Abstract
The transcription factor Kruppel-like factor 4 (KLF4) has the ability, along with other factors, to reprogram somatic cells into induced pluripotent stem (iPS) cells. Here, we determined that KLF4 is expressed in kidney glomerular podocytes and is decreased in both animal models and humans exhibiting a proteinuric. Transient restoration of KLF4 expression in podocytes of diseased glomeruli in vivo, either by gene transfer or transgenic expression, resulted in a sustained increase in nephrin expression and a decrease in albuminuria. In mice harboring podocyte-specific deletion of Klf4, adriamycin-induced proteinuria was substantially exacerbated, although these animals displayed minimal phenotypical changes prior to adriamycin administration. KLF4 overexpression in cultured human podocytes increased expression of nephrin and other epithelial markers and reduced mesenchymal gene expression. DNA methylation profiling and bisulfite genomic sequencing revealed that KLF4 expression reduced methylation at the nephrin promoter and the promoters of other epithelial markers; however, methylation was increased at the promoters of genes encoding mesenchymal markers, suggesting selective epigenetic regulation of podocyte gene expression. Together, these results suggest that KLF4 epigenetically modulates podocyte phenotype and function and that the podocyte epigenome can be targeted for direct intervention and reduction of proteinuria.
Introduction
Kruppel-like factor 4 (KLF4; also known as gut-enriched Kruppel-like factor [GKLF]) is a zinc finger–containing transcription factor that is expressed primarily in epithelial cells of organs such as the gut, skin, lungs, and testes (1–3). In the gut, KLF4 is essential for differentiating among goblet cells in the colon (2). KLF4 is highly expressed in the skin, and mice with a deletion in the Klf4 gene die shortly after birth, presumably due to the loss of skin-barrier function (3). KLF4 is also known to induce differentiation and to act as a tumor suppressor in several cancer cell types (4). Currently, few reports detail the role of KLF4 in the kidney, with an exception reporting a putative role in the process of nephron differentiation in embryonic kidneys (5).
The importance of KLF4 in cell biology was underscored by the recent finding that ectopic expression of KLF4 with 3 other pluripotent reprogramming factors (OCT4, SOX2, and c-MYC) was sufficient to drive the reprogramming of somatic cells into induced pluripotent stem (iPS) cells (6–9). Subsequent studies indicated that the 3-factor combination of KLF4, OCT4, and SOX2 was also effective in inducing cell reprogramming (10, 11). It has been suggested that this reprogramming results from the concerted activation of a mesenchymal-epithelial transition (MET) program by pluripotent factors and that KLF4 contributes by activating the expression of epithelial genes during the initial phase of reprogramming (12). Currently, the mechanisms by which KLF4 regulates epithelial marker gene expression and induces phenotypic changes are unknown.
Previously, we reported that transient treatment of high-dose angiotensin receptor blockers (ARBs) caused a sustained decrease in albuminuria in a mouse adriamycin (ADM) nephropathy model (13). To investigate the sustained effects of the transient treatment, we examined the kidney gene expressions of these mice. We found that KLF4 was expressed in adult mouse kidneys and that its expression was altered by the treatment. These results suggested a previously unrecognized function of this reprogramming factor in the adult diseased kidney. Here, we report that KLF4 is highly expressed in kidney podocytes and that its expression is decreased in proteinuric states in both animal models and humans. Restoration of KLF4 expression in diseased glomeruli in vivo by gene transfer or by transgenic expression resulted in restoration of the nephrin expression and the attenuation of proteinuria. Although the deletion of Klf4 in podocytes presented minimal phenotypical changes at baseline, the ADM-induced proteinuria was significantly exacerbated in the Klf4 KO mice, as compared with the control mice. Further studies on the mechanisms of these changes suggest that KLF4 exerts control of the podocyte phenotype by a gene-specific methylation of epithelial and mesenchymal genes. The podocyte epigenome, therefore, may be a potential new therapeutic target for the attenuation of proteinuria.
Results
KLF4 is expressed in glomerular podocytes, and its expression is decreased in proteinuric status in both animal models and humans.
As shown in Figure 1A, KLF4 was expressed at high levels in the mouse renal glomeruli, with the detectable expression in the blood vessels. Renal tubular expression was less prominent. To further characterize the location of KLF4 expression in the glomeruli, double-staining studies were performed, which revealed the colocalization of glomerular KLF4 expression, with the nuclear marker WT1 and the extranuclear marker nephrin, of glomerular visceral epithelial cells (podocytes) in both mouse and human kidneys, suggesting the presence of KLF4 in both the nucleus and the cytoplasm of podocytes (Figure 1B). Immunoelectron microscopy confirmed staining in both the nucleus and the cytoplasm of podocytes (Figure 1C).
