Abstract
IgA nephropathy (IgAN) represents the leading cause of kidney failure among East Asian populations and the most frequent form of primary glomerulonephritis among Europeans. Patients with IgAN develop characteristic IgA1-containing immune complexes that deposit in the glomerular mesangium, producing progressive kidney injury. Recent studies define IgAN as an autoimmune trait of complex architecture with a strong genetic determination. This Review summarizes new insights into the role of the O-glycosylation pathway, anti-glycan immune response, mucosal immunity, antigen processing and presentation, and the alternative complement pathway in the pathogenesis of IgAN.
Introduction
IgA nephropathy (IgAN) represents a common form of primary glomerulonephritis characterized by highly heterogeneous clinical and pathological features (1). The diagnosis of IgAN relies entirely on a histopathologic evaluation of renal biopsy, which demonstrates mesangial hypercellularity and predominant or co-dominant glomerular deposition of IgA, usually with complement C3 and variable amounts of IgG and/or IgM (1). IgAN is a progressive disease in up to 50% of affected adults. Remaining patients have persistent hematuria or proteinuria, and only a minority enter sustained clinical remission. The prognosis is also highly variable and the outcome difficult to predict in individual patients (2). Approximately 30%–50% of patients will develop end-stage renal disease (ESRD) within 20 years of the initial biopsy (3). Kidney transplant is the treatment of choice in individuals that reach ESRD, but the disease recurs in up to 50% of patients with a renal allograft (4). High rates of recurrence following kidney transplantation long ago suggested that the primary pathogenic defect in IgAN may be of extra-renal origin (5).
The hypothesis of extra-renal causes of IgAN is also strongly supported by studies of altered IgA1 O-glycosylation. Galactose-deficient variants of IgA1 (Gd-IgA1) are more common in the sera of patients compared with that of healthy and disease controls (6). These forms also predominate in the glomerular immunodeposits and in the IgA1-containing circulating immune complexes (7–10). IgA1-producing cells have been identified as the source of elevated serum Gd-IgA1 levels (11). Recent studies suggest that some individuals with high levels of Gd-IgA1 are prone to auto-sensitization and production of anti–Gd-IgA1 antibodies with formation of immune complexes (12). Reliable serum assays for Gd-IgA1 (auto-antigen) and anti–Gd-IgA1 IgG or IgA (auto-antibody) have been developed (6, 12), and early studies suggest that these biomarkers may be useful in predicting accelerated progression of kidney disease (13, 14).
The above findings strongly implicate an autoimmune etiology of IgAN, a clear departure from previous models of a simple imbalance between IgA production and clearance. Further support for an autoimmune pathogenesis comes from genome-wide association studies (GWAS), which unequivocally demonstrated that disease susceptibility is greatly affected by several common variants in the antigen processing and presentation pathway (15–17). These variants have overlapping effects on other autoimmune disorders (16). Additionally, GWAS highlighted the involvement of the mucosal defense system and the alternative complement pathway. Here we review evidence for the involvement of these pathways in the pathogenesis of IgAN, highlighting studies of IgA1 glycosylation defects and anti-glycan immune responses as well as evidence arising from gene-mapping efforts. We further discuss the recently proposed unifying pathogenesis model that incorporates both functional and genetic data.
IgA1 O-glycosylation pathway and anti-glycan antibodies in IgAN
Circulating IgA-IgG immune complexes have been implicated for years in the pathogenesis of IgAN and Henoch-Schönlein purpura nephritis, a related kidney disease with accompanying extra-renal manifestations due to systemic IgA vasculitis (18–20). However, the demonstration that these complexes consist of Gd-IgA1 bound by anti–Gd-IgA1 antibodies came only recently, redefining IgAN as an autoimmune disease (9, 11). Here we focus on characterization of the autoantigen, Gd-IgA1, as other aspects have been reviewed elsewhere (21–23).
IgA1 heavy chains contain a hinge region segment, the site of O-glycosylation, with nine potential sites for attachment of O-glycans (Figure 1A). Of these sites, only three to six are usually glycosylated (24–29). Normal human circulatory IgA1 has O-glycans consisting of N-acetylgalactosamine (GalNAc) with β1,3-linked galactose; each saccharide can also be sialylated. The composition of O-glycans on normal serum IgA1 is variable, but prevailing forms include the GalNAc-galactose disaccharide along with its mono- and di-sialylated forms (29, 30–32).
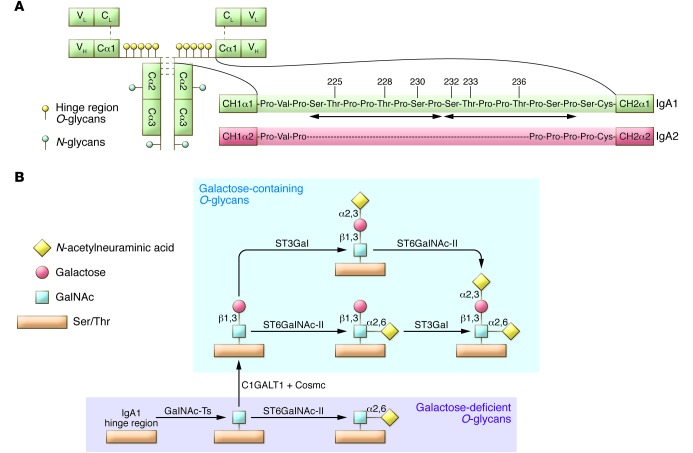
Figure 1. IgA1 structure and O-glycosylation pathway.
(A) Structure of monomeric IgA1 and its hinge region in comparison with IgA2 (A2m(l) allotype) (36). The IgA1 hinge region consists of a short amino acid sequence between CH1 and CH2 of the α1 heavy chain. In contrast to IgA2, the IgA1 hinge region contains two octapeptide repeats composed of Ser, Thr, and proline (Pro) residues (arrows). Of the nine potential Ser and Thr O-glycosylation sites, three to six are usually occupied (numbered Ser/Thr residues) (25–29). (B) Biosynthesis of hinge-region O-glycans of human circulatory IgA1. The possible structures of O-glycans in the hinge region of IgA1 include GalNAc with galactose that may contain up to two N-acetylneuraminic acid residues. Galactose-deficient glycans include terminal or sialylated GalNAc. Enzymes catalyzing the specific steps in this pathway are shown.
Patients with IgAN have a greater fraction of circulatory IgA1 molecules with hinge-region O-glycans without galactose, i.e., consisting of terminal GalNAc or sialylated GalNAc. This galactosylation defect appears to be specific for IgA1, as other O-glycoproteins in serum have a normal content of galactose (33, 34). Normal serum IgA1 had been thought to contain few or no galactose-deficient O-glycans (35), but recent evidence indicates that some galactose-deficient O-glycans may be present at residues threonine 233 (Thr233) and Thr236 (29). The latter observation is consistent with the fact that GalNAc-specific lectins, such as agglutinin from Helix aspersa (HAA), bind small amounts of IgA1 from healthy controls (6, 8, 10). Notably, the other subclass of human IgA, IgA2, has a different hinge region sequence, lacking serine (Ser) and Thr (Figure 1A), and thus is not O-glycosylated (23, 36).
