Abstract
Nontypeable Haemophilus influenzae (NTHi) is a bacterium that resides within the human pharynx. Because NTHi is human-restricted, its long-term survival is dependent upon its ability to successfully colonize new hosts. Adherence to host epithelium, mediated by bacterial adhesins, is one of the first steps in NTHi colonization. NTHi express several adhesins, including the high molecular weight (HMW) adhesins that mediate attachment to the respiratory epithelium where they interact with the host immune system to elicit a strong humoral response. hmwA, which encodes the HMW adhesin, undergoes phase variation mediated by 7-base pair tandem repeats located within its promoter region. Repeat number affects both hmwA transcription and HMW-adhesin production such that as the number of repeats increases, adhesin production decreases. Cells expressing large amounts of HMW adhesins may be critical for the establishment and maintenance of NTHi colonization, but they might also incur greater fitness costs when faced with an adhesin-specific antibody-mediated immune response. We hypothesized that the occurrence of large deletion events within the hmwA repeat region allows NTHi cells to maintain adherence in the presence of antibody-mediated immunity. To study this, we developed a mathematical model, incorporating hmwA phase variation and antibody-mediated immunity, to explore the trade-off between bacterial adherence and immune evasion. The model predicts that antibody levels and avidity, catastrophic loss rates, and population carrying capacity all significantly affected numbers of adherent NTHi cells within a host. These results suggest that the occurrence of large, yet rare, deletion events allows for stable maintenance of a small population of adherent cells in spite of HMW adhesin specific antibody-mediated immunity. These adherent subpopulations may be important for sustaining colonization and/or maintaining transmission.
Keywords: Acute otitis media (AOM), Bacterial adherence, Bacterial colonization, Bacterial transmission, Chronic obstructive pulmonary disease, (COPD)
1. Introduction
Haemophilus influenzae is a Gram-negative coccobacillus that commonly resides within the human pharynx as a commensal and a potential pathogen. Non-encapsulated H. influenzae strains, which are commonly referred to as nontypeable H. influenzae (NTHi), are generally associated with localized infections of the respiratory tract such as pneumonia, sinusitis, and acute otitis media (AOM). AOM is a common childhood disease and in the United States approximately 83% of children have had at least one episode of AOM by the age of three and 45% have suffered three or more AOM episodes (Teele et al., 1989). In adults, NTHi strains are commonly associated with acute exacerbations in patients suffering from chronic obstructive pulmonary disease (COPD) (Garcha et al., 2012; Perotin et al., 2013; Sethi et al., 2002). Both acute exacerbations in COPD patients and AOM often result in antibiotic prescriptions (Lindenauer et al., 2006; Plasschaert et al., 2006). Thus, reducing incidence of NTHi-associated diseases can reduce a significant burden on the healthcare system, antibiotic usage and associated concerns regarding emerging antibiotic resistance.
NTHi strains are spread from person to person via infected respiratory droplets where they establish pharyngeal colonization, with prevalence ranging between 25% and 84% (Bou et al., 2000; Faden et al., 1995; Harabuchi et al., 1994). Since pathogenic NTHi arise from the community of NTHi strains colonizing healthy individuals, colonization marks one of the first steps of NTHi pathogenesis. Consequently, interventions that reduce, or prevent, colonization could potentially decrease the burden of NTHi disease.
Adhesin-mediated attachment to the host epithelium may play a critical role during the early stages of colonization, allowing newly transmitted NTHi cells to overcome host mucociliary clearance mechanisms. Thus, adhesins may confer a fitness advantage, increasing the probability of colonization following transmission. However, adhesins also tend to be antigenic, for example, NTHi colonization stimulates IgG, IgM, and IgA production that specifically targets surface localized adhesins (Barenkamp and Bodor 1990; Pichichero et al., 2010; Barenkamp 1986; Gnehm et al., 1985; Karasic et al., 1985). Thus, as the immunological environment changes, an adhesin that confers a fitness advantage during the early stages of colonization in a naïve host may become a liability when faced with an antibody-mediated immune response.
NTHi adherence to the host epithelium is mediated, in part, by the non-pilin high molecular weight (HMW) adhesins (St. Geme, 1993, 1994), which are present in approximately 40–75% of all NTHi isolates (Barenkamp and Leininger, 1992; Ecevit et al., 2004; Erwin et al., 2005; Erwin et al., 2008; St. Geme et al., 1998; van Schilfgaarde et al., 2000). Functional HMW adhesins are encoded by hmwA (Barenkamp and Leininger, 1992; Dawid et al., 1999), which displays extensive genetic diversity within and between isolates (Dawid et al., 2001; Giufre et al., 2006; Buscher et al., 2004). HMW adhesin amino acid diversity helps to define the tissue tropism of a particular strain (St. Geme et al., 1993; Buscher et al., 2004) and, importantly, generates antigenic diversity (Barenkamp and Bodor, 1990; van Schilfgaarde et al., 2000).
HMW-adhesin expression is phase variable. Tandem arrays of heptanucleotide simple sequence repeats (SSRs) located within the hmwA promoter region form the basis of HMW-adhesin phase variation (Barenkamp and Leininger, 1992; Dawid et al., 1999). During DNA replication, SSRs can be gained or lost by slipped-strand mispairing (Dawid et al., 1999) these changes are reversible and accumulate in a stochastic manner, independent of any external selective pressures. The number of repeats affects both hmwA transcription and translation—as repeat number increases, hmwA-transcript levels, and HMW-adhesin levels, decrease in a graded fashion (Dawid et al., 1999) (Fig. 1a). Ultimately this translates to population-level phenotypic diversity that may be critical for bacterial survival in fluctuating environments as might be experienced, for example, during a transmission event or as a consequence of host immunity.
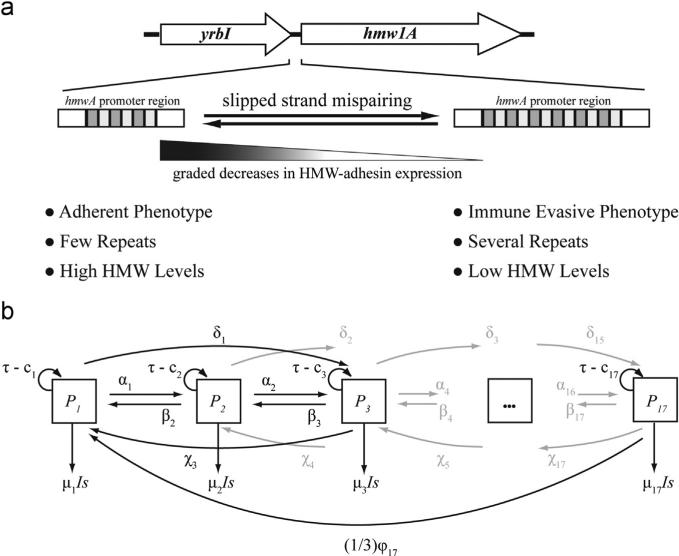
Fig. 1.