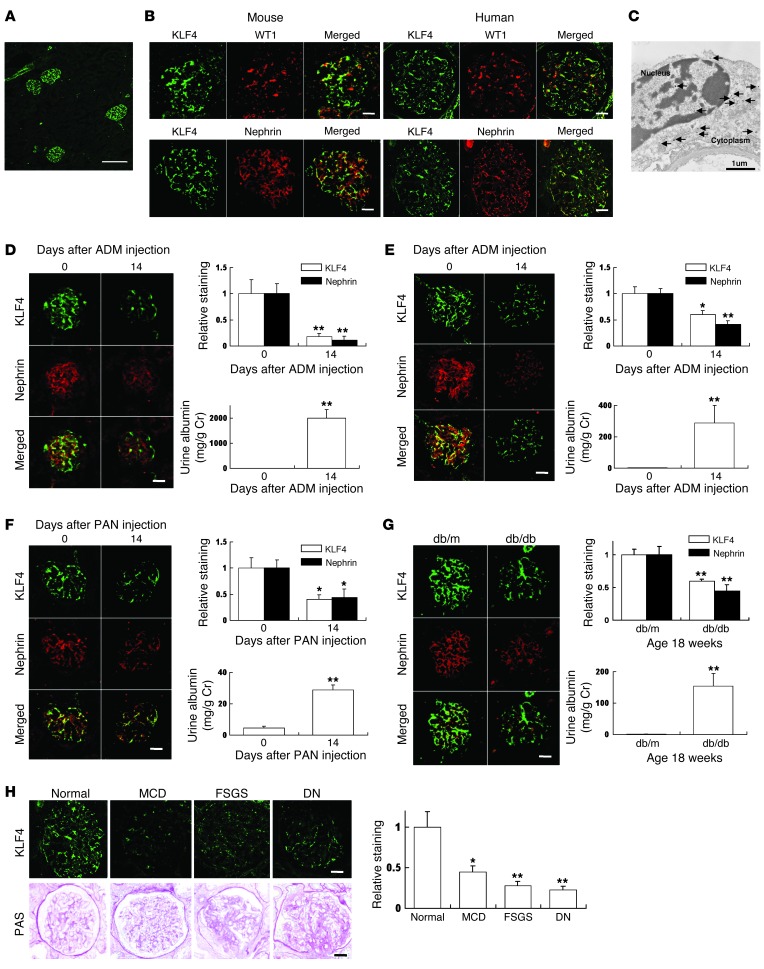
Figure 1. KLF4 is expressed in glomerular podocytes, and its expression is decreased in proteinuric glomerular disease models.
(A) Representative low-power photomicrographs of immunofluorescence staining of KLF4 in kidneys of 8-week-old male C57BL/6J mice. (B) Representative photomicrographs of immunofluorescence double staining of KLF4 (green)/WT1 or nephrin (red) in normal kidney of 8-week-old male C57BL/6J mice or normal human kidney. (C) Immunoelectron microscopy staining of KLF4 in the nucleus and cytoplasm of podocytes in the mouse kidney. Arrows indicate gold particles detecting KLF4 expression. (D–G) KLF4 expression in the ADM nephropathy model in (D) BALB/c mice and (E) C57BL/6J mice, (F) the PAN nephropathy model, and (G) the DN model (n = 5–6 per time point). (Left panels) Representative photomicrographs of immunofluorescence double staining of KLF4 (green)/nephrin (red). (Right panels) Quantification of the immunolabeled area and albuminuria. For the ADM or puromycin nephropathy model, mice were injected with ADM or puromycin at the doses described in Supplemental Methods and sacrificed at the indicated times after injection. For the diabetic model, diabetic (db/db) mice and controls (db/m) were compared at age 18 weeks. (H) (Left panel) Representative immunofluorescence photomicrographs of KLF4 in kidney biopsies from patients with proteinuric glomerular diseases. The bottom row shows the glomerular histology stained with PAS. (Right panel) Relative staining of KLF4 in the biopsy samples compared with control values. Clinical data are presented in Supplemental Table 1. *P < 0.05; **P < 0.01 vs. controls. Scale bars: 100 μm. (A); 25 μm (mouse), 50 μm (human) (B); 25 μm (D–G); 50 μm (H).
We next examined the expression of KLF4 in different animal models of proteinuria. For the ADM nephropathy model, we performed experiments in 2 different strains of mice (BALB/c and C57BL/6J) (Figure 1, D and E, and Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI69557DS1). We titered the dose of ADM, using a dose of 11 mg/kg in the BALB/c strain and 18 mg/kg in the C57BL/6J strain to produce a significant increase in proteinuria (13–16). Serum creatinine (Cr) in the BALB/c strain was significantly increased in ADM-treated mice compared with controls (Cr in control mice: 0.62 ± 0.01 mg/dl; in ADM-treated mice: 0.79 ± 0.02 mg/dl, P < 0.05), whereas the serum Cr in the C57BL/6J strain did not change significantly with ADM treatment (Cr in control mice: 0.48 ± 0.08 mg/dl; in ADM-treated mice: 0.45 ± 0.04 mg/dl). In both strains, we found that the expression of KLF4 was decreased after ADM treatment, which was accompanied by an attenuation in nephrin expression. A similar decrease in KLF4 and nephrin expression and an increase in proteinuria were confirmed in 2 other animal models: the puromycin aminonucleoside (PAN) nephropathy model and the db/db diabetic nephropathy (DN) model (Figure 1, F and G, and Supplemental Figure 1, C and D).
To examine the expression of KLF4 in human glomerular diseases, we performed immunofluorescence staining on renal biopsy samples from patients with proteinuric glomerular diseases (minimal change disease [MCD], focal segmental glomerulosclerosis [FSGS], and DN) (Figure 1H). The clinical data of these patients are shown in Supplemental Table 1. We found decreased glomerular expression of KLF4 in the patients with MCD, FSGS, and DN when compared with normal kidney controls.
Restoration of podocyte KLF4 expression in diseased glomeruli attenuates proteinuria.
We next examined the effects of the restoration of KLF4 expression in diseased glomeruli with reduced KLF4 expression. First, we performed in vivo gene transfer of Klf4-containing plasmids in BALB/c mice that had been pretreated with ADM, using a hydrodynamic-based gene transfer method described by Liu et al. (ref. 17 and Supplemental Figure 2A). This method, using rapid injection of a large volume of DNA solution into the tail vein, is known to produce substantial exogenous DNA in the circulation through an efficient gene transfer into several tissues, including kidney podocytes, with a peak at 8 to 16 hours and substantial clearance after 6 days (15, 17). The efficacy of this gene-transfer method for targeting podocyte injury and attenuating proteinuria has been confirmed by other groups (15, 16, 18). We confirmed that the gene transfer of Klf4 resulted in the increased expression of the podocyte KLF4 at 1 day after injection by performing immunofluorescence studies (Supplemental Figure 2B). By the seventh day, KLF4 expression returned to the same level as before the plasmid injection, but the increased expression of nephrin and the reduction of urine albumin were maintained for longer duration (Supplemental Figure 2, B and C). Serum Cr was also decreased by gene transfer of Klf4 at 2 weeks after plasmid injection (Cr in control mice: 0.78 ± 0.02 mg/dl; in the Klf4 gene transfer mice: 0.70 ± 0.02 mg/dl, P < 0.05). Quantification of foot process effacement revealed a significant decrease in the mice that had received the Klf4 gene transfer compared with vector controls (Supplemental Figure 2D). However, a significant difference could not be detected in the PAS-stained area or in the number of WT1-positive podocytes (Supplemental Figure 2, E and F).
