Abstract
We describe methods for parcellating an individual subject's cortical and subcortical brain structures using resting-state functional correlations (RSFCs). Inspired by approaches from social network analysis, we first describe the application of snowball sampling on RSFC data (RSFC-Snowballing) to identify the centers of cortical areas, subdivisions of subcortical nuclei, and the cerebellum. RSFC-Snowballing parcellation is then compared with parcellation derived from identifying locations where RSFC maps exhibit abrupt transitions (RSFC-Boundary Mapping). RSFC-Snowballing and RSFC-Boundary Mapping largely complement one another, but also provide unique parcellation information; together, the methods identify independent entities with distinct functional correlations across many cortical and subcortical locations in the brain. RSFC parcellation is relatively reliable within a subject scanned across multiple days, and while the locations of many area centers and boundaries appear to exhibit considerable overlap across subjects, there is also cross-subject variability—reinforcing the motivation to parcellate brains at the level of individuals. Finally, examination of a large meta-analysis of task-evoked functional magnetic resonance imaging data reveals that area centers defined by task-evoked activity exhibit correspondence with area centers defined by RSFC-Snowballing. This observation provides important evidence for the ability of RSFC to parcellate broad expanses of an individual's brain into functionally meaningful units.
Keywords: boundary mapping, brain area parcellation, brain networks, individual differences, resting-state functional correlations, snowball sampling
Introduction
The brain is organized at multiple spatial scales ranging from individual neurons to systems of distributed brain areas (Sejnowski and Churchland 1989). The identification of brain areas has been largely based on finding reliable differences in one or more features related to cellular/subcellular architecture, connectivity, topography, or function (e.g., Felleman and Van Essen 1991). Observations anchored on the convergence of these features have facilitated precise delineation of discrete brain areas; for example, the borders of area MT in the macaque (also known as area V5) can be defined by MT's independent representation of the visual field, the presence of neurons with sensitivity to particular properties of visual motion, distinct patterns of incoming and outgoing connections, and the thick band of myelin that is present in layer IV (e.g., Van Essen et al. 1981).
Efforts to parcellate the human brain into a map of areas date back at least to the beginning of the 20th century (e.g., Brodmann 1909; Vogt and Vogt 1919). These seminal studies relied on detailed histological analysis of postmortem brains and have guided our descriptions of human brain organization and function. However, many area parcellations have been found to be either incomplete or inaccurate; accordingly, descriptions of brain areas are continually revised and modified as parcellation methods continue to be developed (Toga et al. 2006; Zilles and Amunts 2010). Most recently, the introduction of neuroimaging has made it feasible to noninvasively parcellate the brain's cortical and subcortical structures.
Many immediate and tangible benefits would result from successful parcellation of the human brain using neuroimaging. A clear picture of the organization of functional areas in individual subjects would allow deeper insights into underlying functional anatomy (e.g., Devlin and Poldrack 2007). Subject-specific parcellation would allow straightforward analyses within a subject by identifying individual regions of interest (e.g., Saxe et al. 2006). Individual parcellations could enhance cross-subject analysis both at the level of regions (cf. (Swallow et al. 2003)) and improve intersubject registration using functional features in addition to anatomical features (e.g., Sabuncu et al. 2010). Most recently, there has been a surge of interest to characterize functional and anatomical “connectomes” (Sporns et al. 2005; Van Essen and Ugurbil 2012); a method that could reliably parcellate the brains of individual subjects could drive rational node definition for the purposes of the analysis of large-scale brain networks (Wig et al. 2011).
Brain parcellation using neuroimaging is still a nascent enterprise, often resulting in parcellations that are either incomplete or incompatible with one another. For example, an effective strategy to parcellate sensory brain areas involves mapping the ordered projection of a sensory surface (e.g., the retina, skin, or cochlea) within brain areas that exhibit a topographic organization of that sensory input (e.g., Sereno et al. 1995; Formisano et al. 2003; for review see Wandell et al. 2007). However, many areas do not map to a specific sensory surface in a clear and discernable way.
Beyond topographic mapping, parcellations have also been obtained using imaging sequences sensitive to cyto-, myelo-, and chemoarchitecture (e.g., Glasser and Van Essen 2011; Caspers et al. 2013; for review see Toga et al. 2006). In many of these cases, the basic strategy has been to identify borders between areas by identifying transitions in the given property. Parcellations have also been derived by examining transitions in the patterns of interareal relationships, including anatomical connectivity via fiber bundles measured using diffusion imaging (e.g., Johansen-Berg et al. 2004, 2005; Behrens et al. 2006; Beckmann et al. 2009; Hua et al. 2009; Mars et al. 2011) and functional connectivity measured by correlations of low-frequency blood oxygenation level-dependent (BOLD) signal acquired during resting wakefulness [i.e., resting-state functional connectivity (RSFC) magnetic resonance imaging (MRI); Cohen et al. 2008; Nelson, Cohen, et al. 2010; Nelson, Dosenbach, et al. 2010; Barnes et al. 2012; Goulas et al. 2012; Kahnt et al. 2012]. As with topographic mapping, parcellation strategies that identify boundaries detected by a specific imaging modality can be limited in their applicability to particular expanses of the brain. For example, imaging sequences sensitive to the myelin content of gray matter can be highly effective in delineating areas with differential levels of myelination [e.g., the stria of Gennari that defines the primary visual cortex (V1; Gennari 1782), or the thick layer of myelin in layer IV of visual area 5 (V5/MT; Allman and Kaas 1971; Tootell and Taylor 1995)], but considerable portions of the cerebral cortex remain undifferentiated by myelin content alone (e.g., Geyer et al. 2011; Glasser and Van Essen 2011).
An alternative possibility is to parcellate brain areas by dissociating them based on their patterns of evoked activity. If a battery of task and stimulus manipulations was available that was capable of eliciting activity in every brain area, brain parcellation might be accomplished by dissociating the brain areas based on their function. However, both the ideal experimental battery and the specific contrasts to identify accurate area divisions are uncertain. Further, collecting a large experimental battery on every subject for which parcellation is required may require unreasonably long data acquisition sessions.
Thus, the development of complementary parcellation strategies is necessary. Here, we present an approach for brain area parcellation inspired by “snowball sampling” methods employed in the social network sciences (e.g., Goodman 1961; Wasserman and Faust 1994). Snowball sampling works on the assumption that members of a population of interest are typically able to identify one another via shared relations (e.g., a social network of musicians in a community). Snowball sampling relies on starting with a potential member of the social network of interest (e.g., a known musician), identifying that individual's acquaintances who share the feature of interest (e.g., playing an instrument), identifying the acquaintances' instrument playing acquaintances, and so forth, until a sample of individuals who reasonably represent a network of musicians have been identified. This general approach has also been used to identify network members in other domains, such as technological networks (e.g., to describe the world-wide web through web-page links (Albert et al. 1999) and even neural networks at the level of synaptic connections (Hoover and Strick 1999).
The application to identification of brain areas with neuroimaging should be apparent: Snowball sampling could be used to identify the areas to which a starting area is related (its neighbors), the areas to which the neighbors are related (neighbors of neighbors), and so forth, until a large collection of related brain areas are revealed. The fundamental requirement of snowball sampling is the ability to measure relationships between objects of interest. For our snowball sampling approach, we use RSFC as a basis for defining relationships between areas of the brain. RSFC is defined by the correlation of low-frequency (e.g., <0.08 Hz) BOLD signal fluctuations obtained in the absence of imposed tasks (Biswal et al. 1995; for reviews see Fox and Raichle 2007; Biswal et al. 2010). Although RSFC relationships are likely mediated by anatomical connectivity, they are not restricted to direct structural connections (Vincent et al. 2007; Honey et al. 2009; for reviews see Deco et al. 2011; Wig et al. 2011).
This report describes the use of snowball sampling using RSFC relationships (RSFC-Snowballing) to identify area centers across broad expanses of the brain. While the term “area” is conventionally restricted to parcellations of the cerebral cortex, for brevity we will use the term more generally throughout this report, in reference to cortical areas as well as subdivisions of subcortical nuclei and the cerebellum. This strategy is complementary to yet distinct from approaches that attempt to identify the locations where a given property transitions (boundary mapping), in that it attempts to highlight the central parts of brain areas rather than the boundaries between them.
To foreshadow what follows, we will start with a description of the general method of RSFC-Snowballing by beginning with a single starting location and by iteratively mapping its functional correlations. We will then describe how this basic process can be applied more broadly to estimate area centers by aggregating the results from numerous starting locations. Following this, we will highlight the utility of RSFC-Snowballing for parcellating cortical and subcortical structures and then demonstrate how the parcellation is largely invariant to numerous parameters [e.g., starting locations, correlation threshold, radius of the seed region of interest (ROI)]. Next, we will describe the application of the RSFC-Boundary Mapping technique (Cohen et al. 2008) to parcellate a subject's cortical surface. We will then perform direct comparisons of our RSFC parcellation methods (RSFC-Snowballing and RSFC-Boundary Mapping), assess their correspondence, and also report measurements of the reliability of parcellations both within and across subjects. We will conclude by describing initial steps in comparing RSFC parcellation with area identification defined by task-evoked data, yielding evidence that our RSFC parcellation methods identify functionally meaningful entities.
Subjects and General Methods
Subjects and Data Acquisition Parameters
Subjects were recruited from the Washington University community and were screened with a self-report questionnaire to ensure that they had no current or previous history of neurological or psychiatric diagnosis. Informed consent was obtained from all subjects. The study was approved by the Washington University School of Medicine Human Studies Committee and Institutional Review Board.
RSFC data were collected from subjects who were instructed to relax while fixating on a black crosshair against a white background. Single-subject analyses focused on 8 subjects (3 females, mean age = 26 years, age range = 24–27 years). For each subject, 4 runs [2.5 s repetition time (TR), 128 steady-state frames (320 s) per run] were acquired in each scan session during 2 separate sessions that were separated by multiple intervening days (average interval of 20 days intervening between the 2 scan sessions; range: 7–53 days).
Data Acquisition Parameters
For the resting-state data sets, structural and functional MRI (fMRI) data were obtained with a 3.0-T Siemens MAGNETOM Trio Tim Scanner (Erlangen, Germany) and a Siemens 12-channel head matrix coil. To help stabilize the head position, each subject was fitted with a thermoplastic mask fastened to holders on the headcoil. A T1-weighted sagittal magnetization-prepared rapid acquisition gradient-echo (MPRAGE) structural image was obtained [echo time (TE) = 3.08ms, TR(partition) = 2.4 s, time to inversion = 1000 ms, flip angle = 8°, 176 slices with 1 × 1 × 1 mm voxels; Mugler and Brookeman 1990]. An auto align pulse sequence protocol provided in the Siemens software was used to align the acquisition slices of the functional scans parallel to the anterior commissure–posterior commissure plane and centered on the brain. This plane is parallel to the slices in the Talairach atlas (Talairach and Tournoux 1988). Functional imaging was performed using a BOLD contrast sensitive gradient-echo echo-planar sequence (TE = 27 ms, flip angle = 90°, in-plane resolution = 4 × 4 mm). Whole-brain echo-planar imaging (EPI) volumes (magnetic resonance frames) of 32 contiguous, 4-mm thick axial slices were obtained every 2.5 s. A T2-weighted turbo spin echo structural image (TE = 84 ms, TR = 6.8 s, 32 slices with 1 × 1 × 4 mm voxels) in the same anatomical planes as the BOLD images was also obtained to improve alignment to the atlas.
Data Preprocessing
Standard fMRI Preprocessing
Functional images were first processed to reduce artifacts (Miezin et al. 2000). These steps included: 1) Correction of odd versus even slice intensity differences attributable to interleaved acquisition without gaps, 2) correction for head movement within and across runs, and 3) across-run intensity normalization to a whole-brain mode value of 1000. Atlas transformation of the functional data was computed for each individual using the MP-RAGE scan. Each run was then resampled to an isotropic 3-mm atlas space (Talairach and Tournoux 1988), combining movement correction and atlas transformation in a single cubic spline interpolation (Lancaster et al. 1995; Snyder 1996). This single-interpolation procedure avoids blurring that would be introduced by multiple interpolations. All subsequent operations were performed on the atlas-transformed volumetric time series.