(a) The HMW adhesin phase variation mechanism illustrated with NTHi 86028-NP hmw1A. Phase variation is mediated by the gain and loss of heptanucleotide repeats (shaded rectangles), located within the hmw1A promoter region between yrbI and hmw1A that are gained or lost during DNA replication by slipped-strand mispairing (Dawid et al., 1999). Differences in repeat number affect hmwA transcription, and HMW-adhesin levels, in a graded fashion such that fewer repeats are associated with increased hmwA transcription and HMW-adhesin production (Dawid et al., 1999). Phenotypically, cells can be characterized as “adherent” cells or “immune evasive” cells. (b) Schematic representation of the within-host NTHi population model. The total population was divided into 17 subpopulations, P1 to P17, designated by boxes. Each NTHi subpopulation is defined by the exact number of repeats located within the hmwA promoter. Arrows represent the rates at which NTHi cells transition between subpopulations or are cleared from the system; model equations, parameter names, and their baseline values are provided in the Appendix.
A NTHi population within a host is thus composed of individual subpopulations that possess varying numbers of hmwA SSR repeats and, therefore, produce varying amounts of surface-expressed HMW adhesins. Since the amount of HMW adhesin that a cell produces is correlated with its adherence capacity and its immunogenicity (Barenkamp and Bodor, 1990; Cholon et al., 2008; Barenkamp, 1996), individual NTHi cells can be arranged along a spectrum of phenotypes ranging from “adherent” to “immune evasive”, corresponding to high or low amounts of HMW adhesin (Fig. 1a). At one end of spectrum, adherent cells expressing high HMW-adhesin levels might be important for transmission or for maintaining colonization. At the other end, cells displaying low amounts of HMW adhesin, and thus potentially capable of evading antibody-mediated killing, may be favored during colonization. The distribution of NTHi cells across the spectrum of HMW-adhesin levels likely reflects an evolutionary tradeoff between the population's ability to transmit between hosts versus its ability to maintain colonization despite antibody-mediated immunity. Thus, an open question centers on how a colonizing NTHi population retains HMW-adhesin mediated adherence, when within-host selective pressures favor a non-adherent, immune evasive population.
We hypothesize that the occurrence of large, yet rare, deletions within the repetitive DNA region that controls hmwA transcription allows for the maintenance of a population of adherent NTHi cells even when the population is faced with a robust antibody-mediated host immune response. We explore this hypothesis by developing a simple mathematical model describing a within-host NTHi population and an antibody mediated immune response of a naïve host. Our conclusions suggest that this hypothesis can indeed explain the spectrum of phenotypes and their role in survival within the human host.
2. The model
Our goal was to represent NTHi within-host population dynamics. We divided the total NTHi population into 17 subpopulations, each defined by the exact number of 7-bp SSRs within the hmwA promoter (Fig. 1b). We chose to incorporate 17 sub-populations because that captures the range in repeat numbers, i.e., 12 to 28 repeats, commonly observed in population-level studies (van Schilfgaarde et al., 2000; Giufre et al., 2006; Dawid et al., 1999; Barenkamp, 1992; Ecevit et al., 2005; Giufre et al., 2008) each subpopulation was described by a single ordinary differential equation (ODE). We assume that changes in repeat number affect HMW-adhesin expression in a graded fashion, as repeats are gained the amount of surface-exposed HMW adhesin decreases in a stepwise fashion (Dawid et al., 1999).
2.1. Bacterial sub-populations and parameterization
We used SSR number as a proxy for the amount of surface-expressed HMW adhesin and assumed that repeat number was proportional to a bacterial cell's ability to adhere to the host epithelium. We defined cells as either “adherent” (i.e., subpopulations P1 to P3 encoding few repeats and producing high levels of HMW adhesin) or “immune evasive” (i.e., subpopulations P15 to P17 encoding several repeats and producing low levels of HMW adhesin); importantly, while we emphasize the extremes of the phenotypic spectrum, there exists a continuum of decreasing HMW-adhesin levels as SSR repeat numbers increase from subpopulations P1 to P17 (Fig. 1a). Furthermore, we assume that each bacterial cell possessed a single hmw locus. Below we present a representative equation, the full set of equations is provided in the Appendix.
Rate of change in NTHi Subpopulation 1 (P1)
| (1) |
Eq. 1 describes the rate of change in the NTHi subpopulation (P1) of bacterial cells that encode 12 SSRs in their hmwA promoter region. Several of the model parameters are subpopulation-specific, designated by subscripts. The first term in Eq. 1 represents a logistic growth term. We assume that the effective growth rate of P1 cells is a function of the effective replication rate, τ, and the rate that cells are cleared from the pharynx by the host mucociliary clearance mechanism, c1. NTHi effective replication rate, τ, is a function of bacterial replication rates and subpopulation-independent death rates. We assumed that SSR number does not affect NTHi replication rate; thus, the same replication rate was used for each subpopulation. We divided bacterial death rates into subpopulation-independent and subpopulation-dependent mechanisms; for example, innate immunity would confer subpopulation-independent killing whereas targeted killing of NTHi mediated by HMW-adhesin specific antibodies was considered subpopulation-dependent. We incorporated mucociliary clearance into our model by specifying subpopulation-specific clearance rates, ci (where i = subpopulation 1 to 17), ci<ci+1 (Appendix). The subpopulation-specific clearance rate (ci) was subtracted from the effective replication rate (τ) to yield subpopulation-specific growth rates, (τ–ci). Finally, we constrained the maximal NTHi population size (i.e., sum of all subpopulations) within a single host by the pharyngeal carrying capacity, K. This is defined by availability of nutrients, adherence locations, etc. N represented the total population size.
2.2. Mutational events
During DNA replication the number of SSRs in the hmwA promoter region can remain unchanged, or a mutational event can occur and one or more repeats can be gained or lost via slipped-strand mispairing (Dawid et al., 1999). Following a mutational event, resultant daughter cells move into a new subpopulation defined by their SSR number. Phase variation rates for hmwA-associated 7-bp repeats have not been determined. However, there are empirical data for phase variation rates for the NTHi mod gene, which are mediated by tetranucleotide SSRs (De Bolle et al., 2000)2. Analysis of mod phase variation reveals that: (1) the most common type of event is the gain or loss of a single repeat, (2) repeats are more likely lost than gained, and (3) mutation rates increase as the number of SSRs increases (De Bolle et al., 2000). Importantly, De Bolle et al. (2000) documented two large deletion events in strains with long SSR tracts. Since the number of SSRs in the hmwA promoter affects HMW adhesin expression, the simultaneous deletion of several SSRs provides a mechanism for the generation of adherent daughter cells from immune evasive parental-cells. We refer to the simultaneous loss of several repeats as a “catastrophic loss” event.
Based on the above study by De Bolle et al. (2000), we defined five different phase variation events in our model, each with a unique rate. During DNA replication, daughter cells transition out of sub-population P1 by gaining one SSR, at a rate equal to α1, or two SSRs, at a rate equal to δ1; these events are represented by loss terms in Eq. (1). Likewise, daughter cells can transit into subpopulation P1 as a result of P2 cells losing one SSR, at a rate equal to β2, or P3 cells losing two SSRs, at a rate equal to χ3. Cells with extended SSR tracts (i.e., subpopulation P15, P16, and P17) experience larger deletion events (i.e., catastrophic losses) during replication at rates of φ15, φ16, and φ17; we assume that one third of the daughter cells resulting from catastrophic losses enter subpopulation P1.