To confirm the reproducibility of these findings, we also performed gene-transfer experiments in the C57BL/6J strain of mice (Supplemental Figure 3A). As in the BALB/c strain, the podocyte KLF4 expression in both the protein and the mRNA was increased at 1 day after injection, after which it decreased to basal levels. Increased expression of nephrin and other epithelial markers (podocin, synaptopodin) and the attenuation of proteinuria were similarly observed after KLF4 expression had normalized (Supplemental Figure 3, B–F).
Confirmatory experiments were also performed in the db/db mouse model of DN (Supplemental Figure 4A). Gene transfer of Klf4 resulted in an increase in nephrin expression and an attenuation of proteinuria (Supplemental Figure 4, B and C) without a significant change in the PAS-stained area and the number of WT1-positive podocytes (Supplemental Figure 4, D and E), which was consistent with the results for the ADM nephropathy model.
To confirm that the transient upregulation of KLF4 exerts prolonged effects on podocyte function, we next constructed mice with inducible and podocyte-specific overexpression of KLF4, using the tetracycline-inducible Tet-On gene induction system (Figure 2A). The mice were generated by intercrossing podocin-rtTA TetO mice (herein referred to as Pod/–) with pTRE-Klf4 mice to produce a double-transgenic strain, which responds to doxycycline (Dox) administration with increased podocyte-specific expression of KLF4 (podocin-rtTA TetO Klf4). Experiments established that Dox administration caused the increased expression of KLF4, which was colocalized with the podocyte marker nephrin, but not the endothelial marker vWF or the mesangial marker desmin. This confirmed the podocyte-specific overexpression of KLF4 in these transgenic mice (Figure 2B). It was found that the transient induction of KLF4 in glomerular podocytes in ADM-treated mice for a period of 4 weeks resulted in an increase in nephrin expression and a decrease in albuminuria, which continued even after the return of KLF4 protein and mRNA expression to basal levels by cessation of Dox administration (Figure 2, C–E). Further experiments confirmed that a sustained increase in the epithelial markers podocin and synaptopodin was also observed after Dox administration, with an opposite trend for the mesenchymal marker vimentin (Figure 3A and Supplemental Figure 5). These results suggested a long-term shift from a mesenchymal phenotype toward an epithelial phenotype of podocytes in these Klf4-transgenic mice. Furthermore, long-term follow-up revealed a sustained decrease in podocyte foot process effacement in the podocytes of the transgenic mice compared with controls (Figure 3B) without a significant change in PAS-stained area (Figure 3C).
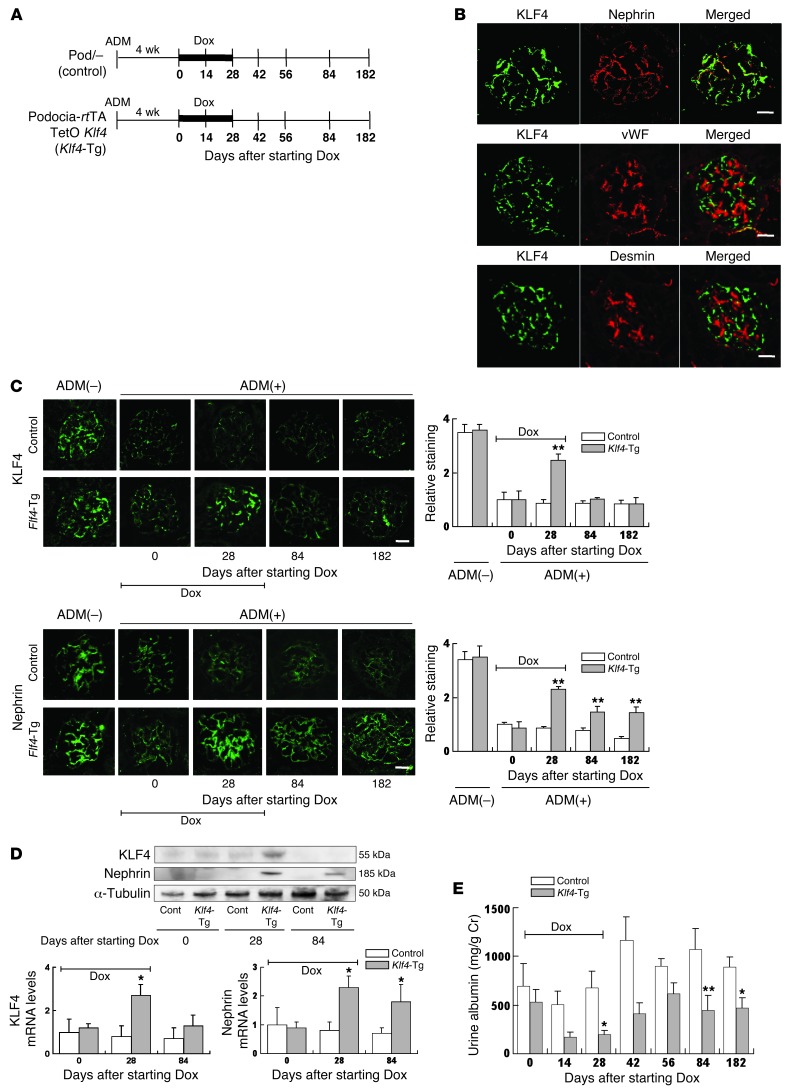
Figure 2. Restoration of podocyte KLF4 expression in diseased glomeruli attenuates proteinuria.