Progress in the analysis of IgA1 O-glycosylation in IgAN initially came from the use of O-glycan–specific lectins (9, 33, 37, 38). Lectin-based assays progressed through a careful screening of GalNAc-specific binding (39) to the development of a quantitative ELISA (6). This assay was used for analyses of IgA1 in several clinical cohorts to demonstrate that most patients with IgAN have elevated serum levels of Gd-IgA1, including children and adults of various ethnicities (6, 13, 14, 40–42).
In addition to lectin-based assays, gas-liquid chromatography and mass spectrometry have been applied to elucidate the O-glycosylation patterns of IgA1 molecules (7, 9, 43–46). Direct localization of O-glycan attachment sites on IgA1 can be accomplished with electron capture dissociation (ECD) or electron transfer dissociation (ETD) tandem mass spectrometry, using hinge region glycopeptides generated by proteolytic cleavage (25–29). Individual hinge region glycoforms are identified by their molecular mass, and the sites of O-glycan attachment are determined by ECD or ETD fragmentation, a process that generates fragments from glycopeptides by breaking the chains of amino acids without removing the attached glycans. These approaches are sensitive enough to distinguish different O-glycoform isomers, i.e., IgA1 isoforms with the same number of glycans but different sites of attachment. Using these techniques, galactose-deficient glycans were found most commonly at Ser230, Thr233, and/or Thr236 in different combinations in normal serum IgA1 and in IgA1 myeloma proteins (see ref. 21 and 23 for a more detailed discussion). This technology is now ready to be applied extensively to analyses of IgA1 from patients with IgAN.
Studies of immortalized IgA1-secreting cells derived from the circulating cells of subjects with IgAN and healthy controls provided additional insights into the origin of Gd-IgA1, including identification of the key enzymes involved in the IgA1 O-glycosylation pathway (Figure 1B). The initiation of O-glycosylation on glycoproteins is catalyzed by GalNAc transferases (GalNAc-Ts) (for more information, see refs. 47, 48). This process is thought to be mediated primarily by GalNAc-T2, but other GalNAc-Ts are abundantly expressed in IgA1-producing cells and may contribute as well (47, 49). Specifically, GalNAc-T14, the closest structural relative of GalNAc-T2, is among the top GalNAc-Ts transcribed in IgA1-producing cells, and its expression is several-fold greater in the cells from IgAN patients compared with those of healthy controls. Not only do cells from IgAN patients secrete IgA1 with a higher degree of galactose deficiency, but the degree of IgA1 galactose deficiency also mimics that of circulatory IgA1 from the corresponding donors (10). The level of secreted Gd-IgA1 correlates with decreased expression and activity of core-1 β1,3-galactosyltransferase (C1GalT1), which adds galactose to GalNAc, as well as higher expression and activity of α-N-acetylgalactosaminide α-2,6-sialyltransferase 2 (ST6GalNAc-II), which adds sialic acid to GalNAc (10, 50, 51). Moreover, the expression of C1GalT1-specific chaperone, necessary for stability of the nascent C1GalT1 protein, is reduced (52, 53).
Thus, significant progress has been made in defining the role of IgA1 O-glycosylation in IgAN pathogenesis; evidence demonstrates that Gd-IgA1 originates from IgA1-secreting cells, and clinical studies have validated serum Gd-IgA1 as a reliable disease biomarker. While determination of the precise origin and nature of the cells producing Gd-IgA1 in vivo requires further work, circulating IgA1-secreting cells appear to recapitulate this biochemical defect in vitro. The pathogenic process leading to the production of Gd-IgA1 involves dysregulated expression of several key glycosyltransferases. The Gd-IgA1 is recognized by glycan-specific autoantibodies, a pathogenic process that leads to formation of nephritogenic immune complexes. Further definition of the nature of abnormal O-glycosylation of IgA1 and the pathways enhancing autoantigen production may uncover potential disease-specific therapeutic targets in future studies.
Additional insights from genetic studies
Numerous lines of evidence support a strong genetic contribution to IgAN (54). First, significant geographic and ethnic differences exist in the prevalence of IgAN: it occurs with greatest frequency in East Asians, is relatively common in Europeans, and is infrequent in individuals of African ancestry (55, 56). Second, familial aggregation of the disease has been well recognized (54). In most reported families, IgAN follows autosomal dominant transmission with incomplete penetrance. The recognition of familial disease has many clinical implications, particularly for the selection of living, related donors for kidney transplantation. In familial IgAN, high serum levels of Gd-IgA1 frequently co-segregate with a biopsy diagnosis of nephritis. Importantly, IgA1 glycosylation defects have been demonstrated to have high heritability in family-based studies (estimated at 50%–70%), further supporting an important role of this biomarker in genetic predisposition to IgAN (57, 58).
Despite numerous efforts at gene mapping using linkage approaches, Mendelian defects responsible for familial IgAN remain elusive (59–61). Linkage studies of IgAN have been limited because definitive diagnosis requires an invasive procedure, which often precludes reliable screening of asymptomatic relatives of affected individuals and increases chances of phenotype misclassification. Newer approaches, such as linkage analysis with phenotyping enhanced by determination of serum Gd-IgA1 levels, in combination with exome or genome sequencing may prove more fruitful (62).
The GWAS approach has emerged as a powerful alternative to family-based studies for complex traits and has been successfully applied to IgAN. By design, the major limitation of GWAS is that they detect only common susceptibility variants, and these typically have relatively small effects (63). Nevertheless, a well-executed GWAS may provide unbiased insights into the biology of human disease that could be clinically relevant regardless of the fraction of risk explained (64). In the case of IgAN, this approach is further motivated by the success of GWAS in the field of autoimmune and inflammatory diseases, with well over 500 genome-wide significant susceptibility loci discovered to date. For example, 193 independent GWAS loci were found for inflammatory bowel disease (IBD), explaining over 25% of familial risk (65) and 110 GWAS loci were identified for multiple sclerosis (MS), explaining nearly 30% of sibling recurrence (66). New biology arising from GWAS allowed for the identification of previously unrecognized pathways that are shared between different autoimmune disorders, shedding light on the common underlying mechanisms that can now be targeted pharmaceutically (64).