2.3. Immune response
In addition to the subpopulation-independent mechanisms captured by the effective growth rate term, cells can also be cleared by antibody-mediated immunity. HMW adhesins stimulate a host-mediated bactericidal antibody response (Barenkamp and Bodor, 1990). This imposes a fitness cost on HMW adhesin-producing cells such that cells displaying greater amounts of adhesin are killed at a higher rate than are cells displaying less adhesin. Empirical studies demonstrate that hmwA SSR number is inversely proportional to HMW-adhesin production (Dawid et al., 1999), thus we used SSR number as a proxy for the amount of surface expressed HMW adhesin. The exact relationship between repeat number and hmwA transcript is μi = 333.79 e 0.387(repeat number)
Next, we defined the scaling factor s, which describes the relationship between HMW-adhesin levels (μ1) and the rate of antibody-mediated NTHi killing; s is analogous to antibody avidity. Finally, we incorporated an immune response, I, that captured the dynamics of a primary antibody response in a naïve host.
Rate of change in the immune response, I(t)
| (2) |
The growth rate of the antibody response was chosen to capture the dynamics of a primary antibody response in a naive host, Fig. A.1(Janeway et al., 2005) and is mathematically similar to what was done previously for a H. pylori model of immunity (Kirschner and Blaser, 1995). The intrinsic growth rate for the immune response is equal to r and Ab_max defines the maximal antibody level. We based the value of Ab_max on data from Pichichero et al. (2010) in which serum concentrations of IgG directed against the NTHi outer membrane protein D were quantified in healthy children colonized with NTHi. The immune response is autonomous, and is phenomenologically represented (Fig. A.1), and, while it affects the NTHi population, the NTHi population does not affect the immune response in this simplified representation. Simulations were initiated with a negligible immune response of 0.001; this resulted in 0.001% of the total NTHi population being killed on day one.
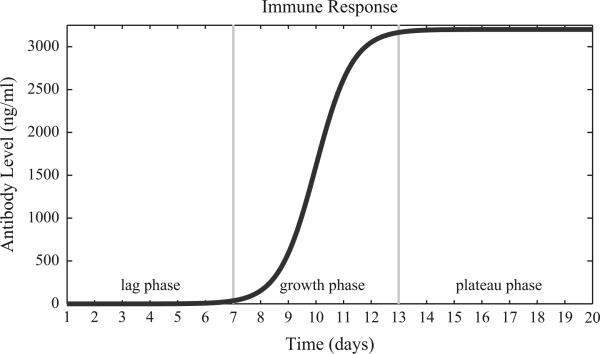
Fig. A1.
Antibody mediated immune response. This graph describes the development of antibody-mediated immune response over the course of a 20-day simulation. The maximal antibody level, defined as Ab_max, was 3200 ng/ml and was chosen based on a study of serum IgG levels against protein D in children colonized with NTHi (Pichichero et al., 2010). The immune response can be divided into three phases, the lag phase, growth phase, and plateau phase, which have been demarcated with the gray vertical lines. The immune response increased from approximately 1% of Ab_max on day 7 to 98.9% of Ab_max by day 13.
2.4. Parameter estimation and uncertainty and sensitivity analyses
Once the model was developed, baseline rates were estimated for each parameter. When available, rates estimates were taken from published literature. In the absence of published data, we employed uncertainty analysis (Marino et al., 2008) to define baseline parameter estimates that gave rise to biologically reasonable outputs. All parameter values, and details of how each parameter was estimated, are summarized in the Appendix.
Sensitivity analysis (SA) is a method for quantifying uncertainty in any type of complex model. The objective of SA is to identify critical inputs (parameters and initial conditions) of a model and to quantify how input uncertainty affects model outcome(s).
We use Latin Hypercube Sampling (LHS) as a sampling scheme and Partial Rank Correlation Coefficient (PRCC) as a sensitivity index. Uncertainty and sensitivity analysis methodologies are described in detail in Marino et al. (2008) and briefly below.
LHS is a so-called stratified sampling without replacement technique, where the random parameter distributions are divided into N equal probability intervals, which are then independently sampled. N represents the sample size, which here is set to 5000. A matrix of 5000 rows and k number of columns (k is equal to the number of parameters varied in the analysis) is generated. Each row of the matrix serves as input for a single model simulation. The outputs of N model simulations are then saved and used for calculating sensitivity indexes. Since our model is a dynamical system, 20-day time course outputs are generated. PRCC was then calculated for each parameter and statistical significance was assessed (Marino et al., 2008). Our outcome of interest for sensitivity analyses was the total number of adherent cells (Table A.1), defined as the sum of the number NTHi cells in subpopulations P1 to P3. In the sensitivity analysis, the catastrophic loss rates were varied independently of all other mutational events. This was achieved by varying the scaling factor q (Table A.1), which simultaneously varied the y-intercepts of the lines describing the relationship between repeat number and phase-variation rate. In this way, the relationships between mutation rates (i.e., βk>αi>χi>δm) were maintained but catastrophic loss rates (φj) could be varied independently.
Table 1.
Parameters, baseline parameter values, and parameter ranges for sensitivity analyses.
| Parameter | Parameter Description | Baseline Values | Sensitivity Analysis (min, max) | Ref. |
|---|---|---|---|---|
| α a | rate at which a single repeat is gained (mutations day-1 repeat-1) | 5.20E-04-1.12E-03 | [32] | |
| β a | rate at which a single repeat is lost (mutations day-1 repeat-1) | 1.12E-03 - 2.32E-03 | [32] | |
| δ a | rate at which two repeats are gained in a single (mutations day-1 repeat-1) | 1.04E-04 - 2.16E-04 | [32] | |
| χ a | rate at which two repeats are lost in a single event (mutations day-1 repeat-1) | 1.50E-04 - 2.90E-04 | [32] | |
| ψ | “catastrophic” loss rate, several repeats in a single event (mutations day-1 repeat-1) | 5.4E-05 - 5.8E-05 | (10-7,10-3) | [32] |
| μ | hmwA repeat-specific antibody mediated killing rate (day-1) | 0.007 - 1.0 | [26] | |
| τ | effective replication rate (generations day-1) | 19.96 | [52] | |
| intercept_cl | y-intercept of the line describing the linear relationship between repeat number and catastrophic loss rate | 2E-06 | (10-7,10-3) | this study |
| Ab_max | maximum HMW antibody (ng ml-1) | 3200 | (320, 5775) | [11] |
| K | NTHi carrying capacity | 109 | (105,1011) | |
| Ci | rate NTHi is lost due to mucociliary clearance (day- 1) | 0.85 | (0.01, 8.5) | [34] |
| s | scaling factor for the relationship between repeat number and antibody mediated killing rate; similar to antibody avidity | 0.00906 | (0.0023,0.0362) | this study |
| r | growth rate of immune response (ng ml-1 day-1) | 1.5 | this study | |
| q | scaling factor that maintains the relationship between the rate at which repeats are gained and lost during replication | 1 | (0.01, 100) | this study |
| Derived Quantities | ||||
| adherent cells | = sum of NTHi cells in subpopulations P1 to P3 | |||
| proportion of adherent cells | = (sum of NTHi cells in subpopulations P1 to P3)/ (total number of NTHi cells in subpopulations P1 to P17) |
The min and max values for sensitivity analysis were controlled by varying the scaling factor q.