RSFC-Specific Preprocessing
For RSFC analyses, several additional preprocessing steps were utilized to reduce spurious variance unlikely to reflect neuronal activity. RSFC preprocessing was performed in 2 iterations. In the first iteration, RSFC preprocessing included, in the following order: (i) Multiple regression of nuisance variables from the BOLD data, including whole-brain signal (cf. Scholvinck et al. 2010), ventricular signal, white matter signal, 6 detrended head realignment parameters obtained by rigid-body head motion correction, and the first-order derivative terms for all aforementioned nuisance variables, (ii) a temporal band-pass filter (0.009 < f < 0.08 Hz), and (iii) volumetric spatial smoothing (a 6-mm full-width at half-maximum in each direction).
Following the initial RSFC preprocessing iteration, and to ameliorate the effect of motion artifact on RSFCs, data were processed following the recently described “scrubbing” procedure (Power et al. 2012). Temporal masks were created to flag motion-contaminated frames, so that they could be ignored during subsequent nuisance regression and correlation calculations. Motion-contaminated volumes were identified by frame-by-frame displacement (FD, calculated as the sum of absolute values of the differentials of the 3 translational motion parameters and 3 rotational motion parameters) and by frame-by-frame signal change (DVARS). Volumes with FD >0.3 mm or DVARS >3% signal change were flagged. In addition, the 2 frames acquired immediately prior to each of these frames and the 2 frames acquired immediately after these frames were also flagged to account for the temporal spread of artifactual signal resulting from the temporal filtering in the first RSFC preprocessing iteration.
The RSFC preprocessing steps outlined above (steps i–iii; including nuisance regression, temporal filtering, and volumetric smoothing) were applied in the second iteration on RSFC data that excluded volumes flagged during motion scrubbing. Following all RSFC preprocessing steps outlined above, one subject's day 2 data had an insufficient number of frames remaining following movement scrubbing (48 frames) and this session was excluded from all analyses. The mean percent of frames excluded from the remaining subjects was 15.9% (range: 3.8–38.0%; minimum of 327 frames remaining per subject).
Methods/Results 1: Implementation and Demonstration of RSFC-Snowballing
RSFC-Snowballing Basic Methods
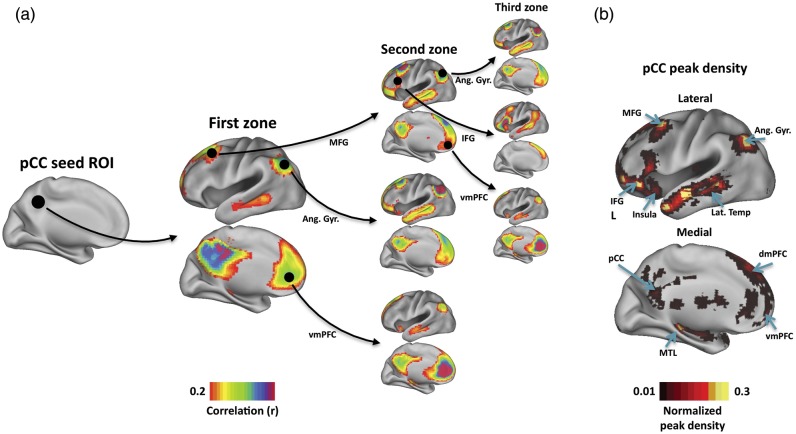
RSFC-Snowballing is an iterative procedure that uses seed-based RSFC to identify locations correlated with a starting seed location (i.e., the “neighbors” of the seed, in a graph theoretic sense), then identifies the neighbors of the neighbors, and so forth. It is important to be clear that a neighbor of a given seed need not be physically adjacent to the seed, but rather is defined by the presence of a RSFC relationship above a given threshold. Adopting the naming convention of snowball sampling in social network analysis (Wasserman and Faust 1994), each iteration of identified neighbors is termed a “zone.” A diagram of the basic process is presented in Figure 1a using an initiating seed ROI placed in the posterior cingulate cortex (pCC; Montreal Neurological Institute coordinates: 0 –45 42 obtained from Wig et al. 2008), an area of the “default system” (Shulman et al. 1997; Raichle et al. 2001). It should be apparent that the locations (neighbors) correlated with a given seed location are typically the clusters of contiguous voxels that can have a varying extent. We can represent each of the clusters as their local maxima (i.e., peak voxel coordinate locations) and then seed these locations to identify neighbors over subsequent zones. By aggregating the peak voxel coordinate locations identified over multiple zones, a whole-brain voxel-wise map can be produced that reflects a spatial distribution of the identified peaks as well as a measure of the number of times each peak was identified (i.e., its magnitude or peak density). Following three zones of snowballing, the pCC's peak density map includes voxels in other areas of the default system, including the angular gyrus, dorsal and medial prefrontal cortex, middle frontal gyrus, and medial and lateral temporal cortex (Fig. 1b).
Figure 1.
RSFC-Snowballing is an iterative procedure that identifies and combines the neighbors (peaks) of seed-based resting-state correlations over multiple zones to produce a peak density map for the starting seed location. (a) RSFC-Snowballing is depicted using the posterior cingulate cortex (pCC) as a starting seed location. While we have restricted our depiction of the procedure to the neighbors of a subset of regions within each zone set, the procedure is replicated across all identified neighbors in every zone of snowballing. (b) The neighbor locations (i.e., voxel coordinates) identified across all zones are tallied in the pCC peak density map, which reflects the spatial distribution of peaks identified following RSFC-Snowballing of the pCC. The pCC peak density map includes regions of the default system (e.g., the angular gyrus, Ang. Gyr.) and the dorsal and ventral medial prefrontal cortex (dmPFC/vmPFC), but also regions not typically associated with the default system [e.g., the inferior frontal gyrus (IFG) and insula cortex (insula)].
Three observations are important to highlight: First, rather than identifying parcels that contain uniform peak density values within the parcel's extent, the pCC's peak density map is a continuous distribution of peak tallies centered around local maxima (this finding is similar to what is observed with voxel-wise distributions of task-evoked activity). We hypothesize that the peak density value is lesser at locations that are transition points (or boundaries) between adjacent areas and greater within an area's interior. Therefore, peak density local maxima may be used to label the inner points of areas. It is worth noting that these inner points will often turn out to be distinct from a putative area's geometric center.
Secondly, in contrast to a typical RSFC map, the pCC peak density map following three zones of snowballing (Fig. 1b) is not limited to the primary neighbors of the pCC (i.e., regions with which the pCC is correlated), nor to regions of the default system. This effect is a result of the iterative nature of RSFC-Snowballing; while many of the neighbors of the pCC are members of the default system (Fig. 1a, first zone), the neighbors of the neighbors need not exhibit the same pattern of correlations. It is perhaps not surprising that some default regions are correlated with regions that are not correlated with the pCC (for further discussion see Wig et al. 2011). For example, the middle frontal gyrus (Fig. 1a, first zone) is correlated with other regions of the default system (e.g., the angular gyrus and ventral medial prefrontal cortex in the second zone), but is also correlated with a region of the inferior frontal gyrus (IFG), that is, neither correlated with the pCC (see neighbors of the IFG ROI in the third zone) nor typically associated with the default system.
Thirdly, while regions outside of the default system have been identified (e.g., in the IFG), the pCC peak density map following 3 zones of snowballing “misses” large expanses of the brain. To identify peaks in the rest of the brain, one approach might be to run the RSFC-Snowballing over additional zones. However, this strategy would be computationally demanding, might still be constrained by the choice of the starting seed location such that locations of sparse connectivity are not identified, and could theoretically result in estimates of area center locations that are biased by the starting location.
Initializing RSFC-Snowballing From Multiple Starting Seed Locations
Based on the reasons noted above, our implementation of RSFC-Snowballing involves initializing RSFC-Snowballing from multiple starting seed locations (i.e., from a predefined set of coordinates or an initialization location set, Fig. 2a), iterating over a series of zones to create a peak density map for each of these starting locations (Fig. 2b), and then combining the peak density maps derived from each starting location to arrive at an aggregate peak density map (Fig. 2c). Aggregating the peak density maps from multiple starting locations should minimize the potential bias of a single starting seed location and provide the estimates of area centers across broad expanses of the brain's cortical and subcortical structures.
Figure 2.
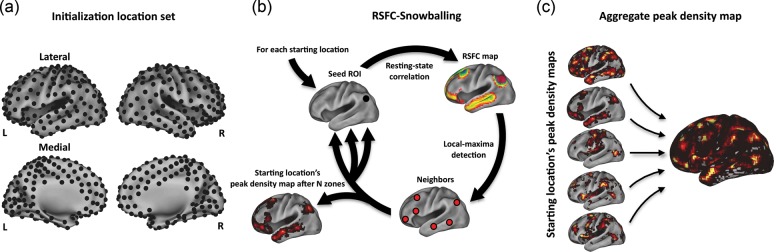
Overview of RSFC-Snowballing using multiple starting seed locations. (a) Initialization location set consisting of seed locations (n = 464) that were regularly spaced across a flattened cortical surface. (b) For each seed location in the initialization location set, RSFC-Snowballing iteratively identifies the neighbors (peaks of RSFC) of seed ROIs over multiple zones and adds these neighbors to a peak density map. (c) The independently derived peak density maps from each of the seed locations of the initialization location set are summed to arrive at an aggregate peak density map.
The ideal starting locations for the tracking patterns of relationships would be a set of known parcellated areas. However, as the goal of RSFC-Snowballing is to derive the set itself, the ideal starting locations for RSFC-Snowballing are presumed to be unknown. To create the initialization location set (Fig. 2a), regularly spaced Cartesian grids were generated on the flattened PALS-B12 average surface of the left and right hemispheres using the Caret software (Van Essen 2005). Each hemisphere's grid covered the entire cortical surface. Grid points were spaced 20 mm apart on the flattened surface; a total of 464 points were created across the 2 hemispheres. The 3-dimensional (3D) stereotactic coordinates from the PALS-B12 average fiducial (midthickness) surface for each grid location were used to obtain the voxel coordinates (3 × 3 × 3 mm resolution) containing that point. Accordingly, the regular surface-grid RSFC-Snowballing seed location set comprised 464 coordinate locations limited to the cortical surface of the left and right hemispheres.
Each coordinate location within the initialization location set served as an independent starting seed ROI for RSFC-Snowballing (Fig. 2b). A spherical ROI (5-mm radius) was created around the starting coordinate location and its average time course was extracted from the subject's resting-state BOLD scan. A Pearson correlation coefficient was computed between this seed ROI's time course and the time course for each voxel across the whole-brain volume. The correlation map was then smoothed with a Gaussian kernel [a 6-mm full-width at half-maximum (FWHM)] and thresholded (r > 0.2) to identify voxels that demonstrated a strong correlation with the seed ROI's time course. To identify the seed ROI's connectivity neighbors (i.e., regions that were most correlated with the seed ROI), the local maxima (peaks) of the contiguous clusters of voxels that both surpassed the correlation threshold and had a minimum distance of 10 mm between peaks were identified. These neighbors corresponded to the first zone of RSFC-Snowballing. The location of each of the identified neighbors from the first zone was then used as a center for a new seed ROI (5-mm radius), and the process was run again to identify the second zone of neighbors (zone 2). Finally, the neighbors of a third zone (zone 3) were identified by seeding all neighbors identified in the second zone. This resulted in a list of all peak coordinate locations (neighbors) identified across seed ROIs from each zone. Importantly, a given peak could be identified multiple times and by multiple seed ROIs, and the full peak coordinate list was retained for each zone. All peak location coordinates identified in each ROI's coorelation map across the 3 zones were combined to generate a voxel-wise map for each starting seed ROI. The value at each voxel in this peak density map reflected a count of the number of times that voxel was identified as a peak.