(A) Experimental protocol using Dox-inducible podocyte-specific Klf4 transgenic mice (podocin-rtTA TetO Klf4) and controls (podocin-rtTA [Pod/–]). Dox was administered for 28 days to induce KLF4 expression as indicated, and mice were sacrificed at days 0, 28, 84, and 182 after starting Dox (n = 5–6 per time point). (B) Immunofluorescence double staining of KLF4 (green) together with nephrin (red), vWF (red), or desmin (red) in Klf4 transgenic mice kidneys after 28 days Dox treatment. (C) (Left panel) Representative confocal photomicrographs and (right panel) quantification of glomerular KLF4 and nephrin expression without ADM treatment or at the indicated days after starting Dox in ADM nephropathy of Klf4 transgenic mice or controls. (D) (Upper panel) Western blot and (lower panel) real-time RT-PCR analysis of KLF4 and nephrin expression in the kidney cortex of Klf4 transgenic mice or controls at the indicated times after starting Dox treatment. (E) Quantification of albuminuria in Klf4 transgenic mice and controls at the indicated times after starting Dox treatment. Control: results for control mice (Pod/–); Klf4-Tg: results for podocyte-specific Klf4 transgenic mice (podocin-rtTA TetO Klf4). *P < 0.05; **P < 0.01 vs. Pod/– controls. Scale bars: 25 μm.
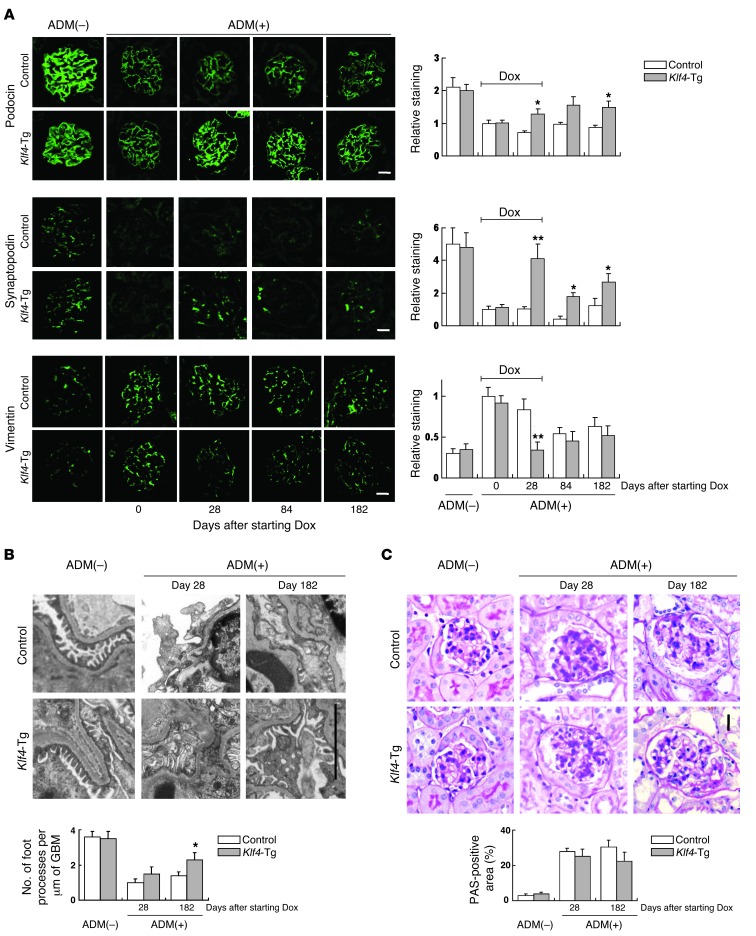
Figure 3. Restoration of podocyte KLF4 expression in diseased glomeruli alters phenotype marker expression.
Details of the experimental protocol are provided in the Figure 2 legend. (A) (Left panel) Representative confocal photomicrographs and (right panel) quantification of podocin, synaptopodin, and vimentin expression at the indicated days after starting Dox in Dox-inducible podocyte-specific Klf4 transgenic mice (podocin-rtTA TetO Klf4) and controls (Pod/–). Scale bars: 25 μm. (B) (Top panel) Representative electron photomicrographs and (bottom panel) quantification of podocyte foot process effacement at the indicated days after starting Dox. Scale bar: 5 μm. (C) Representative photomicrographs of renal histology (PAS staining) without ADM treatment or at the indicated days after starting Dox in ADM nephropathy. Control: results for control mice (Pod/–); Klf4-Tg: results for podocyte-specific Klf4 transgenic mice (podocin-rtTA TetO Klf4). Scale bar: 25 μm. *P < 0.05; **P < 0.01 vs. Pod/– controls.
Podocyte-specific Klf4 deletion mice are more susceptible to ADM nephropathy.
We next generated podocyte-specific Klf4 KO (podicin-Cre Klf4flox/flox, herein referred to as Klf4 KO) mice to determine the physiological role of KLF4 in podocytes (Figure 4A). In the Klf4 KO mice, KLF4 expression was decreased to about half that of the WT mice when we examined the kidney cortex as a whole. Nephrin expression seemed to decrease, but it did not attain a significant difference (Figure 4, B and C). Variances in the gross morphology of the kidney were not detected in the Klf4 KO mice when compared with WT mice. We were unable to detect a significant difference in a basal level of protein in the urine; however, the ADM-induced proteinuria was significantly exacerbated in the Klf4 KO mice (Figure 4D).