2.5. Computer simulations
Once parameters were estimated, we solved the model and tracked NTHi population dynamics over time. All simulations were performed using MATLAB's (ver 7.10, R2010a, Copyright 1984–2010, The MathWorks, Inc.) ode113 solver for non-stiff differential equations. We chose to study colonization dynamics under the assumption that 120 adherent NTHi cells, distributed across the first three NTHi subpopulations, enter the pharynx; our model was flexible and stable to variations in initial conditions (not shown). We considered the total number of adherent bacterial cells (i.e., sum of cell numbers in subpopulations P1 to P3) as a key output measure for the system.
3. Results
We use our model that incorporates hmwA phase variation and antibody-mediated immunity, to explore the potential for large mutational events, i.e., catastrophic losses, to allow for the maintenance of adherent NTHi subpopulations during colonization. We first study these bacterial populations in the absence of immune pressure, and then under various scenarios regarding phase variation and catastrophic loss.
3.1. Phase variation and no immune pressure yields subpopulations of solely adherent phenotypes
In the absence of antibody-mediated immunity, the distribution of NTHi cells among subpopulations was solely a function of phase variation rates. Over the course of a 20-day simulation, the population distribution gradually shifted to the right (i.e., towards cells with increasing SSR numbers) with subpopulations P1 through P8 containing nonzero numbers of bacterial cells (Fig. 2a). Throughout the simulation, the majority (99.6%) of the population remained within the first three subpopulations (i.e., the adherent phenotype) (Fig. 2a). This followed because the mutation rate for subpopulation P1, which contained the fewest SSRs, was nonzero, so there were always NTHi subpopulations with longer repeat tracts. This is not surprising, as examining Eq. (1), one can see that if the immune response is zero, the resulting population distribution for P1 is directly a function of the other populations and their mutation rates, indicating that the most fit (highest growth rates) will win out. In the absence of immunity, catastrophic loss events did not impact the overall NTHi population distribution during the 20-day simulation.
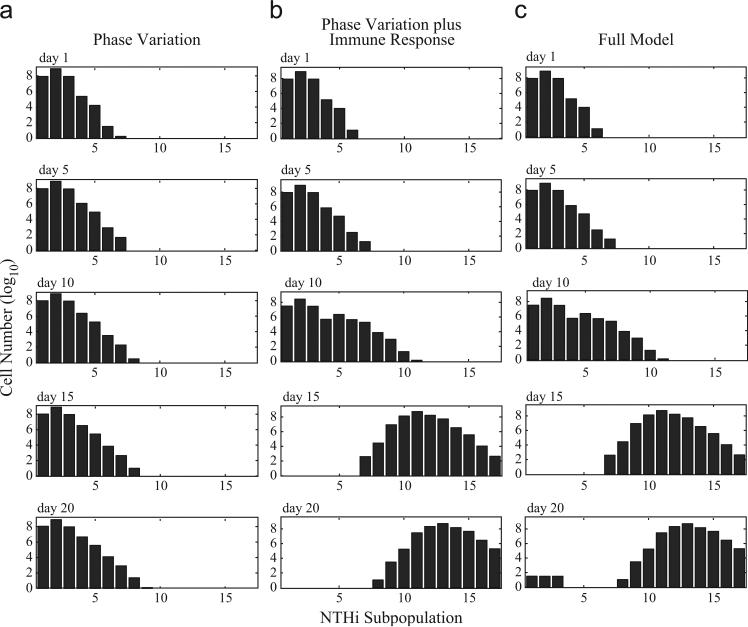
Fig. 2.
Cell numbers of each NTHi subpopulation on days 1, 5, 10, 15, and 20. Each graph represents a specific day during the simulation. Within a graph, each bar represents the number of NTHi cells (log10) in each of the 17 subpopulations as defined by repeat number. (a) Population distribution of NTHi as a result of phase variation alone. (b) Effects of phase variation and immune selection on NTHi population distribution. The immune response imposed a selective pressure that drove the overall NTHi within-host population distribution towards subpopulations with increased repeat numbers (and lower HMW-adhesin production). (c) Full model, including phase variation, immunity, and catastrophic loss events, under baseline conditions (Table A.1). Catastrophic losses allowed for the recovery of a small population of adherent NTHi cells (i.e., those in subpopulations P1 to P3) by day 20.
3.2. Phase variation plus immune pressure yields subpopulations of solely immune-evasive phenotypes
Next we explored how the antibody-mediated immune response (Fig. A.1) affects the NTHi population distribution in the presence of phase variation. The immune response had little impact on population distribution until day ten, at which time the total number of adherent cells dropped from approximately 104 on day 11 to less than a single cell on day 12 (Fig. 2b). As the immune response develops, immune-mediated selective pressure forced the overall NTHi population distribution toward subpopulations with increased repeat number and decreased HMW-adhesin levels (Fig. 2b). In the absence of catastrophic losses, adherent cells were completely eliminated from the population by day 13 (Fig. 2b).
3.3. Catastrophic loss yields subpopulations of both immune-evasive and adherent phenotypes
The inclusion of catastrophic loss events had no effect on overall population distribution through day 16 of our simulation (Fig. 2c). However, a small population of adherent cells was established by day 17 and it increased in size each day until the end of the 20-day simulation (Fig. 2c). This supports the idea that catastrophic losses play a role in the recovery and maintenance of adherent cells. To explore this further, we performed a series of additional simulations. First, we initiated a simulation with 108 cells in subpopulation P17, no catastrophic losses, and no immune response. Under these conditions, there was no immune-mediated selective pressure and mutation rates were maximal because subpopulation P17 has the longest SSR tract. At the end of 20 days, only subpopulations P11 to P17 were populated and it took 690 days until there was a single cell in the adherent-subpopulation P1. Next, we performed a simulation starting with 108 cells in subpopulation P17 and added in a baseline immune response (Eq. (2)), but eliminated the possibility of catastrophic losses. Under these conditions, there were no cells in subpopulation P1 even after 10,000 days; the population was distributed among subpopulations P12 to P17. We believe that these simulations emphasize that catastrophic losses are critical for maintaining adherent NTHi subpopulations (i.e., those in subpopulations P1 to P3). Thus, under the conditions of our model, mutation alone was not sufficient to regain adherent cells (within a biologically-relevant time period) in the presence of an HMW-adhesin mediated immune response.
3.4. Identifying key mechanisms driving model output
To begin the analysis, we explore a simplified scenario where we focus on a single population with no mutations present and immunity at its maximal value, Ab_max. Suppose that we begin with only one population (assume P1) and all mutation rates are zero. If the immune response reaches its maximum value, that would yield,
if we examine the steadystate with I(t) = AB_max,it yields either P1 = (trial) or (with N(t) =P1(t)):
clearly, the parameters that comprise the steady state value are K, Ab_max, μ, c1, and τ. All of these are driving the steady state dynamics in the absence of mutations. In the presence of mutations and other bacterial populations, we turn to the sensitivity and uncertainly analysis to identify parameters that significantly impacted the number of adherent NTHi cells (parameter value ranges from Table A.1). Maximum antibody levels (Ab_max), population carrying capacity (K), catastrophic loss rates (intercept_cl) and antibody avidity (s) were each significantly correlated with the numbers of adherent cells (P<0.01) (Fig. 3). Interestingly, the impact of their effects varied over time and in magnitude. Pharyngeal carrying capacity, K, was strongly correlated with the number of adherent cells (Fig. 3). At all time points, there was a statistically significant (P<0.001) positive correlation between carrying capacity and the number of adherent cells.