The first published GWAS for IgAN was performed in Europeans (15) and was followed by two larger studies performed in Chinese discovery cohorts (16, 17). Notably, the discovery sample size for all three studies combined consisted of only 3,000 IgAN cases and thus is approximately ten-fold smaller compared with the aforementioned studies of IBD and MS, which each included over 30,000 cases. Despite this power limitation, the IgAN GWAS successfully identified seven independent risk loci with genome-wide significance (P < 5 × 10–8) and cumulatively explained approximately 5% of the overall disease variance. Notably, this is a substantially greater fraction compared with other GWAS of kidney phenotypes — for example, a study of 60,000 individuals reported 13 loci that explained only 1.4% of the variance of estimated glomerular filtration rate (67). Nonetheless, a large portion of IgAN risk remains unexplained, and there are likely additional loci that have not yet been discovered. Using a recently proposed method (68), we have modeled this possibility mathematically, and we estimate that doubling the discovery sample size is likely to find up to seven additional loci, while tripling the sample size would identify up to 11 additional loci at a genome-wide significance (P < 5 × 10–8). These observations clearly motivate even larger studies of IgAN.
Although the fraction of the overall risk explained to date by GWAS is relatively small, systematic ethnic differences in IgAN risk allele frequencies correlate well with disease epidemiology (56). Strikingly, East Asians carry the highest average number of risk alleles and have the greatest prevalence of IgAN, while Africans have the lowest burden of risk alleles and are rarely affected. Accordingly, the prevalence of ESRD due to IgAN is nearly ten-fold higher among individuals of East Asian ancestry living in the US when compared with African Americans (56). Because over 85% of the existing GWAS discovery cohorts are composed of individuals of Chinese ancestry, discovery studies in other ethnicities are still needed to better assess the differences in genetic risk profiles among populations. Moreover, careful dissection of ethnicity-specific effects will be important, because the disease appears to have different features in East Asians and Europeans. For example, apart from dramatic differences in prevalence, biopsy series as well as ESRD registries indicate that the disease is equally frequent in males and females in East Asia (69–72). In contrast, IgAN is strikingly more common in European males, with a male-to-female ratio often exceeding 3:1 (73–77). These data suggest a complex interaction between gender and ethnicity on disease risk in IgAN. To date, no one has examined whether these patterns are mediated by a genetic effect. Moreover, prior GWAS showed no significant associations for markers on the sex chromosomes, suggesting that gender differences may be mediated by autosomal SNPs with gender-specific effects. The studies of gender-genotype interactions, as well as careful analysis of sex chromosomes in larger GWAS, may potentially provide some answers as to the origin of these intriguing epidemiologic patterns.
Finally, although GWAS have defined several susceptibility loci, the discovery of specific causal alleles within each locus awaits additional fine mapping or re-sequencing efforts, in addition to functional follow-up studies. Nevertheless, through careful analysis and annotation of the detected loci, several causal candidate genes have been prioritized, providing novel insights into the pathways driving the pathogenesis of IgAN. The implicated pathways include the antigen processing and presentation pathway (three loci on chromosome 6p21 in the MHC region), the mucosal immunity pathway (chromosomes 22q12 HORMAD2 locus, 8p23 α-defensin [DEFA] locus, and 17p13 TNFSF13 locus) and the alternative complement pathway (chromosome 1q32 complement factor H [CFH] locus) (15–17).
Antigen processing and presentation pathway
All three GWAS of IgAN identified strong signals with genome-wide significance within the MHC region. Further dissection of these signals using conditional analyses defined three distinct susceptibility loci on chromosome 6p21: HLA-DRB1/DQB1, HLA-DPB1/DPB2, and TAP1/PSMB9.
The strongest association was observed in the region that included the HLA-DRB1 -DQA1 and -DQB1 genes (16). Imputation of classical HLA alleles from SNP data supported a highly protective effect conferred by the DQB1*0602-DRB1*1501 haplotype. Notably, the same haplotype was reported to also protect from type I diabetes mellitus (78); however, the signal at this locus is complex, and conditional analyses provided support for additional risk variants independent of DQB1*0602-DRB1*1501 (56). This region was also previously associated with several autoimmune phenotypes, including rheumatoid arthritis (79), systemic sclerosis (80), alopecia areata (81), Graves’ disease (82), IgA deficiency (83), systemic lupus erythematosus (84), MS (85), and ulcerative colitis (86).
The second distinct MHC locus was centered over the region of the HLA-DPA1, -DPB1, and -DPB2 genes (also encoding MHC-II molecules), but the causal variant at this locus and its involvement in IgAN are unknown at present. The third MHC locus contained the TAP1, TAP2, PSMB8, and PSMB9 genes. These are interferon-regulated genes involved in antigen processing for presentation by MHC-I molecules, and these genes also play an important role in modulation of cytokine production and cytotoxic T cell response. The top-scoring SNP at this locus (rs2071543) tags a missense variant (Q49K) in exon 2 of PSMB8. It also represents a strong cis- expression quantitative trait locus in PBMCs, where the IgAN risk allele upregulates gene expression of TAP2, PSMB8, and PSMB9 (87). Increased mRNA levels of PSMB8 were detected in PBMCs of patients with IgAN, defining this gene as the top positional candidate (88).
In summary, GWAS support a complex pattern of association in the MHC region and strongly implicate the antigen processing and presentation pathway in the pathogenesis of IgAN.
Mucosal immunity and regulation of IgA production
Secretory IgA is central to intestinal homeostasis between the host and commensal bacteria, playing a critical role in regulating the intestinal microbiome and preventing pathogen overgrowth (Figure 2). Patients with selective IgA deficiency often develop IBD and recurrent intestinal infections (89). The inability of secretory IgA to activate the classical complement pathway may also play a role in creating a more tolerant non-inflammatory host-microbial relationship. Clinical associations of macroscopic hematuria coinciding with mucosal infections led to the hypothesis that defects in the regulation of local IgA response and/or abnormal mucosal antigen handling may trigger IgAN (90).
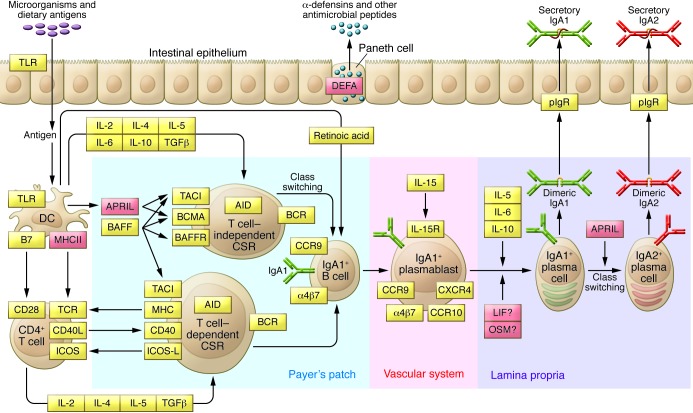
Figure 2. Intestinal network of IgA production.