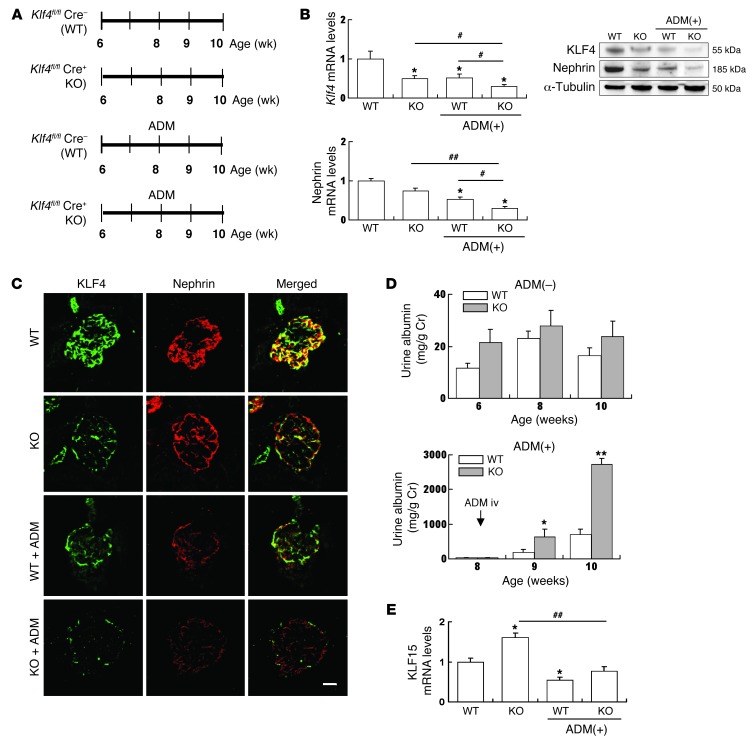
Figure 4. Knockdown of KLF4 in podocytes exacerbates proteinuria in ADM nephropathy.
(A) Experimental protocol. Podocyte-specific Klf4 KO mice (podocin-Cre Klf4flox/flox) and control littermates (Klf4flox/flox) were treated with or without ADM (n = 5–6 per time point). (B) Real-time RT-PCR and Western blotting analysis of nephrin, KLF4, and KLF15 expression in kidney cortex of KOs and controls. (C) Representative photomicrographs of immunofluorescence staining of KLF4 and nephrin in KOs or controls with or without ADM treatment at 2 weeks after ADM injection. Scale bar: 25 μm. (D) Time course of changes in albuminuria in Klf4 KOs and controls without (–) or with (+) ADM treatment. (E) Real-time RT-PCR analysis of Klf15 expression in kidney cortex of KOs and controls. ADM (+): 2 weeks after ADM injection. *P < 0.05, **P < 0.01 vs. controls; #P < 0.05, ##P < 0.01 vs. the respective groups.
To discern why the knockdown of Klf4 did not change the amount of proteinuria at baseline, we investigated KLF15 expression in the Klf4 KO mice, as KLF15 has been recently reported as expressed in podocytes as a transcriptional regulator of podocyte differentiation (19). In the Klf4 KO mice at baseline, KLF15 expression was increased in glomeruli. On the other hand, in the ADM nephropathy of the Klf4 KO mice, a more profound decrease in KLF4 without the increase of KLF15 expression was accompanied by a decrease in nephrin expression and an increase in albuminuria compared with WT controls (Figure 4E).
KLF4 expression induces epithelial cell markers in cultured podocytes.
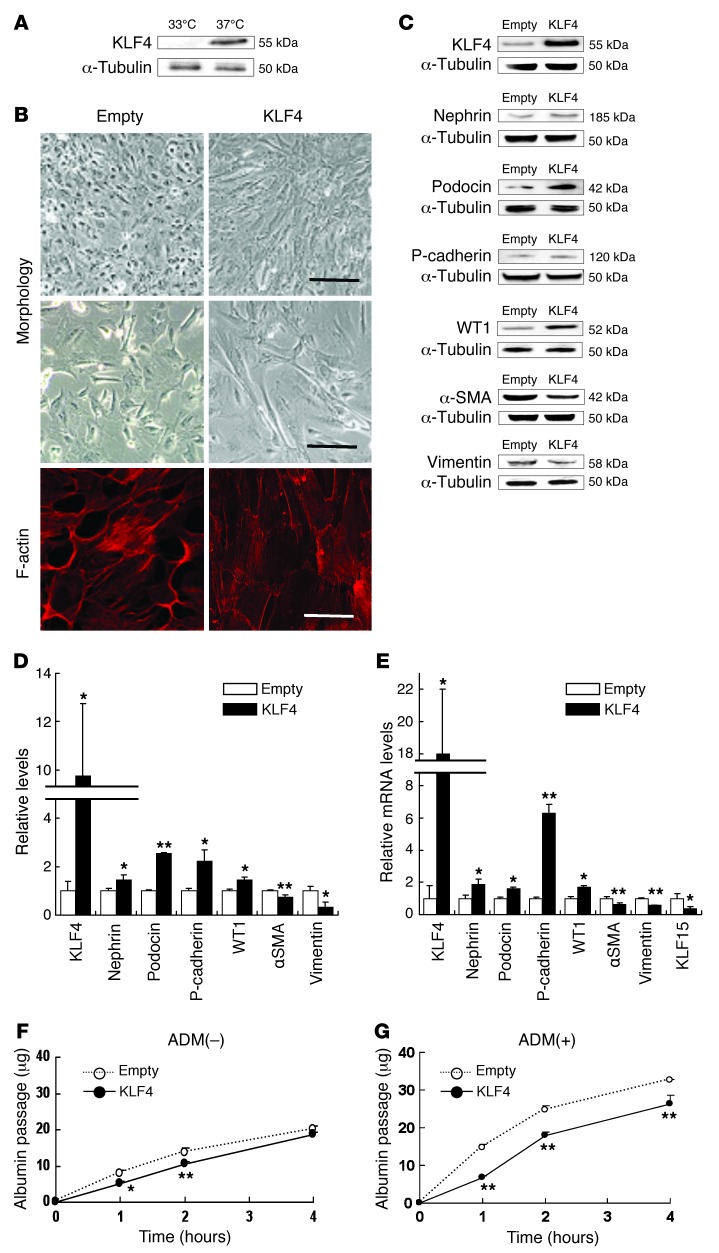
To examine the mechanisms of the KLF4-induced reduction of albuminuria, in vitro studies were performed using cultured human podocytes. Western blot analysis confirmed that KLF4 expression was readily detectable in differentiated podocytes (Figure 5A). In these podocytes, overexpression of KLF4 was found to result in several morphological changes, which included a change to a more arborized morphology together with reorganization of the actin cytoskeleton (Figure 5B). These changes were accompanied by the enhanced protein expression of the epithelial cell markers nephrin, podocin, P-cadherin, and WT1; however, the expression of mesenchymal markers α-SMA and vimentin decreased (Figure 5, C and D). Similar results were found for mRNA expression (Figure 5E).