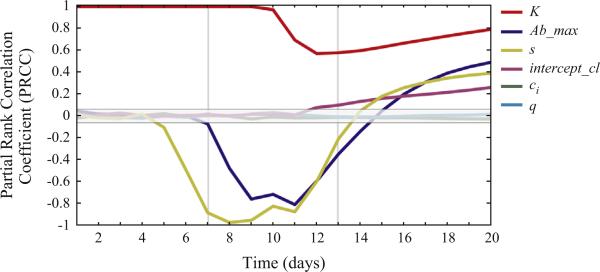
Fig. 3.
Partial rank correlation coefficients (PRCCs) plotted over time. Each line represents the PRCC between a parameter of interest (shown on the right axis) and the total number of adherent cells (i.e., sum of subpopulations P1 to P3) at every time point over the course of a 20-day simulation. Parameter estimates, and the ranges used for sensitivity analysis, are presented in Table A.1 of the Appendix. PRCC values outside of the central gray box, are statistically significant (P-value≤0.001) (equation 7 Marino et al. (2008)). Maximum antibody levels (Ab_max), antibody avidity (s), rate of catastrophic losses (intercept_cl), and pharyngeal carrying capacity (K) were each significantly correlated with the total number of adherent cells, but, the strength and direction of their effects varied over time. The three main phases of the corresponding immune response (Fig. A.1) are demarcated with vertical gray lines to indicate their dramatic effect on parameter dynamics.
The catastrophic loss rate (intercept_cl) also significantly impacted the numbers of adherent cells. The rate of catastrophic loss events was significantly positively correlated (P<0.001) with the number of adherent cells from days 12 to 20 (Fig. 3), confirming our hypothesis that catastrophic loss provides a mechanism allowing for maintenance of adherent cells despite antibody-mediated immunity. As the immune-mediated selective pressure forced the NTHi population to predominantly immune-evasive phenotypes, the PRCCs between the catastrophic loss rates and the numbers of adherent cells increased (Fig. 2b); the PRCC reached its maximal value of 0.26 on day 20 (Fig. 3). This suggests that increased catastrophic-loss rates results in increased numbers of adherent cells.
As in the steady state analysis, the sensitivity analysis also found that Ab_max (the maximum level of anti-HMW-adhesin antibodies produced in response to NTHi colonization) and numbers of adherent cells were strongly correlated (Fig. 3). Ab_max was negatively correlated (P<0.001) with the number of adherent cells from days 7 to 14 (Fig. 3). This time period corresponds to rapidly increasing antibody levels (Fig. A.1), immune-mediated elimination of adherent cells, and a shift in the total population distribution that favored subpopulations with increased repeat number (Fig. 2c). Beginning on day 12, subpopulations P15 to P17 became non-zero and on day 16, catastrophic loss events slowly repopulated the adherent subpopulations P1 to P3 (data not shown). By day 15, the relationship between Ab_max and the number of adherent cells became significantly positively correlated (P<0.001) and continued to increase through the end of the simulation at day 20 (Fig. 3).
Finally, our analysis predicts that s (which is analogous to antibody avidity) and the number of adherent cells were also strongly correlated (Fig. 3). Therefore, increasing s increases the per unit killing rate of bacterial cells by the immune response. The effect of s was evident by day five of the simulation at which time s and numbers of adherent cells was significantly negatively correlated (P<0.001) (Fig. 3). In general, the PRCCs for s tracked closely with those of Ab_max (Fig. 3). This was not surprising since both affect the relationship between the immune response and the rate that NTHi cells are cleared from the population.
4. Discussion
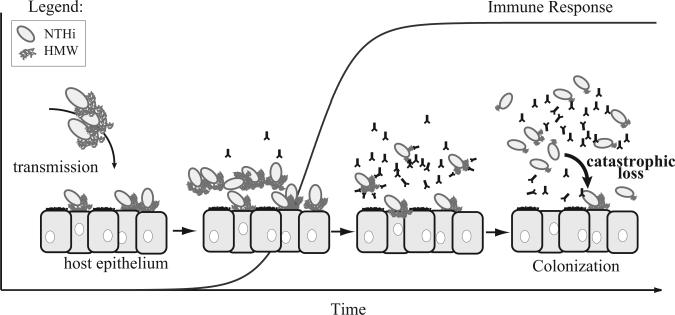
In this paper we presented a mathematical model that explores how phase variation and antibody-mediated immunity can shape the population structure of nontypeable Haemophilus influenzae within a host. Specifically, we focused on phase-variable expression of the HMW adhesin, a surface-exposed immunogenic protein that mediates attachment of NTHi to the respiratory epithelial cells of its host (Barenkamp and Bodor, 1990; St. Geme et al., 1993). Adherence is one of the first steps in bacterial colonization, and numerous studies demonstrate that HMW adhesins mediate the adherence of NTHi to epithelial cells in vitro (St. Geme et al., 1993; van Schilfgaarde et al., 2000; Cholon et al., 2008). Thus, we assumed that in an immunologically naïve host, the HMW adhesins would play a critical role during NTHi transmission and that cells expressing relatively high levels of HMW adhesins would have a fitness advantage over those with lower adhesin levels. However, because HMW-adhesins are highly immunogenic (Barenkamp and Bodor, 1990; van Schilfgaarde et al., 2000), selection pressure from antibody-mediated immunity—targeted to the HMW adhesins—would eventually favor NTHi cells expressing relatively low HMW adhesins levels (Fig. 1). Phase variable regulation of HMW-adhesin production, resulting from random changes in the number of SSRs number located within the promoter region of the adhesin gene hmwA, allows NTHi to balance the fitness costs associated with adherence on the one hand and immune evasion on the other (Fig. 1). We predict that the occurrence of large deletion events, while rare, are critical for the maintenance of adherent NTHi subpopulations in spite of strong selection pressure favoring non-adherent, immune evasive, cells. Furthermore, we speculate that adherent subpopulations may be important for maintaining colonization and/or sustaining NTHi transmission between hosts (Fig. 4).
Fig. 4.
Adherence to the host epithelium, mediated by bacterial adhesins such as the HMW adhesins, is a first step in NTHi colonization. As the NTHi population increases, HMW adhesins stimulate an antibody-mediated immune response in the host that imposes a selective pressure on the NTHi population, favoring “immune evasive” phenotypes. During DNA replication, the long hmwA SSR tracts of immune evasive cells can experience catastrophic losses, which produce “adherent” cells in a single mutation event thus allowing for the maintenance of a small yet stable population of adherent NTHi cells.