IgA is the most abundant antibody isotype in the body, with the majority of IgA found in mucosal secretions. Mucosal IgA production is induced by T cell–dependent or T cell–independent mechanisms. T cell–independent production of IgA is primarily stimulated by IL-6, IL-10, TGF-β, BAFF, and APRIL produced by intestinal epithelial, dendritic, and stromal cells. In this environment, intestinal B cells undergo class switching from IgM to IgA1. IgA1 to IgA2 class switching can also occur and is promoted by APRIL and the mucosal milieu. IgA-secreting plasma cells migrate to lamina propria, where they release dimeric IgA. The dimers are formed through an interaction of two IgA molecules with a joining chain, which is synthesized by plasma cells. IgA dimers can bind to the polymeric Ig receptor (pIgR) on the basolateral surface of the mucosal epithelium and undergo transcytosis to the apical surface, where they dissociate from pIgR and are secreted into the lumen, carrying the secretory component of the receptor. The secretory component protects IgA molecules from proteolytic enzymes in the gut lumen. The bacteriostatic effects of secretory IgA are accompanied by antimicrobial peptides, such as defensins, secreted into the gut lumen by Paneth cells. The key molecules involved in the intestinal network of IgA production are indicated in orange; molecules implicated by GWAS are indicated in red.
This hypothesis is also supported by GWAS. A GWAS locus on chromosome 17p13 contains TNFSF13, which encodes a proliferation-inducing ligand (APRIL). This molecule is involved in T cell–independent generation of IgA-secreting plasma cells, as well as IgA1 to IgA2 class switching (Figure 2 and refs. 91, 92). Serum levels of APRIL are elevated in some patients with IgAN (93), and the 17p23 risk variant was reported to increase total serum IgA (16). Inactivation of Tnfsf13 in mice produces partial IgA deficiency and reduced IgA antibody responses to mucosal immunization (94). Conversely, overexpression of B cell activation factor (BAFF), a related molecule with overlapping functions and receptors with APRIL, results in autoimmune disease with commensal flora-dependent mesangial IgA deposits in mice (93). These data strongly implicate APRIL and BAFF signaling in the pathogenesis of IgAN.
A locus on chromosome 22q12 also influences serum IgA levels and encompasses several genes, including the cytokine-encoding genes LIF and OSM (15). These cytokines are members of the IL-6 family, are expressed in mucosal tissues, and have immunoregulatory properties (95, 96). Interestingly, OSM-deficient mice develop autoimmune disease with positive anti-dsDNA antibodies and glomerulonephritis with mesangial IgG deposits (97). The effects of LIF and OSM on mucosal production of IgA have not been well studied, but the IgAN risk allele at this locus was previously associated with protection against Crohn’s disease (65, 98, 99). We hypothesize that increased IgA production conferred by this allele has protective local anti-inflammatory effects, but the concomitant increase in the circulating IgA results in a higher risk of nephritis. Additional functional studies are clearly needed to test this hypothesis.
The third IgAN GWAS locus implicated in mucosal immunity centers on the DEFA gene cluster on chromosome 8p23. The α-defensin family of genes encodes small, structurally related peptides that are secreted at mucosal surfaces and have microbiocidal and chemoattractant properties (100). Among these peptides, α-defensin 1, 3, and 4 (encoded by DEFA1, DEFA3, and DEFA4) are synthesized in neutrophils, while α-defensin 5 and 6 (DEFA5 and DEFA6) are constitutively released by the intestinal Paneth cells into the gut lumen (Figure 2). Interestingly, α-defensin 1 and 3 differ only by the first amino acid and are encoded by two adjacent genes that are prone to segmental duplications and deletions. High rates of copy number variation are attributable to long runs of repetitive sequence in this region. In Europeans, the copy numbers of DEFA1 vary from two to five per haploid genome, while DEFA3 is occasionally completely deleted. The gene copy is also correlated with the expression level of these peptides (101, 102). Presently, it is not clear whether the IgAN risk allele in this region tags a risk haplotype carrying a specific copy number of DEFA1/3 genes, or if the association is due to variants in the nearby intestinally expressed DEFA5/6 genes.
GWAS strongly implicate dysregulation of mucosal IgA production as one of the causes of IgAN and suggest that the interplay of mucosal immunity with the commensal microbiome may represent an important disease modifier. Future efforts will focus on defining causal alleles at the identified loci by fine mapping, sequencing, and functional studies. Additional work is also needed to test the effects of these loci on the synthesis and O-glycosylation of IgA1.
Alternative complement pathway
Our GWAS identified a common deletion within the CFH locus on chromosome 1q32, which deletes two CFH-related genes (CFHR3 and CFHR1), as protective against IgAN (16). Factor H (FH), encoded by the CFH gene, is the critical regulator of the alternative complement pathway. It binds activated surface-fixed C3b and inhibits formation of C3 convertase while targeting C3b for inactivation by Factor I–mediated cleavage (103). FH-related proteins (FHR1–5) are structurally similar to FH, and are encoded by five genes (CFHR1-5) residing within the same genomic region. Considering the high level of sequence similarity between CFH and CFHRs, these genes are believed to have originated through segmental duplications and, similar to the DEFA locus, this region is prone to recurrent structural rearrangements. Among the known copy number variants in the region, CFHR3,1Δ is the most common, with allelic frequency ranging from 0% to 5% in East Asians and Native Americans, 20% in Europeans, and up to 50% in some African populations (104). In our study, CFHR3,1Δ additively protected against IgAN, with each copy reducing the risk of disease by approximately 40% (16). A similar protective effect of the deletion was reported for age-related macular degeneration (AMD), a complement-mediated retinal disease and a major cause of blindness in the elderly (105, 106).
Rare genomic rearrangements in the CHF region provide insights into the function of FHRs and suggest mechanisms that are likely responsible for the protective effect of CFHR3,1Δ. While CFHR1 haploinsufficiency is protective in IgAN and AMD, extra copies or gain-of-function mutations of CFHR1 cause C3 glomerulonephritis (C3GN), a glomerular disease that results from overactivation of the alternative complement pathway and is characterized by heavy glomerular C3 deposition. Structural studies revealed that FHR1, FHR2, and FHR5 share highly conserved dimerization domains, which enable formation of head-to-tail homo- and heterodimers of different combinations of these proteins (107). The dimeric forms are secreted by FHR-producing cells and are present in circulation in vivo. The dimers become powerful modulators of the activity of C3 convertase by competing with FH for binding to surface-fixed C3b. The binding of dimeric FHRs precludes proper inhibition of C3b by FH and results in increased activation of the convertase. In support of this hypothesis, there are reports of rare rearrangements in CFHR1 and CFHR5, in which the dimerization domains undergo internal duplications (108, 109). The mutant proteins are more likely to form circulating FHR oligomers, which are even more potent competitive antagonists of FH at tissue surfaces. This process results in net overactivity of C3 convertase, increased C3b deposition, and a clinical picture of C3GN. Similarly, individuals with rare duplications with formation of FHR1–3 and FHR2–5 fusion proteins may also present with C3GN, presumably due to enhanced ability of mutant proteins to compete with FH (110, 111). The above data provide a plausible explanation as to why reduced gene dosage of CFHR1 would mediate a protective effect in nephritis. The combined deletion of CFHR3 along with CFHR1 may further modify this protective effect, but FHR3 lacks dimerization domains characteristic of FHR1 and its mechanism of action is presently not well understood.