Figure 5. KLF4 expression causes morphological changes and induces epithelial cell markers in cultured podocytes.
(A) Western blot analysis of KLF4 expression in undifferentiated podocytes cultured at 33°C or differentiated podocytes cultured at 37°C. (B) Representative photomicrographs of (upper/middle panel) cell morphology at high/low confluence and (lower panel) immunofluorescence labeling with F-actin in KLF4-overexpressing podocytes and empty vector–transfected controls. Scale bars: 100 μm (upper panel); 50 μm (middle panel); 10 μm (lower panel). (C) Representative Western blots and (D) quantification of epithelial or mesenchymal markers expression in KLF4-overexpressing podocytes and controls (n = 4). (E) Real-time RT-PCR analysis of mRNA expression in KLF4-overexpressing podocytes and controls (n = 6). (F and G) In vitro permeability of FITC-labeled albumin through podocyte monolayers in KLF4-overexpressing podocytes and controls without (F) or with (G) ADM (0.3 μg/ml) for 48 hours (n = 6). The passage of albumin was measured at indicated hours after FITC-albumin administration. *P < 0.05; **P < 0.01 vs. controls.
We next performed in vitro albumin permeability experiments using the KLF4-overexpressing podocytes. We found that overexpression of KLF4 resulted in a small but significant decrease in the in vitro passage of fluorescent-labeled albumin (Figure 5F). The difference became more marked in podocytes that had been pretreated with ADM at a dose that caused increased albumin permeability of the podocytes without causing increased cell toxicity (Figure 5G).
Next, we examined why the antiproteinuric effects of KLF4 were maintained after transient overexpression of KLF4, using the TetO transient overexpression system in cultured human podocytes. As shown in Supplemental Figure 6, the addition of Dox to the culture system resulted in enhanced expression of KLF4 accompanied by increased expression of the epithelial marker nephrin, whereas the expression of mesenchymal markers α-SMA and vimentin was decreased. Interestingly, the removal of Dox reduced the expression of KLF4, but did not change the levels of marker proteins. Moreover, the arborized morphology of the KLF4-induced podocytes persisted after the removal of Dox.
Knockdown of KLF4 expression caused an increase in albumin permeability in cultured podocytes with ADM treatment.
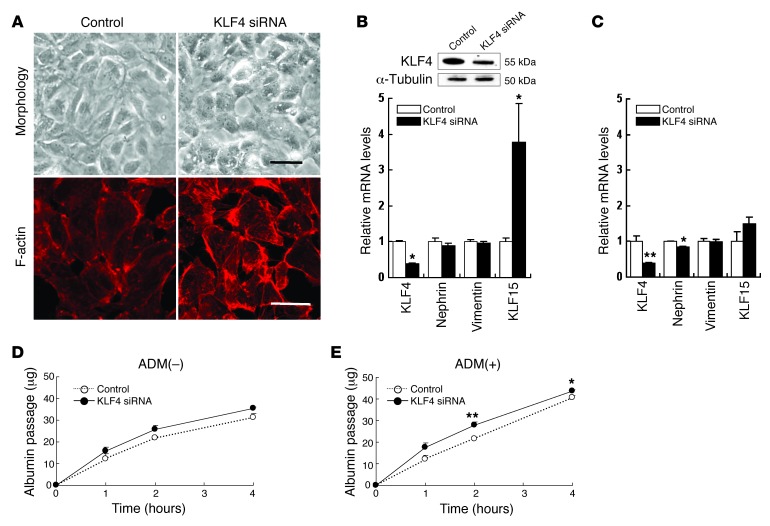
To examine the effect of KLF4 knockdown in cultured podocytes, we performed experiments using siRNA transfection. Reduction of KLF4 expression resulted in a marginal change to a less arborized morphology (Figure 6A). However, the changes in gene expression of the epithelial and mesenchymal markers did not attain statistical significance. KLF15 was significantly increased in podocytes with KLF4 knockdown (Figure 6B). In ADM-treated podocytes, KLF4 knockdown caused decreased expression of KLF4 and nephrin without an increase in KLF15 (Figure 6C). In the fluorescence-labeled albumin permeability assay, no significant differences were observed in KLF4 knockdown podocytes without ADM treatment, whereas increased albumin permeability was observed in KLF4 knockdown podocytes with ADM treatment (Figure 6, D and E).
Figure 6. Knockdown of KLF4 expression caused an increase in albumin permeability in cultured podocytes with ADM treatment.
(A) Representative photomicrographs of (upper panel) cell morphology and (lower panel) immunofluorescence labeling with F-actin in cultured human podocytes transfected with KLF4 siRNA or control RNA for 72 hours. Scale bars: 50 μm (upper panel); 10 μm (lower panel). (B) (Upper panel) Representative Western blots of KLF4 and (lower panel) real-time RT-PCR analysis of mRNA expression in podocytes transfected with KLF4 siRNA or control RNA for 72 hours (n = 6). (C) Real-time RT-PCR analysis of mRNA expression in ADM-treated (0.3 μg/ml) podocytes transfected with KLF4 siRNA or control RNA for 48 hours (n = 6). (D and E) In vitro permeability of FITC-labeled albumin in podocytes transfected with KLF4 siRNA or control RNA without (D) or with (E) ADM (0.3 μg/ml) for 48 hours (n = 6). *P < 0.05; **P < 0.01 vs. controls.