In the absence of HMW-adhesin phase variation, our results suggest that antibody-mediated immunity would eliminate an adherent NTHi population from the host (data not shown). However, phase variable changes in HMW-adhesin expression are capable of mitigating the selective pressure imposed by antibody-mediated immunity. The role of phase variation in mediating immune escape in vivo is supported by the results of a HMW-targeted NTHi vaccine study in which chinchillas were immunized with either HMW1 or HMW2 from NTHi strain 12 (Barenkamp, 1996). Following development of immunity, the chinchillas were challenged with an isogenic NTHi strain expressing both HMW adhesins (Barenkamp, 1996). Despite being immunized, approximately 60% of the challenge animals developed middle ear inflammation (Barenkamp, 1996). The NTHi strains recovered from these animals had increased numbers of SSRs within the hmwA promoter that corresponded to the specific HMW adhesin used for immunization; that is, NTHi recovered from HMW1-immunized animals had a greater number of SSRs upstream of hmw1A specifically, and this was correlated with reduced HMW1-adhesin production (Dawid et al., 1999; Barenkamp, 1996). In another study, genetic analysis of serial NTHi isolates, collected longitudinally, from the sputum of COPD patients revealed increases in SSR number, and decreases in HMW-adhesin levels, that were associated with anti-HMW antibody titers in the patient's serum (Cholon et al., 2008). Taken together, these results demonstrate that phase variable reductions in HMW-adhesin production contribute to NTHi's ability to escape antibody-mediated immunity.
In our simulations, phase variable expression of the HMW adhesin allowed the NTHi population to persist despite increasing antibody levels. Antibody-mediated selective pressure was greatest for cells expressing large amounts of HMW-adhesin (i.e., adherent cells) and decreased as HMW-adhesin levels declined (i.e., SSR number increased). As a consequence, antibody-mediated selective pressure forced the overall NTHi population distribution towards subpopulations with increased repeat number and decreased HMW-adhesin levels. By day 13 of the simulation, adherent cells were completely eliminated from the population. Since NTHi are human-restricted, their long-term survival is dependent upon their ability to colonize new hosts. Thus, if HMW adhesins play a critical role in transmission, then the elimination of adherent NTHi cells during colonization could reduce their ability to transmit, potentially driving the population to an evolutionary dead-end.
How can we use these results to make relevant predictions regarding NTHi colonization and, potentially, pathogenesis? Sensitivity analyses identified four parameters, Ab_max (maximal antibody level), s (antibody avidity), intercept_cl (the rate of catastrophic loss events), and K (carrying capacity), that significantly impacted NTHi population dynamics and were strongly correlated with the total number of adherent NTHi cells (Fig. 3). The effect of Ab_max and s, varied both in time and magnitude (Fig. 3). Both of these parameters directly affect the immune mediated killing rate, increasing Ab_max increases the amount of antibody present at any given time during the simulation whereas increasing s increases the per unit killing rate for a given antibody level. An effective vaccine could increase both of these parameters simultaneously and there is experimental evidence to suggest these immune parameters may act synergistically (Hetherington and Lepow, 1992).
Our model results suggest, however, that the impact of increasing Ab_max (maximal antibody level) on the number of adherent cells is complex. On the one hand, increasing Ab_max increases the rate at which adherent NTHi cells are eliminated from the host during the early days of colonization. On the other hand, bacterial populations subjected to increased immune pressure regained adherent cells earlier during the course of our simulation than did populations subjected to less intense immune selection (i.e., lower Ab_max). The earlier appearance of adherent cells resulted from increased selection pressure rapidly driving the NTHi population to the immune evasive, non-adherent phenotype from which adherent progeny were generated by catastrophic losses. This finding suggests that individuals with a more robust HMW-adhesin-specific immune response may be more likely to harbor adherent NTHi cells during the first 20 days of colonization. If, as we hypothesize, adherent cells are critical for NTHi transmission, then this result has two important, and counterintuitive, implications that warrant further exploration. First, it suggests that the production of highly immunogenic HMW adhesins may actually benefit HMW-adhesin-producing NTHi strains by increasing their fitness (i.e., transmission). Secondly, it suggests that a successful vaccine targeting HMW adhesins, via boosting of the host immune response, could potentially increase NTHi transmission.
Finally, our model revealed a positive correlation between pharyngeal carrying capacity, K, and the number of adherent cells (Fig. 3). This result is interesting because during otitis media, the pharyngeal NTHi community, which in healthy individuals commonly supports multiple different NTHi strains, is dominated by the disease-causing isolate that is simultaneously present in the middle ear (Berrens et al., 2007). This would have the same effect as increasing the carrying capacity in our model since a larger portion of the NTHi in the pharynx could be represented by a single strain, thus, allowing that strain to achieve a larger overall population size. The observation that HMW-adhesins are more prevalent among AOM than commensal isolates from the throats of healthy children suggests that they may play a role in NTHi-pathogenesis. However, mucociliary clearance mechanisms are impaired during AOM due to Eustachian tube obstruction, thus, HMW-adhesin mediated adherence would not be expected to confer a fitness advantage in the middle ear. If HMW-adhesin mediated adherence is critical for NTHi transmission, and disease results in a larger population size, then our finding provides a potential link between a NTHi strain's ability to cause AOM and its transmissibility. Furthermore, this result suggests that therapeutic agents, or preventive measures, that reduce pharyngeal carrying capacity could potentially eliminate adherent cells and thereby reduce NTHi transmission and disease.
NTHi adherence is a multifaceted process mediated by both pilin and non-pilin adhesins. Several studies suggest that HMW adhesins play a significant role in adherence (St. Geme, 1994, 1993; Dawid et al., 2001; Cholon et al., 2008). However, NTHi possesses a variety of cell-surface structures with a demonstrated role in adherence (Carruthers et al., 2012; Jurcisek et al., 2007; Kubiet et al., 2000; Krasan et al., 1999; van Ham et al., 1993; Meng et al., 2011; Fink et al., 2002; St. Geme et al., 1994; Hendrixson and St Geme, 1998; Reddy et al., 1996; Avadhanula et al., 2006). Thus, despite strong selection against HMW adhesins, multiple different adhesins and processes (e.g., biofilm formation) ultimately contribute to NTHi adherence and colonization.
Recently, Palmer et al. (2013), simulated phase variable changes in a bacterial population under varying selective pressure (s) and duration of time in each of two environments (T), where each individual cell encodes a single SSR locus that controls the expression of a selectable phenotype with two states (on or off). Similar to our analysis, mutation rates varied depending on the length of the SSR tract and were based on H. influenzae mod gene tetranucleotide SSR mutation rates (De Bolle et al., 2000). Their analysis revealed that on-off phase variation is favored when the product of the selection coefficient (s) and the duration of time spent in each environment (T) is large enough that the favored phenotype reaches fixation before an environmental switch. Simple sequence phase variation rates were also favored when s × T was low, but equivalent, in both environments. Rather than on–off phase variation, the HMW adhesins display a spectrum of phenotypes resulting from graded changes in adhesin production. Furthermore, instead of abrupt changes between two environments (e.g., as might be experienced during a transmission event), we considered an increasing selective pressure, consistent with the evolution of a primary antibody response of an immunologically naïve host. Thus, our analysis contributes to our understanding of how a population of colonizing NTHi can maintain subpopulations expressing high levels of HMW-adhesin in an immunologically hostile environment.
Acknowledgments
This work was supported with funding from the Interdisciplinary Program in Infectious Diseases (TA32A1049816), the Molecular Mechanisms of Microbial Pathogenesis Training Program (AI007528), and the University of Michigan, School of Public Health, Department of Epidemiology. All authors report no conflicts of interest. This research was additionally supported by the following grants: NIH R01 HL106804, R01 EB012579, and R01 HL 110811 (all 3 awarded to DEK).