Surprisingly, CFHR3,1Δ has two additional phenotypic associations that oppose the protective effect observed for IgAN: CFHR3,1Δ is associated with increased risk of SLE (112) and increased risk of atypical hemolytic uremic syndrome (aHUS) (113), a systemic disorder caused by uncontrolled activation of the alternative complement pathway at endothelial surfaces leading to a characteristic clinical triad of hemolytic anemia, thrombocytopenia, and renal failure. Higher risk of aHUS is observed primarily in CFHR3,1Δ homozygotes and appears to be mediated through formation of inactivating anti-FH antibodies. Interestingly, a rare combined deletion of CFHR1 and CFHR4 genes (CFHR1,4Δ) has also been implicated in this phenomenon (114). Together, these observations suggest that FHR1 may have tolerogenic activity, and its deficiency may lead to increased autoimmune response that is independent of competition with FH for C3b binding (115, 116).
Multi-hit pathogenesis model
We previously formulated a multi-hit disease pathogenesis model that integrates findings from the genetic, biochemical, and clinical studies and highlights the autoimmune nature of IgAN (117). We hypothesize that the disease occurs in the setting of a genetically determined increase in circulating levels of Gd-IgA1 (Hit 1). This is likely due to dysregulation of IgA1 production at mucosal surfaces and alterations in posttranslational modification of O-glycans within IgA1-producing cells. The high heritability of circulating Gd-IgA1 suggests a critical role of inherited factors in this process. Because the 22q12 (HORMAD2), 8p23 (DEFA), and 17p13 (TNFSF13) loci appear to modulate mucosal immunity and production of IgA1, common variations at these loci may explain at least some of the heritability of this trait.
Our family-based studies demonstrated that high serum levels of Gd-IgA1 are not sufficient to produce IgAN and additional cofactors are required to trigger formation of immune complexes. Antiglycan antibodies that recognize GalNAc-containing epitopes on Gd-IgA1 have recently been identified. Thus, the formation of such antibodies may represent one of the essential cofactors or second hits for disease initiation (Hit 2). The anti-glycan response may be incited by exposure to infectious or dietary antigens in the setting of permissive MHC-II haplotypes and favorable genetic variants in the antigen-processing pathway.
The elevation of both Gd-IgA1 and antiglycan autoantibodies leads to formation of immune complexes (Hit 3). This step is critical for the nephritogenicity of Gd-IgA1, but additional molecules may also modulate this process. For example, based on recent animal studies, binding of soluble CD89 to circulating polymeric IgA1 has been proposed as a cofactor that may increase the nephritogenicity of immune complexes (118, 119).
The immune complexes that deposit in the kidney induce mesangial cell proliferation and secretion of extracellular matrix cytokines and chemokines and activate the alternative complement pathway (Hit 4). This process may be further exacerbated by interactions of immune complex–bound CD89 with mesangial transglutaminase-2 and transferrin receptor-1, which in turn may contribute to mesangial stimulation and activation of the complement pathway (118). The alternative pathway activation step is inhibited by FH but can be exacerbated in the presence of competitive FH antagonists, such as FHR1 dimers. The individuals who inherit common deletion of CFHR1 and CFHR3 (CFHR3,1Δ) have a lower concentration of FHR1-containing dimers and are therefore protected from overactivation of the complement system by immune complexes in kidney tissue.
We also hypothesize that mesangial deposition of Gd-IgA1 could occur in the absence of antiglycan antibodies, but this process may be clinically silent. This hypothesis might potentially explain the relatively high frequency of lanthanic IgA deposits observed among kidney allograft donors and in autopsy series of individuals with no known kidney disease (120–122). Unfortunately, this hypothesis remains extremely difficult to test, considering that asymptomatic individuals with isolated Hit 1 (high Gd-IgA1 levels) usually have no indication for a renal biopsy. The lanthanic deposition of Gd-IgA1 could represent the first phase in the disease process (123), while sequential acquisition of additional hits would trigger a full-blown clinical phenotype in a subset of genetically susceptible individuals. Longitudinal assessment of serum Gd-IgA1 and anti-glycan antibody levels along with renal outcomes in prospective population-based cohorts may be able to confirm this hypothesis.
Lastly, because IgAN represents an end-organ manifestation of what we believe is a highly heterogeneous disease process, we recognize that our simple model may not fit all cases, and alternative models may exist in parallel. The above model will undoubtedly continue to evolve as newer experimental and clinical data become available.
Acknowledgments
K. Kiryluk is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK) grants K23-DK090207 and R03-DK099654. J. Novak is supported in part by the NIH/NIDDK grants DK078244 and DK082753, NIH/National Institute of General Medical Sciences (NIGMS) grant GM098539, and a gift from the IGA Nephropathy Foundation of America Inc.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(6):2325–2332. doi:10.1172/JCI74475.