In the final step, an aggregate RSFC-Snowballing peak density map for the initialization location set was derived by summing all the starting location's peak density maps (Fig. 2c). This aggregate peak density map represents the number of times a voxel was identified as a peak across all ROI correlation maps, across all zones, and across all starting seed ROI locations. The aggregate peak density map was spatially smoothed using a 6-mm FWHM Gaussian kernel and normalized relative to the maximum peak value to facilitate viewing and comparison across subjects.
RSFC-Snowballing Results
RSFC-Snowballing Reveals a Nonuniform Distribution of Correlation Peaks across Cortical and Subcortical Structures in an Individual Subject
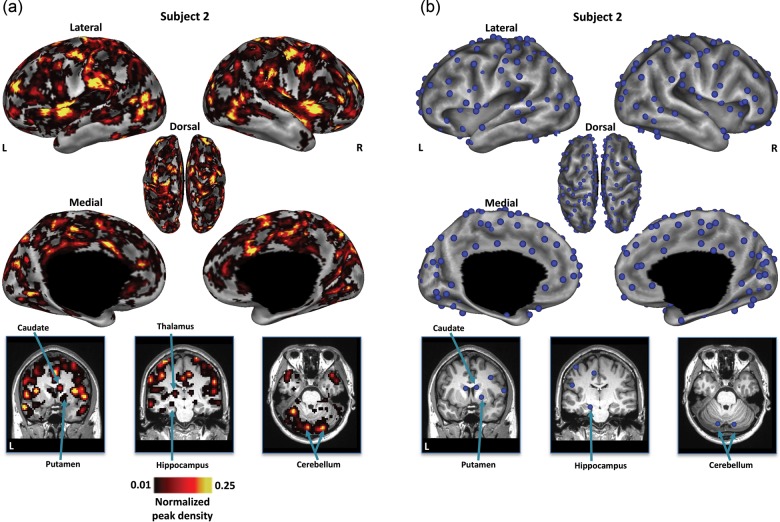
For each subject, the RSFC-Snowballing aggregate peak density map exhibited a nonuniform distribution of peak counts across the brain. (Fig. 3a; for additional subjects see Supplementary Fig. 1). A subset of voxels exhibited relatively high peak counts. The locations of voxels with high RSFC-Snowballing peak counts were distinct from the locations of the starting points in the initialization location set. For example, while the initialization location set was derived from a Cartesian grid placed on the flattened cortical surface, distinct clusters of circumscribed peaks were also identified in noncortical structures, including the head of the caudate, putamen, subnuclei of the thalamus, hippocampus, and regions throughout the lateral and medial cerebellum.
Figure 3.
RSFC-Snowballing of a single subject's resting-state data reveals the locations of area centers across cortical and subcortical structures. (a) The distribution of peak counts identified by RSFC-Snowballing is nonuniform across the brain. Locations of high peak counts are prevalent throughout cortical and subcortical structures. While all analyses are conducted on unthresholded aggregate peak density maps, the aggregate depicted peak density map has been normalized and thresholded (1%) relative to the maximum peak value to facilitate viewing. (b) Local maxima were identified on the subject's unthresholded aggregate peak density map to highlight the inner points of the areas. The subject's aggregate peak density map and locations of the local maxima of this map are displayed on the subject's inflated cortical surfaces (top) and coronal and axial views of the subject's anatomical image for subcortical and cerebellar structures, respectively.
For display purposes, the aggregate peak density maps presented throughout this report have been thresholded (for an illustrative unthresholded map see Supplementary Fig. 2). It is important to note that while thresholding was carried out for visualization, this procedure would bias area parcellation toward only identifying areas that exhibit a higher peak value and potentially missing areas that exhibit sparse connectivity. As such, although we present aggregate peak density maps following thresholding in the figures, all analyses on the RSFC-Snowballing aggregate peak density maps (e.g., the detection of local maxima, comparison across subjects, comparison with task data, etc.) were computed on subjects' unthresholded maps.
Figure 3b highlights the voxels identified as local maxima of our illustrative subject's unthresholded RSFC-Snowballing peak density map. For maxima detection, we imposed a minimum maxima-to-maxima distance criterion of 10 mm. As such it is theoretically possible that we identified multiple maxima satisfying this distance criteria that were all located within a single area. We attempted to test this possibility directly (see Methods/Results 3). In the illustrative subject depicted in FIgure 3, a total of 374 cortical and subcortical local maxima were identified across both hemispheres (mean of 8 subjects: 415 and range across 8 subjects: 374–455).
The Peak Distribution Revealed by RSFC-Snowballing Is Invariant to Initialization Location Set
To determine whether a subject's RSFC-Snowballing aggregate peak density map was sensitive to the starting locations, we ran RSFC-Snowballing using a second independent initialization location set for each subject. This second initialization set consisted of locations defined from meta-analysis of task-evoked data and thus may constitute a set of more rational starting points for capturing functional area relationships. The meta-analysis was performed on a collection of studies where independent groups of subjects performed different tasks with different stimuli. Specifically, the task-defined area centers were identified by searching a large fMRI dataset, acquired in a single scanner (a 1.5-T Siemens MAGNETOM Vision MRI scanner) for brain regions that reliably displayed significant activity when certain tasks were performed (e.g., button pressing) or certain signal types were expected (e.g., error-related activity). The task-defined area centers consisted of 151 coordinate locations across the cerebral cortex and subcortical structures, including the basal ganglia, thalamus, and cerebellum (for the details of analysis see Power et al. 2011) and were distinct from initialization locations defined by the regularly spaced surface-grid (Fig. 4a).
Figure 4.
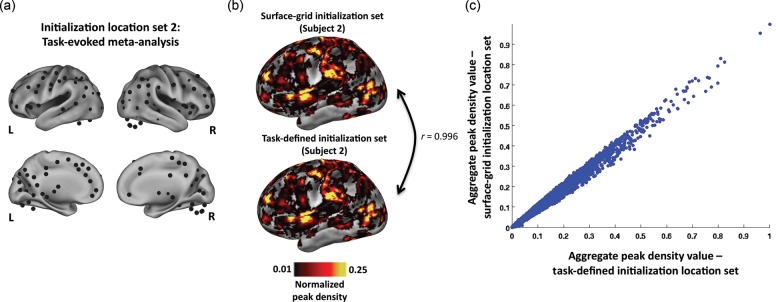
The RSFC-Snowballing aggregate peak density map is highly similar across initialization location sets within a subject. (a) Task-defined initialization location set (including subcortical locations along midline views and cerebellar locations outside of the cortical representation). (b) Lateral inflated views of the left hemisphere depict the high similarity in the distribution of RSFC-Snowballing peaks using two different initialization location sets. Peak density maps are normalized and thresholded (1%) relative to the maximum peak value to facilitate viewing. (c) Voxel-wise scatter plot demonstrates the similarity in the aggregate peak density maps produced by independent initialization location sets. Voxels of high and low peak values are comparable across the 2 aggregate peak density maps.
The final peak map derived from the task-derived location set was strikingly similar to the peak map derived from the surface-grid location set (Fig. 4b). To corroborate the qualitative observation, a spatial correlation was computed between the aggregate peak density maps generated from each initialization location set. The spatial distribution of the peaks was highly similar (Pearson's correlation coefficient: r = 0.996; mean of 8 subjects: r = 0.981; range across 8 subjects: r = 0.942 to 0.997; Fig. 4c).
Many Features Revealed by RSFC-Snowballing Are Relatively Insensitive to Numerous Parameter Settings
There are a number of parameters that can be varied in implementing RSFC-Snowballing (e.g., the radius of the ROI, correlation threshold, and the number of zones). Parameter selection was based, in part, by a motivation to utilize similar parameters as standard seed-based RSFC analyses (e.g., radius of ROI). However, we also systematically tested the impact of different parameter settings on the estimation of area centers derived from RSFC-Snowballing within a subject.
The results presented throughout this report were obtained using a ROI seed radius of 5 mm for each seed-based correlation map (i.e., each instance of neighbor identification). As it is possible that the radius of the seed ROI may have an impact on the neighbors that are identified, we varied the seed radius size and recomputed the RSFC-Snowballing peak density maps using the regular surface-grid starting location sets.
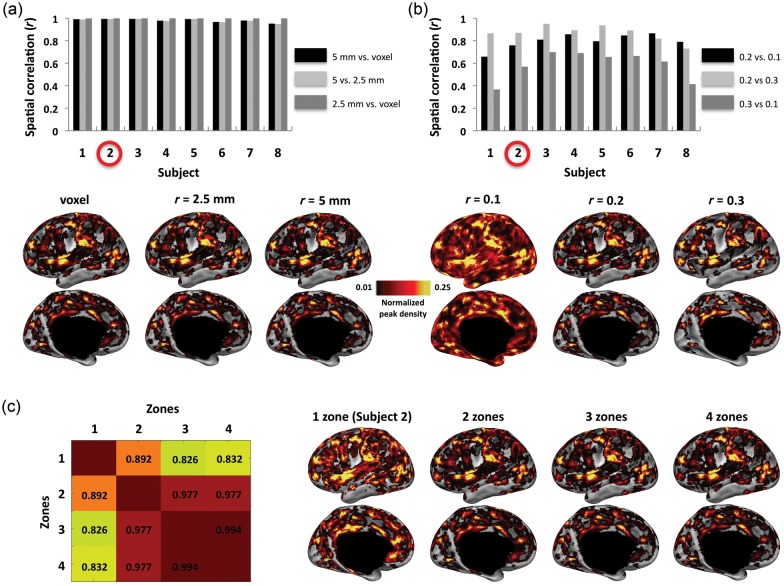
We used ROI seeds with a radius of 2.5 mm and also ROI seeds that were limited to individual voxels corresponding to each of the coordinate locations. The RSFC-Snowballing aggregate peak density maps were highly similar across the different ROI radius sizes, suggesting that this parameter has minimal impact on the final result (the range of spatial correlation between each subject's aggregate peak density map using 5-mm radius spherical ROIs vs. using 2.5-mm radius spherical ROIs: r = 0.948–0.995; the range of spatial correlation between each subject's aggregate peak density map using 5-mm radius spherical ROIs vs. using voxel ROIs: r = 0.952–0.995; Fig. 5a).
Figure 5.
Many of the features of the RSFC-Snowballing aggregate peak density map are similar across a range of parameter settings. (a) The size of the seed ROI (a single voxel vs. a 2.5-mm radius ROI built around the voxel vs. a 5-mm radius ROI built around the voxel) used throughout RSFC-Snowballing had minimal impact on the peak density maps. An illustrative subject's (subject 2) aggregate peak density maps are displayed below the bar plot. (b) Similarity in the aggregate peak density maps was more variable when examining the impact of different correlation thresholds for neighbor identification. The aggregate peak density maps following more stringent thresholds (r > 0.2 and r > 0.3) were more similar than the aggregate peak density maps produced using each of these thresholds relative to the aggregate peak density map produced using a lower threshold (r > 0.1) across the majority of subjects. An illustrative subject's (subject 2) aggregate peak density maps are displayed below the bar plot. (c) A single subject's data are displayed to demonstrate the similarity in aggregate peak density maps following a variable number of zones of RSFC-Snowballing. Aggregate peak density maps were created following 1–4 zones of RSFC-Snowballing, and the spatial similarity of the maps was computed between the 4 independent analyses (spatial correlation r values in matrix). The similarity of peak density maps appears to asymptote following 3 zones of RSFC-Snowballing (left; i.e., compare similarity following 4 zones of RSFC-Snowballing with that following 1, 2, and 3 zones of RSFC-Snowballing). This observation can be further appreciated by examining the distribution of peaks across the number of zones of RSFC-Snowballing on the subject's left hemisphere (right). All peak density maps are normalized and thresholded (1%) relative to the maximum peak value to facilitate viewing, and are displayed on the subject's inflated cortical surface.