Appendix A. Parameter estimation
NTHi effective growth rate
The baseline NTHi effective growth rate of 19.96 day 1 was based upon an “effective mean generation time” of 50 min (Moxon, 1992). NTHi subpopulations with low HMW adhesin levels, were assumed to incur a fitness cost in the form of an increased mucociliary clearance rate (Buscher et al., 2004; Cholon et al., 2008; Farley et al., 1986; Bailey et al., 2012). The fitness advantage associated with adherence was incorporated into the effective growth rate term as a subpopulation-specific mucociliary clearance rate (ci) that was subtracted from the effective replication rate, τ. Clearance rates increased linearly as repeat numbers increased. The maximal clearance rate, c17, of 0.85 day 1 was based upon estimates from a model of Helicobacter pylori dynamics (Kirschner and Blaser, 1995).
Phase variation rates
Based on the on-off switching rate for the tetranucleotide phase variation system described by De Bolle et al. (2000), we defined five different hmwA phase variation events in our model: single repeat gain or loss at rates αi and βk, respectively, where i designates subpopulations P1 to P16 and k designates subpopulations P2 to P17; two repeat gain or loss in a single event with rates δm and χw, respectively, where m designates subpopulations P1 to P15 and w designates subpopulations P3 to P17. Finally, bacterial cells with the largest number of SSRs could experience catastrophic losses, that is, lose several repeats in a single mutation event, during DNA replication at rate φj, where j = subpopulation P15, LP16, or P17.
The relationship between repeat number and phase variation rate, measured as mutations repeat 1day 1 was described by the linear function: mutation rate=(7 × 10 6)(SSR number) – (7 × 10 5). To determine the rate for each type of mutational event, we first estimated the proportion of each event using Table 2 of De Bolle et al. (2000), next we multiplied that proportion by the SSR-specific mutation rate, and then transformed the rates to reflect the mutation rate repeat 1day 1. Consistent with the results of De Bolle et al. (2000), for each event type, the rate at which repeats were lost was assumed to be greater than the rate at which they were gained and the mutation rate increased as the number of SSRs increased. Thus, SSR number determined the level of HMW-adhesin expression and the phase variation rate. We assumed that, βk<αi<χw<δm<φj when k=i=w=m=j.
Relationship between HMW adhesin levels, 7-bp repeat number, and antibody mediated killing rates
HMW adhesin levels and hmwA transcription decrease in a graded fashion as repeat number increases (Dawid et al., 1999; Cholon et al., 2008; Giufre et al., 2008). The relationship between SSR number and hmwA transcription was estimated by fitting a curve to published data quantifying the relationship between SSR number and hmwA transcription (Dawid et al., 1999). There were no published data relating cells containing 12 to 14 SSRs with hmwA transcription; however, strain with 6 SSRs generated the same amount of hmwA transcript as did a strain possessing 15 repeats (Dawid, unpublished data). Thus, we assumed that the relationship between repeat number and HMW adhesin level was the same for cells encoding 12 to 15 repeats; that is, there was a threshold affect beyond which decreasing the SSR number did not further increase hmwA transcription. HMW adhesin levels for cells containing between 23 and 28 repeats were estimated from the curve fitted to the empirical data. The relationship between hmwA transcript level, μ, and repeat number is μi = 333.79 e–0.387repeat number) for repeat tracts encoding 15 to 28 SSRs.
Immune response parameters Ab_max and r
The growth rate of the antibody response (Fig. A.1) was chosen to capture the dynamics of a primary antibody response in a naive host (Janeway et al., 2005). The Ab_max value of 3200 ng IgG ml –1 was based upon a study of serum IgG levels directed against NTHi outer membrane protein D during colonization (Pichichero et al., 2010).
Scaling factors
Uncertainty analysis was used to select the parameter estimate for s=0.00906 which was chosen so that approximately 1.0% of the total NTHi population was killed at day 5. Our model was flexible to variations in this number.
Appendix B. Model equations
Subpopulation 1 (12 SSRs)
Subpopulation 2 (13 SSRs)
Subpopulation 3(14 SSRs)
Subpopulation 4 (15 SSRs)
Subpopulation 5(16 SSRs)
Subpopulation 6(17 SSRs)
Subpopulation 7 (18 SSRs)
Subpopulation 8 (19 SSRs)
Subpopulation 9 (20 SSRs)
Subpopulation 10(21 SSRs)
Subpopulation 11 (22 SSRs)
Subpopulation 12 (23 SSRs)
Subpopulation 13 (24 SSRs)
Subpopulation 14 (25 SSRs)
Subpopulation 15 (26 SSRs)
Subpopulation 16(27 SSRs)
Subpopulation 17 (28 SSRs)
Immune response
Footnotes
This assumption may not hold, for example, differences in the rate of DNA repair for tetranucleotide and heptanucleotide repeats could bias our rate estimates. However, phase variation rates of both tetranucleotide repeats and hmwA heptanucleotide repeats are recA-independent (Dawid et al., 1999; Carruthers et al., 2012). Thus, while it is possible that tetranucleotide and heptanucleotide repair mechanisms differ, we believe that any such differences would have little impact our model outcome based on our analysis.
References
- Avadhanula V, et al. Nontypeable Haemophilus influenzae adheres to intercellular adhesion molecule 1 (ICAM-1) on respiratory epithelial cells and upregulates ICAM-1 expression. Infect. Immun. 2006;74(2):830–838. doi: 10.1128/IAI.74.2.830-838.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KL, et al. Non-typeable Haemophilus influenzae decreases cilia beating via protein kinase C epsilon. Respir. Res. 2012;13(1):49. doi: 10.1186/1465-9921-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp SJ. Protection by serum antibodies in experimental nontypable Haemophilus influenzae otitis media. Infect. Immun. 1986;52(2):572–578. doi: 10.1128/iai.52.2.572-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp SJ. Outer membrane proteins and lipopolysaccharides of nontypeable Haemophilus influenzae. J. Infect. Dis. 1992;165(Suppl 1):S181–S184. doi: 10.1093/infdis/165-supplement_1-s181. [DOI] [PubMed] [Google Scholar]
- Barenkamp SJ. Immunization with high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae modifies experimental otitis media in chinchillas. Infect. Immun. 1996;64(4):1246–1251. doi: 10.1128/iai.64.4.1246-1251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp SJ, Bodor FF. Development of serum bactericidal activity following nontypable Haemophilus influenzae acute otitis media. Pediatr. Infect. Dis. J. 1990;9(5):333–339. doi: 10.1097/00006454-199005000-00006. [DOI] [PubMed] [Google Scholar]
- Barenkamp SJ, Leininger E. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 1992;60(4):1302–1313. doi: 10.1128/iai.60.4.1302-1313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrens ZJ, et al. Genetic diversity of paired middle-ear and pharyngeal nontypeable Haemophilus influenzae isolates from children with acute otitis media. J. Clin. Microbiol. 2007;45(11):3764–3767. doi: 10.1128/JCM.00964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou R, et al. Prevalence of Haemophilus influenzae pharyngeal carriers in the school population of Catalonia. Working Group on invasive disease caused by Haemophilus influenzae. Eur. J. Epidemiol. 2000;16(6):521–526. doi: 10.1023/a:1007632122570. [DOI] [PubMed] [Google Scholar]