References
- 1.Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med. 2013;368(25):2402–2414. doi: 10.1056/NEJMra1206793. [DOI] [PubMed] [Google Scholar]
- 2.D’Amico G. Natural history of idiopathic IgA nephropathy: role of clinical and histological prognostic factors. Am J Kidney Dis. 2000;36(2):227–237. doi: 10.1053/ajkd.2000.8966. [DOI] [PubMed] [Google Scholar]
- 3.Schena FP. A retrospective analysis of the natural history of primary IgA nephropathy worldwide. Am J Med. 1990;89(2):209–215. doi: 10.1016/0002-9343(90)90300-3. [DOI] [PubMed] [Google Scholar]
- 4.Bumgardner GL, Amend WC, Ascher NL, Vincenti FG. Single-center long-term results of renal transplantation for IgA nephropathy. Transplantation. 1998;65(8):1053–1060. doi: 10.1097/00007890-199804270-00008. [DOI] [PubMed] [Google Scholar]
- 5.Berger J, Yaneva H, Nabarra B, Barbanel C. Recurrence of mesangial deposition of IgA after renal transplantation. Kidney Int. 1975;7(4):232–241. doi: 10.1038/ki.1975.35. [DOI] [PubMed] [Google Scholar]
- 6.Moldoveanu Z, et al. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 2007;71(11):1148–1154. doi: 10.1038/sj.ki.5002185. [DOI] [PubMed] [Google Scholar]
- 7.Hiki Y, et al. Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int. 2001;59(3):1077–1085. doi: 10.1046/j.1523-1755.2001.0590031077.x. [DOI] [PubMed] [Google Scholar]
- 8.Allen AC, Bailey EM Brenchley PE, Buck KS, Barratt J, Feehally J. Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: observations in three patients. Kidney Int. 2001;160(3):969–973. doi: 10.1046/j.1523-1755.2001.060003969.x. [DOI] [PubMed] [Google Scholar]
- 9.Tomana M, Matousovic K, Julian BA, Radl J, Konecny K, Mestecky J. Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int. 1997;52(2):509–516. doi: 10.1038/ki.1997.361. [DOI] [PubMed] [Google Scholar]
- 10.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J. Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest. 1999;104(1):73–81. doi: 10.1172/JCI5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, et al. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118(2):629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, et al. Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest. 2009;119(6):1668–1677. doi: 10.1172/JCI38468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berthoux F, et al. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol. 2012;23(9):1579–1587. doi: 10.1681/ASN.2012010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao N, et al. The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int. 2012;82(7):790–796. doi: 10.1038/ki.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feehally J, et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21(10):1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gharavi AG, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43(4):321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu XQ, et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2011;44(2):178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 18.Levinsky RJ, Barratt TM. IgA immune complexes in Henoch-Schönlein purpura. Lancet. 1979;2(8152):1100–1103. doi: 10.1016/s0140-6736(79)92505-4. [DOI] [PubMed] [Google Scholar]
- 19.Coppo R, et al. Circulating immune complexes containing IgA, IgG and IgM in patients with primary IgA nephropathy and with Henoch-Schönlein nephritis. Correlation with clinical and histologic signs of activity. Clin Nephrol. 1982;18(5):230–239. [PubMed] [Google Scholar]
- 20.Czerkinsky C, et al. Circulating immune complexes and immunoglobulin A rheumatoid factor in patients with mesangial immunoglobulin A nephropathies. J Clin Invest. 1986;77(6):1931–1938. doi: 10.1172/JCI112522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novak J, Renfrow MB, Gharavi AG, Julian BA. Pathogenesis of immunoglobulin A nephropathy. Curr Opin Nephrol Hypertens. 2013;22(3):287–294. doi: 10.1097/MNH.0b013e32835fef54. [DOI] [PubMed] [Google Scholar]
- 22.Kiryluk K, Novak J, Gharavi AG. Pathogenesis of immunoglobulin a nephropathy: recent insight from genetic studies. Annu Rev Med. 2013;64:339–356. doi: 10.1146/annurev-med-041811-142014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mestecky J, et al. IgA nephropathy: Molecular mechanisms of the disease. Annu Rev Pathol. 2013;8:217–240. doi: 10.1146/annurev-pathol-011110-130216. [DOI] [PubMed] [Google Scholar]
- 24.Tarelli E, Smith AC, Hendry BM, Challacombe SJ, Pouria S. Human serum IgA1 is substituted with up to six O-glycans as shown by matrix assisted laser desorption ionisation time-of-flight mass spectrometry. Carbohydr Res. 2004;339(13):2329–2335. doi: 10.1016/j.carres.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 25.Renfrow MB, et al. Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation Fourier transform-ion cyclotron resonance mass spectrometry. J Biol Chem. 2005;280(19):19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 26.Renfrow MB, et al. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: Implications for IgA nephropathy. Anal Bioanal Chem. 2007;389(5):1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, et al. Clustered O-glycans of IgA1: Defining macro- and micro-heterogeneity by use of electron capture/transfer dissociation. Mol Cell Proteomics. 2010;9(11):2545–2557. doi: 10.1074/mcp.M110.001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada Y, et al. Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Mol Cell Proteomics. 2010;9(4):719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, et al. Identification of structural isomers in IgA1 hinge-region O-glycosylation using high-resolution mass spectrometry. J Proteome Res. 2012;11(2):692–702. doi: 10.1021/pr200608q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baenziger J, Kornfeld S. Structure of the carbohydrate units of IgA1 immunoglobulin II. Structure of the O-glycosidically linked oligosaccharide units. J Biol Chem. 1974;249(22):7270–7281. [PubMed] [Google Scholar]
- 31.Tomana M, Niedermeier W, Spivey C. Microdetermination of monosaccharide in glycoproteins. Anal Biochem. 1978;89(1):110–118. doi: 10.1016/0003-2697(78)90731-5. [DOI] [PubMed] [Google Scholar]
- 32.Field MC, Dwek RA, Edge CJ, Rademacher TW. O-linked oligosaccharides from human serum immunoglobulin A1. Biochem Soc Trans. 1989;17(6):1034–1035. doi: 10.1042/bst0171034. [DOI] [PubMed] [Google Scholar]
- 33.Allen AC, Harper SJ, Feehally J. Galactosylation of N- and O-linked carbohydrate moieties of IgA1 and IgG in IgA nephropathy. Clin Exp Immunol. 1995;100(3):470–474. doi: 10.1111/j.1365-2249.1995.tb03724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith AC, de Wolff JF, Molyneux K, Feehally J, Barratt J. O-Glycosylation of serum IgD in IgA nephropathy. J Am Soc Nephrol. 2006;17(4):1192–1199. doi: 10.1681/ASN.2005101115. [DOI] [PubMed] [Google Scholar]
- 35.Mattu TS, et al. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J Biol Chem. 1998;273(4):2260–2272. doi: 10.1074/jbc.273.4.2260. [DOI] [PubMed] [Google Scholar]
- 36. Mestecky J, Moro I, Kerr MA, Woof JM. Mucosal immunoglobulins. In: Mestecky J, Bienenstock J, Lamm ME, Mayer L, McGhee JR, Strober W, eds. Mucosal Immunology. 3rd ed. Amsterdam, The Netherlands: Elsevier Academic Press; 2005:153–181. [Google Scholar]
- 37.Andre PM, Le Pogamp P, Chevet D. Impairment of jacalin binding to serum IgA in IgA nephropathy. J Clin Lab Anal. 1990;4(2):115–119. doi: 10.1002/jcla.1860040208. [DOI] [PubMed] [Google Scholar]