We next tested the choice of correlation threshold for identifying neighbors. Increasing the correlation threshold (r > 0.3) for neighbor identification had a marginal impact on the aggregate peak density map (the range of spatial correlation between each subject's aggregate peak density map using r > 0.2 vs. >0.3 correlation threshold for neighbor identification: r = 0.731–0.951; Fig. 5b). However, while results obtained by decreasing the correlation threshold (r > 0.1) retained many of the features identified using higher thresholds, this manipulation also resulted in peaks in regions of the white matter and ventricles. These peak density maps also lacked some of the specificity seen in peak density maps derived from other parameter settings.
Finally, we tested whether the choice of the number of RSFC-Snowballing zones has an impact on the peak density maps. While the peak density map following 1 and 2 zones of RSFC-Snowballing differed from one another and from the peak density map obtained following 3 zones of RSFC-Snowballing, we found a high degree of similarity between peak density maps following 3 or 4 zones of RSFC-Snowballing. This result suggests that the spatial similarity of the peak density maps asymptotes after 3 zones of RSFC-Snowballing when initiated using the present starting location sets (illustrative subject shown in Fig. 5c).
As an alternate test of the impact of the parameter selection on RSFC-Snowballing parcellation, we computed voxel-wise comparisons of subject's RSFC-Snowballing aggregate peak density maps. Specifically, for each of the parameters that were tested (initialization location set, radius of seed ROI, correlation threshold for neighbor identification, and the number of zones), a whole-brain voxel-wise repeated-measures analysis of variance (ANOVA) was computed on subject's aggregate peak density maps treating subject as the random factor (P < 0.05 corrected for the false discovery rate (FDR), minimum cluster size of 100 surface vertices). This analysis revealed whether and which voxels were sensitive to one of the tested levels of a given parameter. Consistent with the correlation analyses (Fig. 5), while the initialization location sets and the radius of seed ROI had a minimal outcome on RSFC-Snowballing aggregate peak density map values (no voxels exhibited a significant main effect), the choice of the number of zones and threshold for neighbor correlation had a greater impact (Fig. 6). We take this observation as supporting evidence that correlation threshold for neighbor identification and zone parameters need to be carefully considered when employing RSFC-Snowballing for area parcellation.
Figure 6.
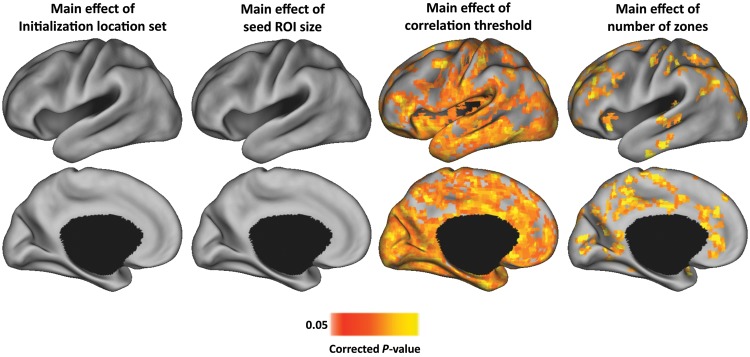
RSFC-Snowballing aggregate peak density maps are most sensitive to the choice of correlation threshold and the number of zones. Maps depict voxel-wise repeated-measures ANOVA main effect maps (treating subject as the random factor) for each of the parameters tested (initialization location set, radius of seed ROI, correlation threshold for neighbor identification, and number of zones). The analysis demonstrates that a considerable number of voxel's aggregate peak density map values are highly sensitive to correlation threshold and the number of RSFC-Snowballing zones, but not the initialization location set or radius of seed ROIs tested. All maps depicted on the partially inflated PALS atlas surface (Van Essen 2005).
Methods/Results 2: Comparison with RSFC-Boundary Mapping
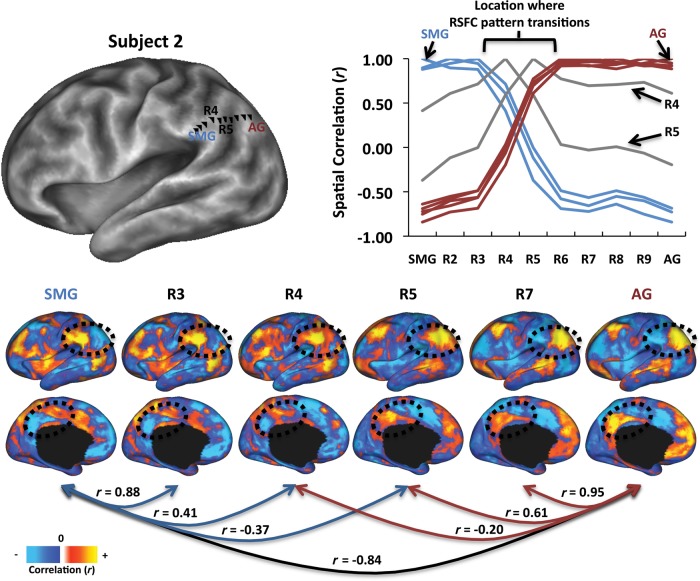
If RSFC-Snowballing identifies the interiors of areas (area centers), then RSFC-Snowballing peak density maps should exhibit an areal parcellation complementary to maps obtained by boundary-detection techniques. In order to compare the area centers derived from RSFC-Snowballing with an alternate method of parcellation that identifies area boundaries, we employed RSFC-Boundary Mapping (Cohen et al. 2008). RSFC-Boundary Mapping rests on the assumption that an area's RSFCs are relatively uniform within the extent of an area and may be distinct from the RSFCs of an adjacent area. Identifying locations where the patterns of RSFC correlations exhibit abrupt transitions provides the estimates of putative boundaries between areas. This abrupt transition can be illustrated by drawing a line across the cortical surface of a single subject and by measuring and comparing the RSFC maps between points along the line. As seen in Figure 7, the RSFC maps do not change smoothly, but exhibit rapid changes. Furthermore, these locations of change are consistent in both directions (i.e., from a region in the supramarginal gyrus to a region in the angular gyrus, or in reverse), suggesting the presence of a functional boundary between 2 adjacent areas similar to that which is observed when measuring connectional anatomy. This basic approach can be extended across the cortical surface with the aid of image-processing tools to create a voxel-wise estimate of the likelihood with which a location is being identified as a boundary between 2 points in the brain.
Figure 7.
Patterns of RSFC exhibit abrupt changes across the lateral parietal cortex in a single subject. RSFC maps were derived for vertices (R2–R9) between a region in the supramarginal gyrus (SMG) and angular gyrus (AG) of a single subject (defined anatomically; vertices are shown as black triangles). The plot depicts the similarity of every vertex's RSFC map with the RSFC map of every other vertex. RSFC maps are strikingly similar from SMG to R3, followed by a location of abrupt change (R4–R5). A similar pattern is revealed in the reverse direction (from AG toward SMG), providing additional support for the identification of a transition point (or area border) at R4–R5. Similarity values have been color coded to denote RSFC similarity with SMG (blue) or AG (maroon). Two vertices (R4 and R5; color-coded gray) have RSFC maps that are not highly similar to either the SMG or AG groups. RSFC maps of a subset of the regions are depicted on the lower panel, along with the spatial similarity of the map to other maps. Regions in the lateral and medial parietal cortex are circled to highlight 2 locations that exemplify the pattern of stability followed by change described.
RSFC-Boundary Mapping Methods
The RSFC-Boundary Mapping method used here represents the evolution of an analysis strategy first described by Cohen et al. (2008) that was designed to highlight transitions in correlation patterns across the surface of the brain. The original technique took advantage of 2-dimensional image processing tools for gradient calculation and edge detection by sampling time courses from a Cartesian grid of ROIs projected onto a patch on a flattened cortical surface (e.g., Nelson et al. 2010). The cuts required for flattening the surface lead to distortions in the surface representation and make gradients near the cut surface's edge more difficult to interpret. We employed a version of the method that avoids these issues by performing all computations directly on the subject's “midthickness” cortical surface, a closed surface mesh that was resampled to reflect the spatial resolution of the RSFC data. Further, rather than restricting boundary mapping to a small patch, we performed the analysis on each hemisphere's whole cortical surface using recently developed caret software tools (http://brainvis.wustl.edu/wiki/index.php/Caret:About).
An overview of the RSFC-Boundary Mapping method we employed is depicted in Figure 8. Following the volumetric registration procedures, the atlas-registered MPRAGE anatomical image for each subject was processed through the Freesurfer analysis pipeline (Freesurfer 4.5; Dale and Sereno 1993; Dale et al. 1999; Fischl, Sereno, et al. 1999; Ségonne et al. 2004, 2005) to generate white matter, pial, and spherical surfaces for each subject. A gray midthickness surface was generated by averaging the pial and white matter surfaces. For each subject, cortical surfaces were then registered to the PALS-B12 atlas (Van Essen 2005). The full-resolution (73 730 vertices) PALS-registered mesh was resampled to a coarser mesh of 18 434 [18K] vertices (∼2.5-mm average vertex spacing on the midthickness) that approximates the spatial resolution of the fMRI data.
Figure 8.
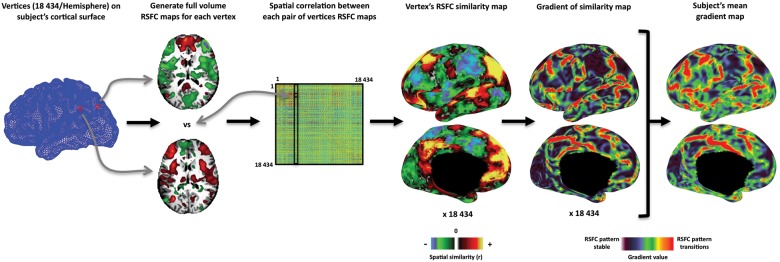
Overview of RSFC-Boundary Mapping. RSFC-Boundary Mapping was implemented on a closed topology. For each hemisphere, an 18 434 (18K) node PALS-registered cortical surface mesh was first generated for the subject. A full-volume RSFC map was computed for each vertex, and the spatial correlation between each pair of vertices RSFC maps was computed to create a correlation matrix (18K × 18K). Each column of this correlation matrix represents the spatial similarity of a given vertex location's RSFC map with the RSFC map of all other vertices and can be represented on the subject's cortical surface. Regions of high transition in the similarity of patterns of RSFC were identified by computing the first spatial derivative of these similarity maps (i.e., spatial gradient magnitude maps). A mean RSFC spatial gradient image for the subject's left hemisphere is computed by averaging across all the spatial gradient maps and depicts the likelihood with which a given location is being identified as an areal boundary.
The time courses of each subject's RSFC BOLD volume were then projected to each subject's 18K PALS-registered cortical surface mesh using trilinear interpolation. Full-volume correlation maps were generated for each surface vertex by correlating their time courses with the time courses of every voxel in the brain (18K vertices × 65 549 voxels). Full-volume correlation maps were used at this stage rather than surface correlation maps in order to retain the contribution of subcortical structures to each surface vertices overall correlation pattern. The similarity of each vertex's correlation map to every other correlation map was then found by computing their spatial correlation. This approach generated an 18K-by-18K correlation matrix, each row of which could be displayed on the surface and where the intensity at each vertex indicated how similar the correlation map at that position was to the reference vertex's correlation map. These 18K spatial similarity images were smoothed along the surface with a Gaussian kernel (FWHM = 6 mm) and then the first spatial derivative was computed across the surface to generate 18K gradient maps. High gradient magnitudes in these maps represent rapid transitions in the underlying full-volume correlation patterns. The 18K gradient maps were combined to generate an average spatial gradient map. The algorithm used for calculating gradients along a cortical surface is a general use Caret function available with Caret 5.65 and has been used to find surface gradients in other data types (Glasser and Van Essen 2011). This average spatial gradient map reflects the likelihood with which each location was identified as representing a point of rapid change in the RSFC maps between 2 adjacent locations of the brain. Finally, to facilitate the identification of sharp edges in the average gradient image for area boundary definition, we employed a nonmaxima suppression procedure by suppressing vertices that were not local maxima with respect to at least 2 pairs of spatially adjacent vertex gradient values in the average gradient image.
