.Din N, Gilkes NR, Tekant B, Miller RC, Warren AJ, Kilburn DG. Non-Hydrolytic Disruption of Cellulose Fibers by the Binding Domain of a Bacterial Cellulase. Bio-Technol. 1991;9:1096–1099. [Google Scholar]
- 36.Teeri T, Reinikainen T, Ruohonen L, Jones TA, Knowles JKC. Domain Function in Trichoderma-Reesei Cellobiohydrolases. J Biotechnol. 1992;24:169–176. [Google Scholar]
- 37.Esteghlalian AR, Srivastava V, Gilkes NR, Kilburn DG, Warren RA, Saddle JN. Do cellulose binding domains increase substrate accessibility? Appl Biochem Biotechnol. 2001;91–93:575–592. doi: 10.1385/abab:91-93:1-9:575. [DOI] [PubMed] [Google Scholar]
- 38.Valjamae P, Sild V, Nutt A, Pettersson G, Johansson G. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur J Biochem. 1999;266:327–334. doi: 10.1046/j.1432-1327.1999.00853.x. [DOI] [PubMed] [Google Scholar]
- 39.Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttila M, Ando T, Samejima M. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science. 2011;333:1279–1282. doi: 10.1126/science.1208386. [DOI] [PubMed] [Google Scholar]
- 40.Forsberg Z, Vaaje-Kolstad G, Westereng B, Bunaes AC, Stenstrom Y, Mackenzie A, Sorlie M, Horn SJ, Eijsink VG. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20:1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohmine K, Ooshima H, Harano Y. Kinetic study on enzymatic hydrolysis of cellulose by cellulose from Trichoderma viride . Biotechnol Bioeng. 1983;25:2041–2053. doi: 10.1002/bit.260250813. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, Irwin DC, Escovar-Kousen J, Wilson DB. Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry. 2004;43:9655–9663. doi: 10.1021/bi049394n. [DOI] [PubMed] [Google Scholar]