KLF4 expression causes DNA demethylation of the nephrin promoter and methylation of the vimentin promoter in podocytes.
The sustained effect of KLF4 overexpression suggests the hypothesis that KLF4 can exert epigenetic control over the podocyte phenotype. One potential mechanism of epigenetic regulation is promoter region DNA methylation/demethylation. We performed a genome-wide analysis of promoter methylation using a microarray-based DNA methylation–profiling system, which suggested that KLF4 overexpression was associated with gene-specific changes in promoter methylation (Supplemental Figure 7). In particular, methylation of the promoter regions of epithelial genes such as nephrin was reduced, whereas the DNA methylation of the corresponding regions in mesenchymal genes was increased.
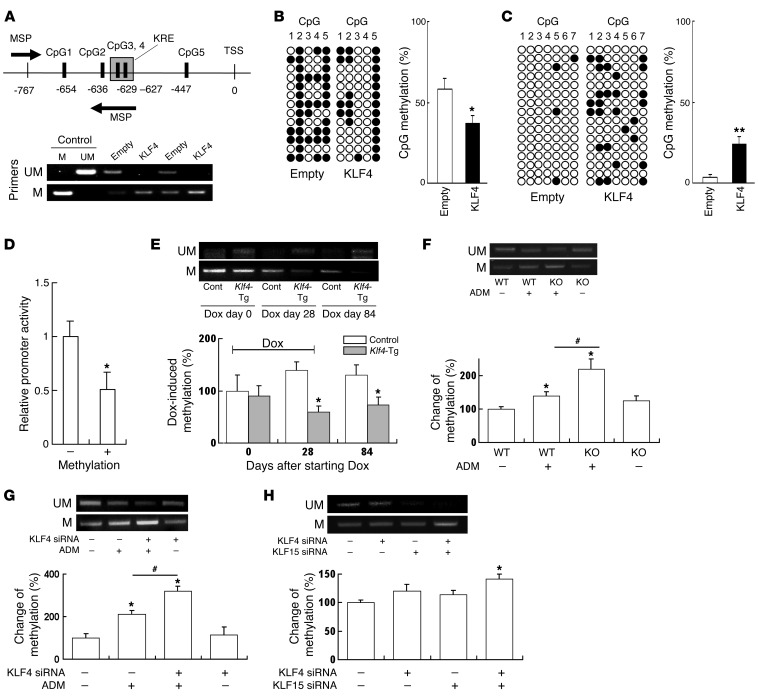
To confirm the results of the methylation-profiling microarray for nephrin, which was the epithelial marker with the greatest changes in promoter methylation, we reexamined the methylation of CpG sites included in the array, qualitatively using methylation-specific PCR (MSP) and quantitatively using the bisulfite genomic sequence (BGS) method. The analyzed region is presented in Supplemental Figure 8, and 5 CpGs (CpG 1–5) were included in the region. CpG sites CpG2 and CpG4 were incorporated in the HumanMethylation450 DNA Analysis BeadChip used in the methylation-profiling microarray. Interestingly, examination of the promoter DNA sequence adjacent to these sites revealed a sequence (designated KLF4-response element [KRE]) that was almost identical to the putative KRE consensus sequence (G/A)C(G/A)CC(C/T)(C/T) (20). Both methylation assays confirmed that there was increased demethylation of the CpG nucleotides in the nephrin promoter region of the KLF4-overexpressing cells, particularly at CpG sites 2–4, which were adjacent to the KRE (Figure 7, A and B). For comparison, we also performed BGS analysis of the promoter region of the mesenchymal gene vimentin in which 7 CpG sites (CpG 1–7) were included (Supplemental Figure 8) and confirmed that promoter methylation was increased in a region in the vicinity of the KRE in clear contrast to the results for nephrin (Figure 7C).
Figure 7. KLF4 expression causes demethylation of the nephrin promoter and methylation of the vimentin promoter in podocytes.
(A) (Upper panel) Map of nephrin promoter MSP primers and 5 CpG sites analyzed by BGS. (Lower panel) Results of MSP using primers for unmethylated (UM, top) and methylated (M, bottom) nephrin promoter. (B and C) Results of BGS of (B) nephrin and (C) vimentin promoter in KLF4-overexpressing podocytes and controls. The columns correspond to the CpG sequences shown in Supplemental Figure 8. Each row represents a single sequenced clone. White dots, unmethylated CpGs; black dots, methylated CpGs. The bar graph shows the net methylation of the analyzed CpG sites. KLF4, KLF4-overexpressing podocytes, empty: empty vector–transfected control podocytes. (D) Relative promoter activity of pCpG-free promoter plasmid containing nephrin promoter before (–) or after (+) methylation with SssI (n = 6). (E–H) (Upper panels) Results of MSP using primers for unmethylated (UM, top) and methylated (M, bottom) nephrin promoter. (Lower panels) Changes of methylation (%) using real-time MSP analysis in (E) laser-microdissected glomeruli of Klf4-transgenic mice or controls at the indicated times after starting of Dox in the protocol used for Figure 2A (n = 4), (F) laser-microdissected glomeruli of Klf4 KO mice or controls with or without ADM in the protocol used for Figure 4A (n = 4), (G) podocytes treated with KLF4 siRNA or control RNA with or without ADM in the protocol of Figure 6 (n = 6), and (H) podocytes with KLF4 and/or KLF15 siRNA for 48 hours (n = 6). *P < 0.05, **P < 0.01 vs. controls; #P < 0.05 versus the respective groups.