Comparison with RSFC-Boundary Mapping Results
RSFC-Snowballing Peak Voxel Locations Fall Within Area Borders Defined by RSFC-Boundary Mapping
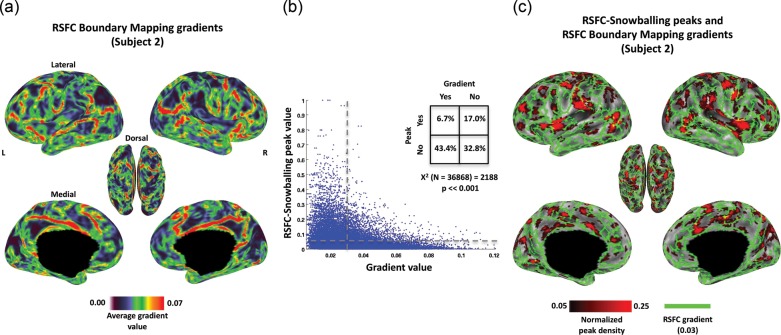
As was observed in the RSFC-Snowballing peak distribution maps, there was a nonuniform distribution of gradient values across subject's RSFC-Boundary Mapping maps (e.g., Fig. 9a; for additional subjects see Supplementary Fig. 3). We compared the peak values identified using RSFC-Snowballing with gradient values identified using RSFC-Boundary Mapping within the same individuals by projecting RSFC-Snowballing peak values to each subject's cortical surface. This comparison was limited to the cortical gray matter, as the implementation of RSFC-Boundary Mapping used here did not extend to subcortical structures or the cerebellum.
Figure 9.
Peaks defined by RSFC-Snowballing are surrounded by gradients defined by RSFC-Boundary Mapping in an illustrative subject. (a) Voxel-wise distribution of mean gradient values defined by RSFC-Boundary Mapping reveal nonuniformity in the average gradient value (i.e., likelihood of an area boundary) across the cortical surface for an individual subject (presented on the subject's native 18K vertex surface mesh). (b) A scatter plot demonstrates the relationship between the 2 independent methods of RSFC area parcellation. As an additional illustration of the correspondence, dashed lines indicate the thresholds applied to compute the chi-square test for independent samples (inset), and the corresponding map of these thresholded values is depicted in (c). Voxels with high peak values as defined by RSFC-Snowballing are largely surrounded by voxels with high gradient values as defined by RSFC-Boundary Mapping, demonstrating that the 2 methods of area parcellation complement one another closely.
Vertices with a higher likelihood of being an area center as defined by RSFC-Snowballing tended not to have a higher likelihood of being an area boundary as defined by RSFC-Boundary Mapping (Fig. 9b). This observation was confirmed by computing Pearson's product–moment correlation coefficient between the vertices of each of the 2 RSFC parcellation maps for each subject. For all subjects, there was a significant negative correlation between their RSFC-Snowballing aggregate peak density values and their RSFC-Boundary Mapping average gradient values (mean of 8 subjects: r = −0.23; range across 8 subjects: r = −0.17 to −0.32, all 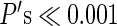 ). The negative relationship was reliably different from 0 across subjects (t(7) = −14.0, P < 0.00001).
). The negative relationship was reliably different from 0 across subjects (t(7) = −14.0, P < 0.00001).
As noted earlier, the thresholding procedure could bias parcellation. For the purposes of illustration, however, a threshold was applied to each map (RSFC-Snowballing aggregate peak density map and RSFC-Boundary Mapping gradient map following nonmaxima suppression) to identify the vertices with the highest peak values (threshold of 0.05 which corresponds to >5% of maximum peak value) and highest gradient values (mean gradient value >0.03), respectively. A chi-square test for independence revealed that vertices identified as having a high likelihood of being a center of an area versus a border between areas came from non-overlapping populations [X2 (1, N = 36868) = 2188, P ≪ 0.001; mean of 8 subjects: X2 (1, N = 36868) = 1777; range across 8 subjects: X2 (1, N = 36868) = 800 to 3006, all P's ≪ 0.001; Figure 9b inset]. Figure 9c depicts the thresholded maps together for an illustrative subject. The majority of the strongest peaks identified using RSFC-Snowballing fall in between the strong gradients identified using RSFC-Boundary Mapping.
While the analysis focused on examining the independence of the highest peak and gradient values by converting each variable into a discrete outcome (i.e., the presence or absence of an area center or area border), and the choice of threshold may seem somewhat arbitrary in this regard, the statistical test was also computed over a range of RSFC-Snowballing threshold values (0.01 of the maximum peak density value to 0.1 in steps of 0.01). For all subjects, across the complete range of tested thresholds, the results of the chi-square tests were consistent and revealed a high degree of nonoverlap in vertices identified as centers by RSFC-Snowballing versus vertices identified as borders by RSFC-Boundary Mapping (all 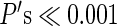 ).
).
Area Centers Defined by RSFC-Snowballing Exhibit Distinct Patterns of RSFC
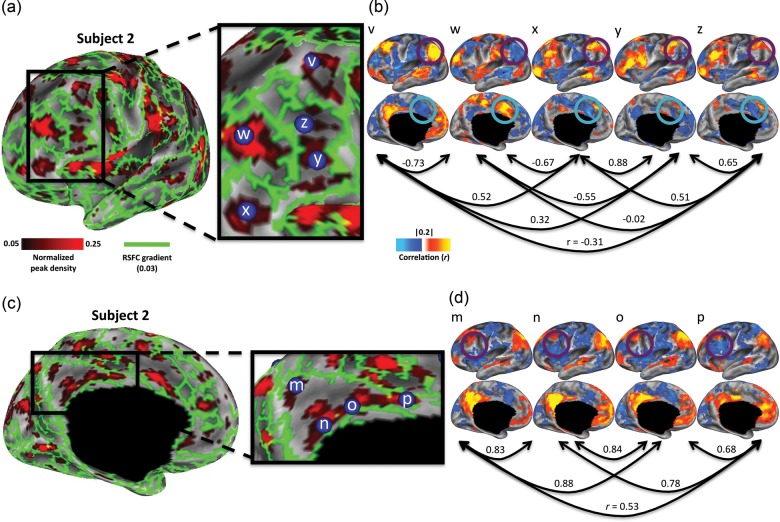
To further illustrate the complementarity of RSFC-Snowballing and RSFC-Boundary Mapping, we zoom in on a portion of the middle and inferior frontal gyrus of the left hemisphere in our illustrative subject (Fig. 10a). A number of strong boundaries defined by RSFC-Boundary Mapping separate the vertices with high peak values defined by RSFC-Snowballing. The local maxima of the RSFC-Snowballing aggregate peak density map were identified (using an unthresholded map), and seed-based voxel-wise resting-state correlation maps were computed for spherical seed ROIs built around each of the local maxima (a 5-mm radius).
Figure 10.
Adjacent peaks identified by RSFC-Snowballing exhibit distinct patterns of resting-state correlations. (a) Focusing on the middle and inferior frontal gyrus of the left hemisphere of a subject reveals the clusters of high peak count as identified by RSFC-Snowballing that are separated by strong gradients as identified by RSFC-Boundary Mapping. (b) Area centers defined by RSFC-Snowballing exhibit distinct patterns of resting-state correlations. Local maxima of the RSFC-Snowballing aggregate peak density map have dissimilar resting-state correlation maps (arrows denote the spatial correlation of the RSFC maps) and different patterns of resting-state correlations with specific regions (e.g., the angular gyrus highlighted with a purple circle and the anterior cingulate gyrus highlighted with a teal circle). (c) RSFC-Snowballing identifies 4 area centers that are not separated by strong gradients in the medial parietal cortex of a subject, providing evidence that RSFC-Snowballing may be able to identify unique parcellations of brain areas. (d) The area centers identified in the medial parietal cortex exhibit subtle, yet distinct patterns of resting-state correlation (e.g., the lateral frontal cortex highlighted with a purple circle), providing evidence that they may be functionally distinct areas. RSFC-Snowballing and RSFC-Boundary Mapping parcellation maps are depicted as thresholded maps as in Figure 9c, to facilitate viewing.
The ROIs separated by strong gradients typically exhibit different patterns of functional connectivity, suggesting that they are portions of distinct cortical areas (Fig. 10b). For example, the ROI in the dorsal portion of the middle frontal gyrus (ROI “v”: −43 19 46) exhibits positive resting-state correlations with regions of the default network, including the pCC, angular gyrus (highlighted with a purple circle), and medial prefrontal cortex. An adjacent ROI (ROI “w”: −39 39 21), located more anteriorly on the middle frontal gyrus, sits on the other side of a set of strong gradients (i.e., the putative boundary between 2 areas). This region's pattern of resting-state correlations is quite different: Positive correlations are observed with the anterior cingulate gyrus (highlighted with a teal circle), supramarginal gyrus, and posterior insula cortex. Moreover, this anterior middle frontal gyrus ROI (“w”) exhibit strong negative correlations with regions positively correlated with the dorsal middle frontal gyrus ROI. To confirm these qualitative observations, we computed spatial correlations between each pair of ROIs' RSFC maps. Adjacent ROI pairs defined by RSFC-Snowballing that are separated by strong gradients tend to have very dissimilar RSFC maps.
RSFC-Snowballing Provides Unique Information to Inform RSFC-Boundary Mapping
The presence of a non-perfect negative correlation between RSFC-Snowballing and RSFC-Boundary Mapping parcellations (Fig. 9b) suggests that the methods are not completely redundant. RSFC-Snowballing and RSFC-Boundary Mapping may each provide unique cortical parcellation information in certain locations. This observation may be due to a practical as opposed to a conceptual limitation of each of the methods—operationally, the thresholds that are most useful for a given method of parcellation may miss distinctions in another method of parcellation. For example, adjacent areas that share very similar patterns of RSFC would have a weak boundary between them, yet the centers might be highlighted by RSFC-Snowballing.
In Figure 10c, we focus on a medial portion of the posterior parietal cortex to demonstrate this feature of RSFC-Snowballing. Four area centers identified by RSFC-Snowballing were enclosed by a strong border identified by RSFC-Boundary Mapping with no obvious border between them (based on the threshold that revealed prominent and reasonable borders throughout other locations in the cortex). Three of these locations fall along the posterior- and midcingulate cortex, and the fourth location was in the precuneus. Although the 4 ROIs exhibit similar patterns of RSFC connectivity (Fig. 10d), a specific feature of their patterns of functional connectivity provides evidence that they may be distinct. In particular, the presence of a negative RSFC with the lateral frontal cortex serves to distinguish between the ROIs (highlighted with a purple circle). It is important to note that the observed pattern would not be present if the 4 ROIs were all subclusters within a single area whose correlation pattern progressively changed with a march toward its boundary. The presence of a strong negative correlation with the lateral frontal cortex is only observed in the most inferior (ROI “n”: 0–47 36) and the most anterior (ROI “o”: 0–16 41) of the 4 parietal regions. It is also worth noting that the spatial correlation between the RSFC maps is opposite to that which would be observed if the spatial similarity between the maps were related to geometric distance between the ROIs (e.g., the correlation maps of ROIs “n” and “p” are more similar to one another than ROIs “o” and “p”).