- Buscher AZ, et al. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol. 2004;186(13):4209–4217. doi: 10.1128/JB.186.13.4209-4217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers MD, et al. Biological roles of nontypeable Haemophilus influenzae type IV pilus proteins encoded by the pil and com operons. J. Bacteriol. 2012;194(8):1927–1933. doi: 10.1128/JB.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholon DM, et al. Serial isolates of persistent Haemophilus influenzae in patients with chronic obstructive pulmonary disease express diminishing quantities of the HMW1 and HMW2 adhesins. Infect. Immun. 2008;76(10):4463–4468. doi: 10.1128/IAI.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid S, Barenkamp SJ, St Geme JW., 3rd Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA. 1999;96(3):1077–1082. doi: 10.1073/pnas.96.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid S, Grass S, St. Geme JW., 3rd Mapping of binding domains of nontypeable Haemophilus influenzae HMW1 and HMW2 adhesins. Infect. Immun. 2001;69(1):307–314. doi: 10.1128/IAI.69.1.307-314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle X, et al. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 2000;35(1):211–222. doi: 10.1046/j.1365-2958.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Ecevit IZ, et al. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia Genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 2004;42(7):3065–3072. doi: 10.1128/JCM.42.7.3065-3072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecevit IZ, et al. Identification of new hmwA alleles from nontypeable Haemophilus influenzae. Infect. Immun. 2005;73(2):1221–1225. doi: 10.1128/IAI.73.2.1221-1225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AL, et al. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 2005;73(9):5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AL, et al. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J. Bacteriol. 2008;190(4):1473–1483. doi: 10.1128/JB.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faden H, et al. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J. Infect. Dis. 1995;172(1):132–135. doi: 10.1093/infdis/172.1.132. [DOI] [PubMed] [Google Scholar]
- Farley MM, et al. Pathogenesis of IgA1 protease-producing and -nonproducing Haemophilus influenzae in human nasopharyngeal organ cultures. J. Infect. Dis. 1986;154(5):752–759. doi: 10.1093/infdis/154.5.752. [DOI] [PubMed] [Google Scholar]
- Fink DL, Green BA, St. Geme JW., 3rd The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect. Immun. 2002;70(9):4902–4907. doi: 10.1128/IAI.70.9.4902-4907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcha DS, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67(12):1075–1080. doi: 10.1136/thoraxjnl-2012-201924. [DOI] [PubMed] [Google Scholar]
- Giufre M, et al. Conservation and diversity of HMW1 and HMW2 adhesin binding domains among invasive nontypeable Haemophilus influenzae isolates. Infect. Immun. 2006;74(2):1161–1170. doi: 10.1128/IAI.74.2.1161-1170.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giufre M, et al. Variation in expression of HMW1 and HMW2 adhesins in invasive nontypeable Haemophilus influenzae isolates. BMC Microbiol. 2008;8:83. doi: 10.1186/1471-2180-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnehm HE, et al. Characterization of antigens from nontypable Haemophilus influenzae recognized by human bactericidal antibodies. Role of Haemophilus outer membrane proteins. J. Clin. Invest. 1985;75(5):1645–1658. doi: 10.1172/JCI111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham SM, et al. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73(6):1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- Harabuchi Y, et al. Nasopharyngeal colonization with nontypeable Haemophilus influenzae and recurrent otitis media. Tonawanda/Williamsville pediatrics. J. Infect. Dis. 1994;170(4):862–866. doi: 10.1093/infdis/170.4.862. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, St Geme JW., 3rd The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell. 1998;2(6):841–850. doi: 10.1016/s1097-2765(00)80298-1. [DOI] [PubMed] [Google Scholar]
- Hetherington SV, Lepow ML. Correlation between antibody affinity and serum bactericidal activity in infants. J. Infect. Dis. 1992;165(4):753–756. doi: 10.1093/infdis/165.4.753. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Travers P, Walport M, Shlomchik M. Immunobiology. 6th ed. Garland Science Publishing; New York: 2005. [Google Scholar]
- Jurcisek JA, et al. The PilA protein of non-typeable Haemophilus Influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 2007;65(5):1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- Karasic RB, et al. Modification of otitis media in chinchillas rechallenged with nontypable Haemophilus Influenzae and serological response to outer membrane antigens. J. Infect. Dis. 1985;151(2):273–279. doi: 10.1093/infdis/151.2.273. [DOI] [PubMed] [Google Scholar]
- Kirschner DE, Blaser MJ. The dynamics of Helicobacter pylori infection of the human stomach. J. Theor. Biol. 1995;176(2):281–290. doi: 10.1006/jtbi.1995.0198. [DOI] [PubMed] [Google Scholar]
- Krasan GP, et al. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 1999;67(1):449–454. doi: 10.1128/iai.67.1.449-454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiet M, et al. Pilus-mediated adherence of Haemophilus influenzae to human respiratory mucins. Infect. Immun. 2000;68(6):3362–3367. doi: 10.1128/iai.68.6.3362-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenauer PK, et al. Quality of care for patients hospitalized for acute exacerbations of chronic obstructive pulmonary disease. Ann. Intern. Med. 2006;144(12):894–903. doi: 10.7326/0003-4819-144-12-200606200-00006. [DOI] [PubMed] [Google Scholar]
- Marino S, et al. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 2008;254(1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, et al. Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30(18):3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon ER. Molecular basis of invasive Haemophilus influenzae type b disease. J. Infect. Dis. 1992;165(Suppl 1):S77–S81. doi: 10.1093/infdis/165-supplement_1-s77. [DOI] [PubMed] [Google Scholar]
- Palmer ME, et al. Broad conditions favor the evolution of phase-variable Loci. MBio. 2013;4:1. doi: 10.1128/mBio.00430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotin JM, et al. Detection of multiple viral and bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a pilot prospective study. J. Med. Virol. 2013;85(5):866–873. doi: 10.1002/jmv.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichichero ME, et al. Antibody response to Haemophilus influenzae outer membrane protein D, P6, and OMP26 after nasopharyngeal colonization and acute otitis media in children. Vaccine. 2010;28(44):7184–7192. doi: 10.1016/j.vaccine.2010.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert AI, et al. Trends in doctor consultations, antibiotic prescription, and specialist referrals for otitis media in children: 1995–2003. Pediatrics. 2006;117(6):1879–1886. doi: 10.1542/peds.2005-2470. [DOI] [PubMed] [Google Scholar]
- Reddy MS, et al. Binding between outer membrane proteins of nontypeable Haemophilus influenzae and human nasopharyngeal mucin. Infect. Immun. 1996;64(4):1477–1479. doi: 10.1128/iai.64.4.1477-1479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schilfgaarde M, et al. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 2000;68(8):4658–4665. doi: 10.1128/iai.68.8.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, et al. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 2002;347(7):465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- St Geme JW., 3rd The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 1994;62(9):3881–3889. doi: 10.1128/iai.62.9.3881-3889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Geme JW, 3rd, Falkow S, Barenkamp SJ. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA. 1993;90(7):2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Geme JW, 3rd, de la Morena ML, Falkow S. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 1994;14(2):217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- St. Geme JW, 3rd, et al. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 1998;66(1):364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teele DW, Klein JO, Rosner B. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J. Infect. Dis. 1989;160(1):83–94. doi: 10.1093/infdis/160.1.83. [DOI] [PubMed] [Google Scholar]