- 38.Mestecky J, Tomana M, Crowley-Nowick PA, Moldoveanu Z, Julian BA, Jackson S. Defective galactosylation and clearance of IgA1 molecules as a possible etiopathogenic factor in IgA nephropathy. Contrib Nephrol. 1993;104:172–182. doi: 10.1159/000422410. [DOI] [PubMed] [Google Scholar]
- 39.Moore JS, et al. Reactivities of N-acetylgalactosamine-specific lectins with human IgA1 proteins. Mol Immunol. 2007;44(10):2598–2604. doi: 10.1016/j.molimm.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau KK, Gaber LW, Delos Santos NM, Fisher KA, Grimes SJ, Wyatt RJ. Pediatric IgA nephropathy: clinical features at presentation and outcome for African-Americans and Caucasians. Clin Nephrol. 2004;62(3):167–172. doi: 10.5414/cnp62167. [DOI] [PubMed] [Google Scholar]
- 41.Shimozato S, Hiki Y, Odani H, Takahashi K, Yamamoto K, Sugiyama S. Serum under-galactosylated IgA1 is increased in Japanese patients with IgA nephropathy. Nephrol Dial Transplant. 2008;23(6):1931–1939. doi: 10.1093/ndt/gfm913. [DOI] [PubMed] [Google Scholar]
- 42.Camilla R, et al. Oxidative stress and galactose-deficient IgA1 as markers of progression in IgA nephropathy. Clin J Am Soc Nephrol. 2011;6(8):1903–1911. doi: 10.2215/CJN.11571210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwase H, et al. Estimation of the number of O-linked oligosaccharides per heavy chain of human IgA1 by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF) analysis of the hinge glycopeptide. J Biochem. 1996;120(2):393–397. doi: 10.1093/oxfordjournals.jbchem.a021425. [DOI] [PubMed] [Google Scholar]
- 44.Odani H, et al. Evaluation of the specific structures of IgA1 hinge glycopeptide in 30 IgA nephropathy patients by mass spectrometry. J Nephrol. 2010;23(1):70–76. [PubMed] [Google Scholar]
- 45.Takahashi K, et al. Structural analyses of O-glycan sugar chains on IgA1 hinge region using SELDI-TOF with various lectins. Biochem Biophys Res Commun. 2006;350(3):580–587. doi: 10.1016/j.bbrc.2006.09.075. [DOI] [PubMed] [Google Scholar]
- 46.Novak J, et al. Heterogeneity of O-glycosylation in the hinge region of human IgA1. Mol Immunol. 2000;37(17):1047–1056. doi: 10.1016/S0161-5890(01)00019-0. [DOI] [PubMed] [Google Scholar]
- 47.Stuchlova Horynova M, Raska M, Clausen H, Novak J. Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell Mol Life Sci. 2013;70(5):829–839. doi: 10.1007/s00018-012-1082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerken TA, et al. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J Biol Chem. 2013;288(27):19900–19914. doi: 10.1074/jbc.M113.477877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wandall HH, et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology. 2007;17(4):374–387. doi: 10.1093/glycob/cwl082. [DOI] [PubMed] [Google Scholar]
- 50.Raska M, et al. Identification and characterization of CMP-NeuAc:GalNAc-IgA1 α2,6-sialyltransferase in IgA1-producing cells. J Mol Biol. 2007;369(1):69–78. doi: 10.1016/j.jmb.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki H, et al. Cytokines alter IgA1 O-glycosylation by dysregulating C1GalT1 and ST6GalNAc-II enzymes. J Biol Chem. 2014;289(8):5330–5339. doi: 10.1074/jbc.M113.512277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc Natl Acad Sci U S A. 2002;99(26):16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ju T, Cummings RD. Protein glycosylation: chaperone mutation in Tn syndrome. Nature. 2005;437(7063):1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- 54.Kiryluk K, et al. Genetic studies of IgA nephropathy: past, present, and future. Pediatr Nephrol. 2010;25(11):2257–2268. doi: 10.1007/s00467-010-1500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall YN, Fuentes EF, Chertow GM, Olson JL. Race/ethnicity and disease severity in IgA nephropathy. BMC Nephrol. 2004;5:10. doi: 10.1186/1471-2369-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiryluk K, et al. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet. 2012;8(6):e1002765. doi: 10.1371/journal.pgen.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gharavi AG, et al. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J Am Soc Nephrol. 2008;19(5):1008–1014. doi: 10.1681/ASN.2007091052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiryluk K, et al. Aberrant glycosylation of IgA1 is inherited in both pediatric IgA nephropathy and Henoch-Schönlein purpura nephritis. Kidney Int. 2011;80(1):79–87. doi: 10.1038/ki.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gharavi AG, et al. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22-23. Nat Genet. 2000;26(3):354–357. doi: 10.1038/81677. [DOI] [PubMed] [Google Scholar]
- 60.Paterson AD, et al. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol. 2007;18(8):2408–2415. doi: 10.1681/ASN.2007020241. [DOI] [PubMed] [Google Scholar]
- 61.Bisceglia L, et al. Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet. 2006;79(6):1130–1134. doi: 10.1086/510135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiryluk K, Gharavi AG, Izzi C, Scolari F. IgA nephropathy — the case for a genetic basis becomes stronger. Nephrol Dial Transplant. 2010;25(2):336–338. doi: 10.1093/ndt/gfp593. [DOI] [PubMed] [Google Scholar]
- 63.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 64.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.International Multiple Sclerosis Genetics Consortium (IGC) et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi IJ, et al. An analysis of 4,514 cases of renal biopsy in Korea. Yonsei Med J. 2001;42(2):247–254. doi: 10.3349/ymj.2001.42.2.247. [DOI] [PubMed] [Google Scholar]
- 70.Chou YH, et al. Clinical outcomes and predictors for ESRD and mortality in primary GN. Clin J Am Soc Nephrol. 2012;7(9):1401–1408. doi: 10.2215/CJN.04500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee H, et al. Mortality of IgA nephropathy patients: a single center experience over 30 years. PLoS One. 2012;7(12):e51225. doi: 10.1371/journal.pone.0051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugiyama H, et al. Japan Renal Biopsy Registry: the first nationwide, web-based, and prospective registry system of renal biopsies in Japan. Clin Exp Nephrol. 2011;15(4):493–503. doi: 10.1007/s10157-011-0430-4. [DOI] [PubMed] [Google Scholar]
- 73.D’Amico G. The commonest glomerulonephritis in the world: IgA nephropathy. Q J Med. 1987;64(245):709–727. [PubMed] [Google Scholar]
- 74.Briganti EM, et al. The incidence of biopsy-proven glomerulonephritis in Australia. Nephrol Dial Transplant. 2001;16(7):1364–1367. doi: 10.1093/ndt/16.7.1364. [DOI] [PubMed] [Google Scholar]
- 75.Geddes CC, et al. A tricontinental view of IgA nephropathy. Nephrol Dial Transplant. 2003;18(8):1541–1548. doi: 10.1093/ndt/gfg207. [DOI] [PubMed] [Google Scholar]
- 76.Cattran DC, Reich HN, Beanlands HJ, Miller JA, Scholey JW, Troyanov S. The impact of sex in primary glomerulonephritis. Nephrol Dial Transplant. 2008;23(7):2247–2253. doi: 10.1093/ndt/gfm919. [DOI] [PubMed] [Google Scholar]
- 77.Maixnerova D, et al. The retrospective analysis of 343 Czech patients with IgA nephropathy — one centre experience. Nephrol Dial Transplant. 2012;27(4):1492–1498. doi: 10.1093/ndt/gfr482. [DOI] [PubMed] [Google Scholar]