Methods/Results 3: Reliability of RSFC Parcellation Within and Between Subjects
To test whether the RSFC-Snowballing provides similar parcellation across 2 independent scans collected from the same subject, the consistency of the RSFC-Snowballing aggregate peak density maps was examined across multiple scan sessions that were separated by multiple intervening days. Each of our subjects was scanned on 2 separate days with an average interval of 20 days intervening between the 2 scan sessions (range: 7–53 days). As noted earlier, data from one subject's “day 2” resting state scan had an insufficient amount of data remaining following RSFC preprocessing.
Reliability Results
RSFC-Snowballing Parcellations Exhibit Considerably Greater Similarity Within an Individual Scanned Across Multiple Days than Across Individuals
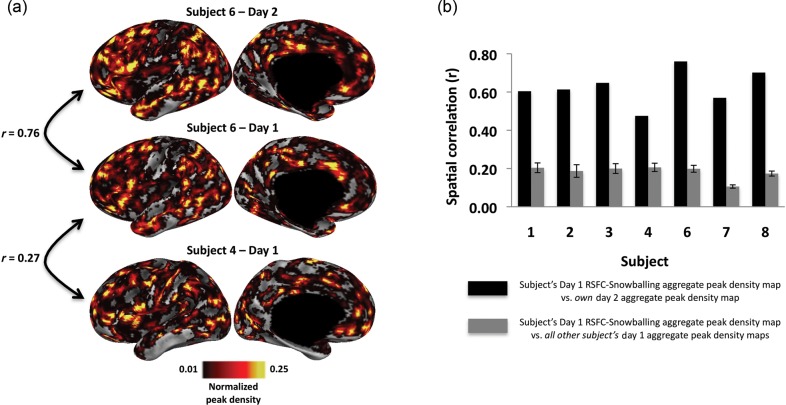
Subjects demonstrated relatively high spatial similarity between their day 1/day 2 RSFC-Snowballing parcellation maps. For example, comparing the RSFC-Snowballing aggregate peak density maps for a subject scanned on 2 separate sessions with 13 intervening days between the scans revealed a high degree of similarity (Fig. 11; mean spatial correlation between each subject's day 1 and day 2 RSFC-Snowballing aggregate peak density maps = 0.62, range of all subjects: r = 0.48–0.76).
Figure 11.
RSFC-Snowballing parcellation maps are more similar across independent scans of the same individual than across individuals. (a) An illustrative subject demonstrates higher spatial similarity between their day 1/day 2 RSFC-Snowballing aggregate peak density maps relative to the spatial similarity between their day 1 aggregate peak density map and the day 1 aggregate peak density map of another subject. (b) Spatial similarity for each subject's day 1 RSFC-Snowballing aggregate peak density maps relative to their day 2 aggregate peak density map is higher than the average of the similarity between each subject's day 1 aggregate peak density map relative to all other subject's day 1 aggregate peak density maps (error bars denote standard error of the mean).
While the day 1/day 2 similarities in RSFC-Snowballing parcellation maps for subjects were relatively high, the spatial similarity between subjects' parcellation maps was lower (mean spatial correlation between each subject's day 1 RSFC-Snowballing aggregate peak density map and all other subject's day 1 RSFC-Snowballing aggregate peak density maps: r = 0.18, range: r = 0.11–0.21). These qualitative observations were confirmed by a direct comparison: The within-subject day 1/day 2 spatial similarity was significantly greater than the between-subject spatial similarity (t(6) = 12.5, P < 0.0001). Comparable patterns were observed when comparing the subject's RSFC-Boundary Mapping average gradient maps within and across subjects (Supplementary Fig. 4). These observations collectively highlight the variability in the spatial distribution of RSFC area parcellation across subjects despite being registered to a common space.
RSFC-Snowballing Parcellation Maps Exhibit Several Features that Are Consistent Across Subjects
While the day 1/day 2 comparison of the RSFC parcellation maps revealed greater variability between subjects than within subjects, many of the voxels with the highest peak values seemed to be in similar locations across subjects (see Supplementary Fig. 1). To statistically examine the reliability of the RSFC-Snowballing parcellation maps across subjects, RSFC data from an additional 40 subjects (20 females, mean age = 25 years, age range = 21–30 years) were processed, parcellated with RSFC-Snowballing, and analyzed using a voxel-wise one-sample t-test across subject's average RSFC-Snowballing aggregate peak density maps [One-sample t-tests were computed across the median of the average RSFC-Snowballing aggregate peak density map. This test would normally be performed against the mean of the average map, but since the distribution of peak density values is non-normally distributed (there is a slight skew toward higher peak values), we performed the statistical test against the median as it reflects a more accurate metric of central tendency of the dataset.]. The observed P-value distribution was used to calculate a FDR threshold, controlling for the expected proportion of false positives among suprathreshold locations (Genovese et al. 2002). Only voxels associated with a P-value <0.05 are reported.
Figure 12 depicts the locations exhibiting consistent RSFC-Snowballing parcellation features across subjects. The presence or absence of significance at a particular location is related to variability in the peak density value, anatomical location, or a combination of these factors. The locations of the voxels with the reliable aggregate peak density map values include a number of the area centers highlighted earlier (i.e., regions along the middle frontal gyrus and posterior regions of the medial parietal cortex). Other locations with the high peak density value consistently revealed by RSFC-Snowballing include regions of the anterior insular cortex, angular gyrus, supramarginal gyrus, IFG, and anterior cingulate/medial-superior frontal cortex.
Figure 12.
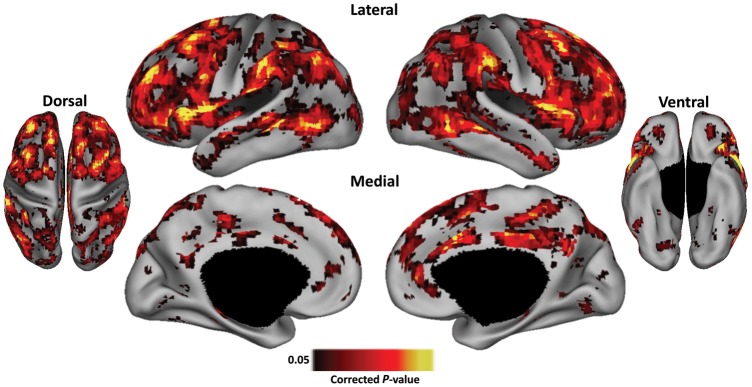
RSFC-Snowballing aggregate peak density maps exhibit consistent features across subjects. Statistical map revealing locations that exhibit reliable peak density values across the RSFC-Snowballing parcellation maps of 40 subjects (P < 0.05, corrected for FDR). Hot values represent peak values statistically higher than the overall distribution. Statistical maps are depicted on the partially inflated PALS surface (Van Essen 2005).
Methods/Results 4: Comparison with Area Centers Defined From Task-Evoked Data
If RSFC-Snowballing parcellation reveals functionally plausible area centers, then area centers defined by task-evoked activity should overlap with area centers defined by RSFC-Snowballing. Similarly, area centers defined by task-evoked activity should exhibit higher RSFC-Snowballing peak counts than would be expected by chance.
A large battery of task-evoked data was not available for each of our subjects. As a preliminary test of overlapping parcellation, we used an alternative strategy in which area centers were first defined by a large meta-analysis of task-evoked data. For this purpose, we used the same locations identified in the task-evoked meta-analysis described earlier (for locations, see Fig. 4a).
Comparison With Task-Evoked Data Results
Task-Defined Area Centers Exhibit Relatively Higher RSFC-Snowballing Peak Counts
The task-defined method of area parcellation is quite distinct from the subject-specific methods of area parcellation highlighted throughout the present report. Task-evoked area centers were derived from a meta-analysis across many subjects performing many different tasks, and the data for this meta-analysis were collected on a different scanner than the scanner on which RSFC data were collected.
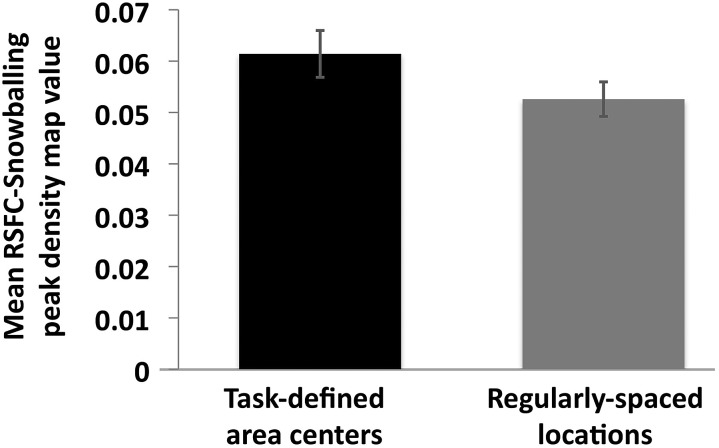
Despite the differences in the methods of area parcellation noted above, we directly quantified the agreement between the 2 methods of parcellation by determining whether task-defined area centers exhibited higher RSFC-Snowballing peak counts than would be observed at random locations. Analysis was conducted using subject's average day 1/day 2 aggregate peak density maps (with the exception the one subject for which only a day 1 RSFC-Snowballing aggregate peak density map was available). The mean RSFC-Snowballing aggregate peak density map value at each task-defined area center location was computed for every subject. For comparison, we also evaluated the mean RSFC-Snowballing aggregate peak density map value at each location in the regularly spaced initialization location set since these locations were generated with no reference to task activity. Across subjects, the mean peak value was significantly greater at task-defined area centers than at the surface-grid locations (t(7) = 3.81, P = 0.007; Fig. 13). Voxels that were identified as area centers through a meta-analysis of task-evoked data across many subjects had higher RSFC-Snowballing peak values across our individual subjects' aggregate peak density maps than voxels distributed regularly across the cortical surface.
Figure 13.
Task-defined area centers (obtained from a meta-analysis of task-evoked data in an independent collection of subjects) have significantly greater mean RSFC-Snowballing peak values than regularly spaced locations. The mean RSFC-Snowballing peak density value was computed across all task-defined area center locations and regularly-spaced locations for each subject's aggregate peak density map independently. Error bars denote the standard error of the mean.
As a second set of analyses to determine the overlap between area centers defined by RSFC-Snowballing and task-defined data, we performed chi-square tests for independence using every subject's RSFC-Snowballing aggregate peak density maps (average of day 1/day 2 data). Each subject's RSFC-Snowballing aggregate peak density maps were first thresholded to identify voxels identified as area centers, and the incidence of overlap for the task-defined and regular-grid locations was determined. As noted earlier, a thresholding procedure can bias area detection, so we performed this test over a range of thresholds (0.01 of the maximum peak density value to 0.1 in steps of 0.01). The chi-square tests revealed that the task-defined locations had a greater proportion of overlap with area centers defined by RSFC-Snowballing than the regular-grid locations. This was the case for every subject across all thresholds [range across subjects X2 = 4.95–493.67, all P's < 0.0001 with one P < 0.05 (with Yate's correction for continuity)]. Collectively, these observations provide important initial evidence that peaks defined by RSFC-Snowballing converge with parcellation defined by task-evoked functional activity in individual subjects.
Discussion
RSFC-Snowballing can parcellate an individual's brain into cortical area centers (or specifically, the interiors of areas) and subdivisions of subcortical structures with relatively high reliability. This parcellation complemented the parcellation generated with RSFC-Boundary Mapping, but also provided additional distinctions across cortical and subcortical structures not revealed by RSFC-Boundary Mapping. Importantly, estimates of area centers defined by RSFC-Snowballing showed some overlap with peaks defined by meta-analysis of task-evoked data, providing preliminary evidence that the RSFC parcellations are functionally meaningful.
Distinct brain areas often have distinct properties related to function, architectonics, and connectivity. By focusing on transitions in these properties, boundary mapping techniques can identify borders between adjacent brain areas. While this general approach has been applied across various modalities for both invasive (e.g., Brodmann 1909; Vogt and Vogt 1919; Tootell and Taylor 1995; Amunts et al. 1999; Caspers et al. 2006) and noninvasive brain parcellations (e.g., Johansen-Berg et al. 2004; Cohen et al. 2008; Glasser and Van Essen 2011; Caspers et al. 2013), there are important limitations to the approach. The first limitation is practical: Detecting transitions in a given property is highly contingent on the measuring tool's sensitivity to that property and the thresholds used to define the presence or absence of a border; in general, more subtle transitions are going to be more difficult to detect. A second, greater, limitation is conceptual: Adjacent areas need not exhibit distinct fingerprints for a given property. Exclusively relying on the transitions of a single property (e.g., architectonics or connectivity) for the purposes of parcellation may miss divisions between areas that may be distinguishable by different properties or methods. For these reasons, it is necessary to develop complementary techniques and methods for parcellation, such as RSFC-Snowballing.
Convergence of RSFC-Snowballing, RSFC-Boundary Mapping, and Parcellation from Task-Evoked Data
RSFC-Snowballing is conceptually complementary to boundary mapping techniques in that it attempts to identify the strongest estimates of “nonborders”', or area centers. To test the complementary nature of RSFC-Snowballing and boundary mapping using patterns of RSFC (RSFC-Boundary Mapping), we first described a method to extend the RSFC-Boundary Mapping method across the whole cortical surface and then directly compared the parcellation derived from this method with RSFC-Snowballing parcellation in the same subjects. Surface vertices that exhibited high aggregate peak density map values (i.e., more likely to be area centers) were less likely to exhibit high gradient map values (i.e., less likely to be an area boundary; Fig. 9). Closer examination confirmed that many adjacent area centers identified by RSFC-Snowballing were separated by strong borders identified by RSFC-Boundary Mapping, and exhibited highly distinct patterns of RSFCs. The non-perfect negative correlation suggests that the 2 methods are more than just complementary.
We highlighted some locations where RSFC-Snowballing provided informative parcellation to complement RSFC-Boundary Mapping. For example, RSFC-Snowballing revealed putative divisions in cortical regions surrounded by a strong RSFC-Boundary Mapping gradient (i.e., Fig. 10c). The failure to find contiguous borders between the RSFC-Snowballing parcellations within the medial parietal cortex may be related to the fact that each method (RSFC-Boundary Mapping and RSFC-Snowballing) is sensitive to different acquisition, preprocessing, and analysis choices of RSFC. It is worth noting that similar divisions of the posterior medial parietal cortex have been reported using postmortem histological analysis of cytoarchitecture (Vogt et al. 1995), autoradiography (Palomero-Gallagher et al. 2009), and connectivity (Beckmann et al. 2009; Leech et al. 2011) garnering additional support for our RSFC-Snowballing based parcellation of this part of the brain. We were also able to leverage RSFC-Snowballing to parcellate structures currently inaccessible by the present RSFC-Boundary Mapping methods. Here, RSFC-Boundary Mapping was restricted to the cortical surface, although in principle, gradients could be applied to subcortical gray matter structures as well. The ability of RSFC-Snowballing to easily operate across the volume of the brain makes it useful for parcellating structures not in the cerebral cortex, such as the cerebellum and various subcortical nuclei (Fig. 3).
In addition to demonstrating the complementarity of RSFC parcellations using the different methods (RSFC-Snowballing and RSFC-Boundary Mapping), it is important to both examine convergence between parcellations derived from fundamentally different data types and also to determine whether the RSFC parcellations are functionally meaningful. Although a large battery of task-evoked data was not available for each of our subjects, task-defined area centers from a large meta-analysis of task-evoked data across subjects were available. We recognize that this comparison is not ideal—area centers defined over groups of subjects performing different tasks need not correspond to an individual subject's area centers. Nevertheless, our preliminary test revealed evidence for convergence in the methods of area parcellation: Area centers defined by task-evoked activity exhibited significantly higher RSFC-Snowballing peak counts in individual subject's RSFC-Snowballing peak density maps than that observed at random locations. Subsequent work will be necessary to explore the compatibility between the methods of parcellation in greater detail and using subject-specific task-defined area parcellation.
Limitations of RSFC Area Parcellations
There are at least several limitations to both the methods described in the present report, and parcellating the brain with RSFC in general. First, while RSFC-Snowballing focuses on identifying the centers/interiors of areas, it may be poorly suited to identifying the entire extent of an area. Parcellations that fail to include areal extent will likely result in an under-representation of the anatomy and function of areas. Given that RSFC-Boundary Mapping is suited to delineating area borders, yet often identifies discontinuous borders, one appealing idea would be to combine approaches, where possible, and leverage each method's strength to create a more complete and accurate parcellation.
Second, there may be locations in the brain where RSFC, in general, may either fail to identify areal divisions or reveal additional features, independent of the specific method used (i.e., RSFC-Snowballing or RSFC-Boundary Mapping). Typical gradient-echo fMRI scans cannot cover the whole brain at usable signal-to-noise ratio because of signal dropout from magnetic field inhomogeneities created by air-filled sinuses located close to the brain. As such, RSFC-based parcellation will naturally fail in these locations. There is also strong reason to suspect that RSFC may reveal additional features in areas that exhibit the topographic mapping of a sensory surface. For example, RSFC has revealed that visually responsive areas exhibit the patterns of resting-state correlations that follow the lines of visual eccentricity (i.e., foveal vs. eccentric) in addition or as opposed to the areal divisions (i.e., V1 vs. V2; e.g., Vincent et al. 2007). Accordingly, RSFC parcellation in these areas may follow retinotopic divisions in addition to areal divisions. [Along similar lines, distinct patterns of connectivity for face and body subregions (Johnson et al. 1996; Matelli et al. 1998; Tanne-Gariepy et al. 2002) in the somatosensory and motor cortex may result in additional RSFC parcellation than that obtained with architectonic parcellation.]
Last, we anticipate that technological improvements in EPI and improved image processing techniques will reduce the amount and extent of smoothing needed during both preprocessing and the various steps for parcellation methods we've described, thereby enabling visualization of RSFC correlations and parcellations with better spatial resolution and correspondence both within modality and across modality.
In addition to boundary detection and peak finding (e.g., using RSFC-Snowballing or analysis of task-evoked data), other methods of brain analysis are frequently used to examine brain organization. Clustering and source separation techniques can be applied to RSFC time series to identify the groups of voxels that share similar temporal covariance (e.g., Smith et al. 2009; Mumford et al. 2010; Doucet et al. 2011; Power et al. 2011; Yeo et al. 2011). Importantly, as the aim of a clustering tool is to aggregate similar elements and to separate them from other elements, they neither mandate spatially contiguous parcels nor ensure that parcels with similar properties will be separated from one another. For example, depending on the total number of components returned (in the case of an independent component analysis decomposition) or the correlation threshold (in the case of community detection in graph theoretic analysis of RSFC), a single cluster can include multiple disparate areas that are distributed throughout the brain (e.g., a “default system” component or community). Conversely, functionally distinct, but spatially adjacent, areas with similar properties can often be aggregated into a single cluster (e.g., a “visual system” component or community). As such, while clustering tools might be able to parcellate some brain areas and areal borders when applied correctly, it is important to understand their limitations.
Several recently developed RSFC methods have focused on identifying the voxels that are the network “hubs” or locations of greatest “global connectivity” (e.g., Buckner et al. 2009; Cole et al. 2010; Tomasi and Volkow 2011). We have previously argued how using voxels as nodes in a brain network misrepresents the underlying neurobiology and can lead to mischaracterization of basic network properties, such as clustering, path length, and hub identity (Wig et al. 2011). It is quite possible that many of the locations identified as area centers by RSFC-Snowballing converge with locations identified as points of high voxel-wise connectivity by these other methods, and future work will assess the degree of overlap between the different methods. We argue here, however, that the value of this enterprise is not necessarily to identify the location of highest “global connectivity” or “hubs”, but rather to parcellate areas across the brain based on identifying locations where connectivity is relatively greater (e.g., in the interior of an area as compared to at it's border). As conveyed throughout the present report, we hypothesize that these densely connected (correlated) locations are strong candidates for cortical area centers and subdivisions of subcortical structures.
It is not surprising that a single method or modality is unable to parcellate all cortical and subcortical divisions. Furthermore, for any given parcellation method, there are multiple choices that must be made in deciding what constitutes a meaningful division (e.g., the correct threshold or statistical test for deciding there is an area center or boundary). As mentioned earlier, the “ideal” methods and parameter choices may be different for different brain areas. While we have made some decisions for the purposes of demonstration, it is not necessary that the choices that we made were optimal or that the optimal choice will be obvious by focusing on a single method or modality of parcellation. For these reasons, similar to parcellation of brain areas in other species (e.g., Felleman and Van Essen 1991), parcellation of human brain areas will be a product of the convergence of multiple analysis approaches and modalities. The development of novel strategies and approaches that complement previous methods while simultaneously overcoming some of their conceptual and methodological limitations will aid noninvasive parcellation of the brain. To this end, although we have focused our efforts on demonstrating how snowball sampling can be applied to RSFC, this approach could also be applied to alternative modalities of brain imaging (e.g., diffusion imaging).
Future Directions
The area parcellation derived from RSFC-Snowballing was highly invariant across starting location sets and a range of parameter settings, providing evidence for its robustness in identifying area centers. Subjects' RSFC parcellations were fairly similar across repeat sessions of scanning (both RSFC-Snowballing and RSFC-Boundary Mapping), indicating that the area parcellation defined by RSFC is relatively stable. Interestingly, the spatial correlation between RSFC parcellation maps were comparable with the cross-session reliability that has been observed in measurements of single RSFC connections and correlation maps within individual subjects (e.g., Damoiseaux et al. 2006; Shehzad et al. 2009). It will be important to determine whether the residual variance relates to underlying variability in RSFC measurement over time, or signal noise that persists.
While many of the area centers that exhibited high RSFC-Snowballing peak values and high RSFC-Boundary Mapping gradient values overlapped across subjects, there was considerable variability between subject's area parcellations. This observation is consistent with prior reports that have demonstrated within-cohort variability in brain area organization as defined by task-evoked activity (e.g., Dougherty 2003; Fedorenko et al. 2010; Sabuncu et al. 2010), architectonics (e.g., Amunts et al. 2004; Caspers et al. 2006), and anatomical connectivity (e.g., Johansen-Berg et al. 2005). Between-subject registration of RSFC parcellation maps may benefit from alternate atlas registration techniques, such as those that focus on surface-curvature (Fischl, Sereno, Tootell, Dale 1999) or nonlinear registration (Klein et al. 2009). As between-subject differences are more likely to be pronounced across different cohorts of subjects (e.g., across the lifespan and in patient populations), accurately localizing and comparing cognitive operations across individuals and groups of individuals motivate the need for subject-specific brain parcellation. Fortunately, RSFC can be extracted with minimal effort from most of the populations, allowing RSFC-Snowballing and RSFC-Boundary Mapping to parcellate and compare brain areas across individual brains that span ranges of age, mental health, and even species.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
This work was supported by an Institute for Aging Post-doctoral Fellowship from the Canadian Institute for Health Research (G.S.W.), P50NS006833 and P30NS048056 (A.Z.S.), NIH R01HD057076 (B.L.S.), NIH NS61144, and a McDonnell Foundation Collaborative Action Award (S.E.P.), and the Human Connectome Project (1U54MH091657) from the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research. Funding to pay the Open Access publication charges for this article was provided by a McDonnell Foundation Collaborative Action Award (S.E.P.).
Supplementary Material
Notes
The authors thank Malcolm Tobias and the Center for High Performance Computing at Washington University School of Medicine for invaluable technical assistance, and David Van Essen and 2 anonymous reviewers for helpful comments on a previous version of this manuscript. Conflict of Interest: None declared.