- 78.Erlich H, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Radstake TR, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42(5):426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466(7302):113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chu X, et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat Genet. 2011;43(9):897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- 83.Ferreira RC, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42(9):777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 84.Yang W, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;92(1):41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.International Multiple Sclerosis Genetics Consortium et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 86.Okada Y, et al. HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn’s disease. Gastroenterology. 2011;141(3):864–865. doi: 10.1053/j.gastro.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 87.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coppo R, et al. Upregulation of the immunoproteasome in peripheral blood mononuclear cells of patients with IgA nephropathy. Kidney Int. 2009;75(5):536–541. doi: 10.1038/ki.2008.579. [DOI] [PubMed] [Google Scholar]
- 89.Cerutti A, et al. Regulation of mucosal IgA responses: lessons from primary immunodeficiencies. Ann N Y Acad Sci. 2011;1238:132–144. doi: 10.1111/j.1749-6632.2011.06266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bene MC, Faure GC. Mesangial IgA in IgA nephropathy arises from the mucosa. Am J Kidney Dis. 1988;12(5):406–409. doi: 10.1016/s0272-6386(88)80035-0. [DOI] [PubMed] [Google Scholar]
- 91.Stein JV, et al. APRIL modulates B and T cell immunity. J Clin Invest. 2002;109(12):1587–1598. doi: 10.1172/JCI15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26(6):812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 93.McCarthy DD, et al. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J Clin Invest. 2011;121(10):3991–4002. doi: 10.1172/JCI45563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castigli E, et al. Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci U S A. 2004;101(11):3903–3908. doi: 10.1073/pnas.0307348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rockman SP, et al. Expression of interleukin-6, leukemia inhibitory factor and their receptors by colonic epithelium and pericryptal fibroblasts. J Gastroenterol Hepatol. 2001;16(9):991–1000. doi: 10.1046/j.1440-1746.2001.02588.x. [DOI] [PubMed] [Google Scholar]
- 96.Guimbaud R, et al. Leukemia inhibitory factor involvement in human ulcerative colitis and its potential role in malignant course. Eur Cytokine Netw. 1998;9(4):607–612. [PubMed] [Google Scholar]
- 97.Esashi E, Ito H, Minehata K, Saito S, Morikawa Y, Miyajima A. Oncostatin M deficiency leads to thymic hypoplasia, accumulation of apoptotic thymocytes and glomerulonephritis. Eur J Immunol. 2009;39(6):1664–1670. doi: 10.1002/eji.200839149. [DOI] [PubMed] [Google Scholar]
- 98.Imielinski M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41(12):1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lehrer RI, Lu W. α-Defensins in human innate immunity. Immunol Rev. 2012;245(1):84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 101.Aldred PM, Hollox EJ, Armour JA. Copy number polymorphism and expression level variation of the human α-defensin genes DEFA1 and DEFA3. Hum Mol Genet. 2005;14(14):2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- 102.Khan FF, et al. Accurate measurement of gene copy number for human α-defensin DEFA1A3. BMC Genomics. 2013;14:719. doi: 10.1186/1471-2164-14-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Atkinson JP, Goodship TH. Complement factor H and the hemolytic uremic syndrome. J Exp Med. 2007;204(6):1245–1248. doi: 10.1084/jem.20070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holmes LV, et al. Determining the population frequency of the CFHR3/CFHR1 deletion at 1q32. PLoS One. 2013;8(4):e60352. doi: 10.1371/journal.pone.0060352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38(10):1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 106.Raychaudhuri S, et al. Associations of CFHR1-CFHR3 deletion and a CFH SNP to age-related macular degeneration are not independent. Nat Genet. 2010;42(7):553–555. doi: 10.1038/ng0710-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goicoechea de Jorge E, et al. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci U S A. 2013;110(12):4685–4690. doi: 10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tortajada A, et al. C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J Clin Invest. 2013;123(6):2434–2446. doi: 10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gale DP, et al. Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet. 2010;376(9743):794–801. doi: 10.1016/S0140-6736(10)60670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik TH, et al. A hybrid CFHR3-1 gene causes familial C3 glomerulopathy. J Am Soc Nephrol. 2012;23(7):1155–1160. doi: 10.1681/ASN.2012020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen Q, et al. A novel hybrid CFHR2/CFHR5 gene develops MPGN II and provides insights into disease mechanism and therapeutic implications. Immunobiology. 2012;217(11):1131–1132. [Google Scholar]
- 112.Zhao J, et al. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011;7(5):e1002079. doi: 10.1371/journal.pgen.1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zipfel PF, et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3(3):e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moore I, et al. Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood. 2010;115(2):379–387. doi: 10.1182/blood-2009-05-221549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jozsi M, et al. Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood. 2008;111(3):1512–1514. doi: 10.1182/blood-2007-09-109876. [DOI] [PubMed] [Google Scholar]
- 116.Noris M, Remuzzi G. Atypical hemolytic-uremic syndrome. N Engl J Med. 2009;361(17):1676–1687. doi: 10.1056/NEJMra0902814. [DOI] [PubMed] [Google Scholar]
- 117.Suzuki H, et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol. 2011;22(10):1795–1803. doi: 10.1681/ASN.2011050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berthelot L, et al. Transglutaminase is essential for IgA nephropathy development acting through IgA receptors. J Exp Med. 2012;209(4):793–806. doi: 10.1084/jem.20112005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Launay P, et al. Fcalpha receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger’s disease). Evidence for pathogenic soluble receptor-Iga complexes in patients and CD89 transgenic mice. J Exp Med. 2000;191(11):1999–2009. doi: 10.1084/jem.191.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suzuki K, Honda K, Tanabe K, Toma H, Nihei H, Yamaguchi Y. Incidence of latent mesangial IgA deposition in renal allograft donors in Japan. Kidney Int. 2003;63(6):2286–2294. doi: 10.1046/j.1523-1755.63.6s.2.x. [DOI] [PubMed] [Google Scholar]
- 121.Varis J, et al. Immunoglobulin and complement deposition in glomeruli of 756 subjects who had committed suicide or met with a violent death. J Clin Pathol. 1993;46(7):607–610. doi: 10.1136/jcp.46.7.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Waldherr R, Rambausek M, Duncker WD, Ritz E. Frequency of mesangial IgA deposits in a non-selected autopsy series. Nephrol Dial Transplant. 1989;4(11):943–946. doi: 10.1093/ndt/4.11.943. [DOI] [PubMed] [Google Scholar]
- 123.Glassock RJ. The pathogenesis of IgA nephropathy. Curr Opin Nephrol Hypertens. 2011;20(2):153–160. doi: 10.1097/MNH.0b013e3283436f5c. [DOI] [PubMed] [Google Scholar]