References
- Albert R, Jeong H, Barabasi A-L. Internet: diameter of the world-wide web. Nature. 1999;401:130–131. [Google Scholar]
- Allman JM, Kaas JH. A representation of the visual field in the caudal third of the middle temporal gyrus of the owl monkey (Aotus trivirgatus) Brain Res. 1971;31:85–105. doi: 10.1016/0006-8993(71)90635-4. [DOI] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Burgel U, Mohlberg H, Uylings HB, Zilles K. Broca's region revisited: cytoarchitecture and intersubject variability. J Comp Neurol. 1999;412:319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Amunts K, Weiss PH, Mohlberg H, Pieperhoff P, Eickhoff S, Gurd JM, Marshall JC, Shah NJ, Fink GR, Zilles K. Analysis of neural mechanisms underlying verbal fluency in cytoarchitectonically defined stereotaxic space–the roles of Brodmann areas 44 and 45. Neuroimage. 2004;22:42–56. doi: 10.1016/j.neuroimage.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Barnes KA, Nelson SM, Cohen AL, Power JD, Coalson RS, Miezin FM, Vogel AC, Dubis JW, Church JA, Petersen SE, et al. Parcellation in left lateral parietal cortex is similar in adults and children. Cereb Cortex. 2012;22:1148–1158. doi: 10.1093/cercor/bhr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M, Johansen-Berg H, Rushworth MFS. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J Neurosci. 2009;29:1175–1190. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Jenkinson M, Robson MD, Smith SM, Johansen-Berg H. A consistent relationship between local white matter architecture and functional specialisation in medial frontal cortex. Neuroimage. 2006;30:220–227. doi: 10.1016/j.neuroimage.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende lokalisationslehre der grosshirnrinde in ihren prinzipien dargestellt auf grund des zellenbaues. Leipzig: J.A. Barth; 1909. [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers S, Geyer S, Schleicher A, Mohlberg H, Amunts K, Zilles K. The human inferior parietal cortex: cytoarchitectonic parcellation and interindividual variability. Neuroimage. 2006;33:430–448. doi: 10.1016/j.neuroimage.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Caspers S, Schleicher A, Bacha-Trams M, Palomero-Gallagher N, Amunts K, Zilles K. Organization of the human inferior parietal lobule based on receptor architectonics. Cereb Cortex. 2013;23:615–628. doi: 10.1093/cercor/bhs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W. Identifying the brain's most globally connected regions. Neuroimage. 2010;49:3132–3148. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with cortical surface reconstruction: a linear approach. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Devlin JT, Poldrack RA. In praise of tedious anatomy. Neuroimage. 2007;37:1033–1041. doi: 10.1016/j.neuroimage.2006.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet G, Naveau M, Petit L, Delcroix N, Zago L, Crivello F, Jobard G, Tzourio-Mazoyer N, Mazoyer B, Mellet E, et al. Brain activity at rest: a multiscale hierarchical functional organization. J Neurophysiol. 2011;105:2753–2763. doi: 10.1152/jn.00895.2010. [DOI] [PubMed] [Google Scholar]
- Dougherty RF, Koch VM, Brewer AA, Fischer B, Modersitzki J, Wandell BA. Visual field representations and locations of visual areas V1/2/3 in human visual cortex. J Vis. 2003;3:586–598. doi: 10.1167/3.10.1. [DOI] [PubMed] [Google Scholar]
- Fedorenko E, Hsieh PJ, Nieto-Castanon A, Whitfield-Gabrieli S, Kanwisher N. New method for fMRI investigations of language: defining ROIs functionally in individual subjects. J Neurophysiol. 2010;104:1177–1194. doi: 10.1152/jn.00032.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano E, Kim DS, Di Salle F, van de Moortele PF, Ugurbil K, Goebel R. Mirror-symmetric tonotopic maps in human primary auditory cortex. Neuron. 2003;40:859–869. doi: 10.1016/s0896-6273(03)00669-x. [DOI] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Gennari F. Italy. Parma: Regio Typographeo; 1782. Francisci Gennari Parmensis Medicinae Doctoris Collegiati de Peculiari Structura Cerebri Nonnullisque Eius Morbis-Paucae Aliae Anatom. Observat. Accedunt. [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Geyer S, Weiss M, Reimann K, Lohmann G, Turner R. Microstructural parcellation of the human cerebral cortex–from Brodmann's post-mortem map to in vivo mapping with high-field magnetic resonance imaging. Front Hum Neurosci. 2011;5:19. doi: 10.3389/fnhum.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman L. Snowball sampling. Ann Math Stat. 1961;32:148–170. [Google Scholar]
- Goulas A, Uylings HB, Stiers P. Unravelling the intrinsic functional organization of the human lateral frontal cortex: a parcellation scheme based on resting state fMRI. J Neurosci. 2012;32:10238–10252. doi: 10.1523/JNEUROSCI.5852-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover JE, Strick PL. The organization of cerebellar and basal ganglia outputs to primary motor cortex as revealed by retrograde transneuronal transport of herpes simplex virus type 1. J Neurosci. 1999;19:1446–1463. doi: 10.1523/JNEUROSCI.19-04-01446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K, Oishi K, Zhang J, Wakana S, Yoshioka T, Zhang W, Akhter KD, Li X, Huang H, Jiang H, et al. Mapping of functional areas in the human cortex based on connectivity through association fibers. Cereb Cortex. 2009;19:1889–1895. doi: 10.1093/cercor/bhn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc Natl Acad Sci USA. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Sillery E, Ciccarelli O, Thompson AJ, Smith SM, Matthews PM. Functional-anatomical validation and individual variation of diffusion tractography-based segmentation of the human thalamus. Cereb Cortex. 2005;15:31–39. doi: 10.1093/cercor/bhh105. [DOI] [PubMed] [Google Scholar]
- Johnson PB, Ferraina S, Bianchi L, Caminiti R. Cortical networks for visual reaching: physiological and anatomical organization of frontal and parietal lobe arm regions. Cereb Cortex. 1996;6:102–119. doi: 10.1093/cercor/6.2.102. [DOI] [PubMed] [Google Scholar]
- Kahnt T, Chang LJ, Park SQ, Heinzle J, Haynes JD. Connectivity-based parcellation of the human orbitofrontal cortex. J Neurosci. 2012;32:6240–6250. doi: 10.1523/JNEUROSCI.0257-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Glass TG, Lankipalli BR, Downs H, Mayberg H, Fox PT. A modality-independent approach to spatial normalization of tomographic images of the human brain. Hum Brain Mapp. 1995;3:209–223. [Google Scholar]
- Leech R, Kamourieh S, Beckmann CF, Sharp DJ. Fractionating the default mode network: distinct contributions of the ventral and dorsal posterior cingulate cortex to cognitive control. J Neurosci. 2011;31:3217–3224. doi: 10.1523/JNEUROSCI.5626-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Jbabdi S, Sallet J, O'Reilly JX, Croxson PL, Olivier E, Noonan MP, Bergmann C, Mitchell AS, Baxter MG, et al. Diffusion-weighted imaging tractography-based parcellation of the human parietal cortex and comparison with human and macaque resting-state functional connectivity. J Neurosci. 2011;31:4087–4100. doi: 10.1523/JNEUROSCI.5102-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matelli M, Govoni P, Galletti C, Kutz DF, Luppino G. Superior area 6 afferents from the superior parietal lobule in the macaque monkey. J Comp Neurol. 1998;402:327–352. [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL. Characterizing the hemodynamic response: Effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. NeuroImage. 2000;11:735–759. doi: 10.1006/nimg.2000.0568. [DOI] [PubMed] [Google Scholar]
- Mugler JP, III, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15:152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- Mumford JA, Horvath S, Oldham MC, Langfelder P, Geschwind DH, Poldrack RA. Detecting network modules in fMRI time series: a weighted network analysis approach. Neuroimage. 2010;52:1465–1476. doi: 10.1016/j.neuroimage.2010.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Dosenbach NU, Cohen AL, Wheeler ME, Schlaggar BL, Petersen SE. Role of the anterior insula in task-level control and focal attention. Brain Struct Funct. 2010;214:669–680. doi: 10.1007/s00429-010-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero-Gallagher N, Vogt BA, Schleicher A, Mayberg HS, Zilles K. Receptor architecture of human cingulate cortex: evaluation of the four-region neurobiological model. Hum Brain Mapp. 2009;30:2336–2355. doi: 10.1002/hbm.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. Function-based intersubject alignment of human cortical anatomy. Cereb Cortex. 2010;20:130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe R, Brett M, Kanwisher N. Divide and conquer: a defense of functional localizers. Neuroimage. 2006;30:1088–1096. doi: 10.1016/j.neuroimage.2005.12.062. ; discussion 1097–1099. [DOI] [PubMed] [Google Scholar]
- Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci USA. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Ségonne F, Grimson E, Fischl B. A genetic algorithm for the topology correction of cortical surfaces. Inf Process Med Imaging. 2005;19:393–405. doi: 10.1007/11505730_33. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ, Churchland PS. Brain and cognition. In: Posner M, editor. Foundations of cognitive science. Cambridge: MIT Press; 1989. p. 888. [Google Scholar]
- Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, Rosen BR, Tootell RB. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, et al. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE. Common blood flow changes across visual tasks: II. Decreases in cerebral cortex. J Cogn Neurosci. 1997;9:648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, et al. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AZ. Difference image vs. ratio image error function forms in PET-PET realignment. In: Myer R, Cunningham VJ, Bailey DL, Jones T, editors. Quantification of brain function using PET. San Diego, CA: Academic Press; 1996. pp. 131–137. [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow KM, Braver TS, Snyder AZ, Speer NK, Zacks JM. Reliability of functional localization using fMRI. Neuroimage. 2003;20:1561–1577. doi: 10.1016/s1053-8119(03)00436-1. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers, Inc; 1988. [Google Scholar]
- Tanne-Gariepy J, Boussaoud D, Rouiller EM. Projections of the claustrum to the primary motor, premotor, and prefrontal cortices in the macaque monkey. J Comp Neurol. 2002;454:140–157. doi: 10.1002/cne.10425. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Mori S, Amunts K, Zilles K. Towards multimodal atlases of the human brain. Nat Rev. 2006;7:952–966. doi: 10.1038/nrn2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Functional connectivity hubs in the human brain. Neuroimage. 2011;57:908–917. doi: 10.1016/j.neuroimage.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RBH, Taylor JB. Anatomical evidence for MT and additional cortical visual areas in humans. Cereb Cortex. 1995;1:39–55. doi: 10.1093/cercor/5.1.39. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Maunsell JH, Bixby JL. The middle temporal visual area in the macaque: myeloarchitecture, connections, functional properties and topographic organization. J Comp Neurol. 1981;199:293–326. doi: 10.1002/cne.901990302. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K. The future of the human connectome. Neuroimage. 2012;62:1299–1310. doi: 10.1016/j.neuroimage.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anesthetized monkey brain. Nature. 2007;447:46–47. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Nimchinsky EA, Vogt LJ, Hof PR. Human cingulate cortex: surface features, flat maps, and cytoarchitecture. J Comp Neurol. 1995;359:490–506. doi: 10.1002/cne.903590310. [DOI] [PubMed] [Google Scholar]
- Vogt O, Vogt C. Allgemeine Ergebnisse unserer Hirnforschung. J Psychol Neurol. 1919;25:273–462. [Google Scholar]
- Wandell BA, Dumoulin SO, Brewer AA. Visual field maps in human cortex. Neuron. 2007;56:366–383. doi: 10.1016/j.neuron.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Wasserman S, Faust K. Social network analysis: methods and applications. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- Wig GS, Grafton ST, Demos KE, Wolford GL, Petersen SE, Kelley WM. Medial temporal lobe BOLD activity at rest predicts individual differences in memory ability in healthy young adults. Proc Natl Acad Sci USA. 2008;105:18555–18560. doi: 10.1073/pnas.0804546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Ann N Y Acad Sci. 2011;1224:126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centenary of Brodmann's map–conception and fate. Nat Rev. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.