The effect of methylation/demethylation of the nephrin promoter region was examined further using a CpG-free plasmid vector system. First, the nephrin promoter region was cloned into the CpG sequence free promoter plasmid (pCpGfree-promoter), which lacks intrinsic CpG sequences in the vector, allowing direct examination of the effects of promoter methylation on promoter activity. The nephrin promoter plasmid DNA was then treated with or without bacterial methylase SssI to induce promoter methylation at CpG1–5. Analysis of the nephrin promoter activity revealed that the activity was decreased in the methylated samples compared with the unmethylated controls (Figure 7D), suggesting that promoter methylation of the investigated region plays a functional role in the control of nephrin promoter activity.
We then investigated the effect of KLF4 on the methylation status of the nephrin promoter in vivo by MSP analysis of laser-microdissected glomeruli. Transient overexpression of KLF4 induced the demethylation of nephrin promoter, and it was sustained after cessation of KLF4 induction, as shown in Figure 7E. In the Klf4 KO mice, Klf4 deletion did not alter the methylation status of the nephrin promoter; however, the increase in methylation of the nephrin promoter by ADM treatment was significantly potentiated in the Klf4 KO mice (Figure 7F). The results in Klf4 KO mice were confirmed with in vitro KLF4 knockdown experiments (Figure 7G). Moreover, single knockdown of KLF4 or KLF15 in a basal state of cultured podocytes did not induce a significant change in methylation of the nephrin promoter region, whereas knockdown of both KLF4 and KLF15 induced significant methylation of the region (Figure 7H).
To investigate the potential mechanisms of KLF4-induced changes in nephrin and vimentin promoter methylation, we examined the involvement of DNA methyltransferases (DNMTs) in ChIP analysis. It revealed that DNMT1 bound to the nephrin promoter region and was significantly reduced in KLF4-overexpressing podocytes, but binding of DNMT1 was not clearly detectable in the vimentin promoter or control (Supplemental Figure 9A). Binding of DNMT3a and DNMT3b could not be detected in either the nephrin or vimentin promoters using this assay (Supplemental Figure 9B).
We next investigated the expression of the acetylated histone protein acetyl-H3K9, which has been previously reported to be decreased in the glomeruli of mice with ADM nephropathy (21). When we examined the interactions of acetyl-H3K9 with different promoter regions by ChIP, assays revealed that KLF4 overexpression significantly increased acetyl-H3K9 associated with the nephrin promoter region containing the KRE, whereas H3K9 acetylation was clearly decreased in the vimentin promoter (Supplemental Figure 9C).
KLF4 expression increases nephrin promoter activity in podocytes.
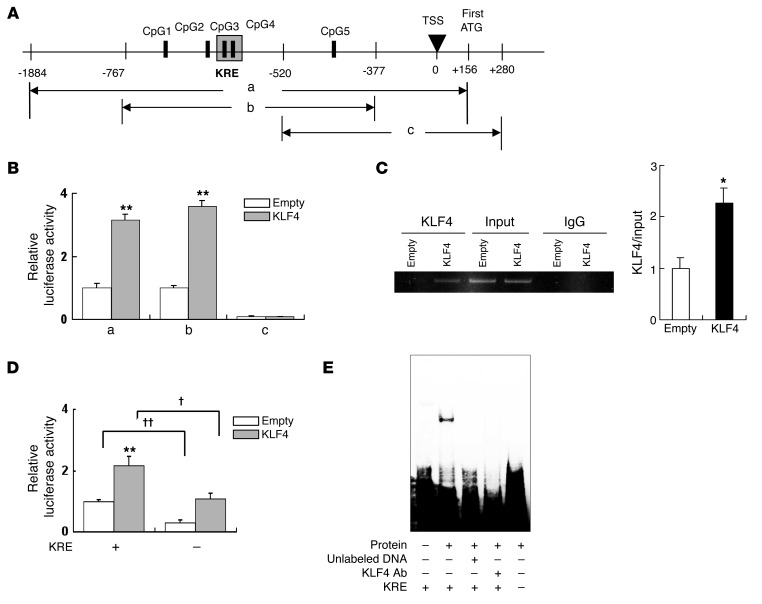
Finally, to investigate the role of KLF4 as a transcriptional factor in the nephrin promoter region containing the KRE, we performed deletion studies using luciferase reporter constructs containing nephrin promoter regions –1884 to +156 (a), –767 to –377 (b) or –520 to +280 (c) relative to the transcription start site (TSS) (Figure 8A). We observed significant increases in luciferase activity in KLF4-overexpressing podocytes with reporter constructs a and b, but not c. This indicates that KLF4 induced a direct increase in nephrin promoter activity within region b, which contains the KRE (Figure 8B). To confirm that this nephrin promoter region was able to bind to KLF4, we performed ChIP analysis using an anti-KLF4 antibody. It was found that KLF4 overexpression was associated with increased binding of KLF4 to the same region (region b) of the nephrin promoter (Figure 8C). Furthermore, deletion studies confirmed that deletion of the KRE resulted in a reduction of nephrin promoter activity (Figure 8D), and attenuation of the DNA-nuclear protein binding activity of this region on EMSA (Figure 8E).
Figure 8. KLF4 expression increases nephrin promoter activity in podocytes.
(A) Map of nephrin promoter regions included in the luciferase constructs. CpG 1–5 correspond to the sites shown in Supplemental Figure 8. (B) Luciferase activity 48 hours after transfection of KLF4-overexpressing podocytes (KLF4) or control podocytes (empty) with constructs containing regions a–c (n = 6). (C) ChIP assay for the presence of KLF4 in the promoter region of nephrin in KLF4-overexpressing podocytes (KLF4) or control podocytes (empty). The amplified fragment corresponds to region B in Figure 8A. The bar graph shows the quantification of KLF4/input intensity (n = 4). (D) Effect of KRE deletion on nephrin promoter activity. Luciferase activity was assayed 48 hours after transfection of luciferase constructs containing region B with (KRE[–]) or without (KRE[+]) specific deletion of the KRE (shown boxed in the upper panel of Supplemental Figure 8) into KLF4-overexpressing podocytes or control podocytes (n = 6). (E) Effect of KRE deletion on DNA binding to podocyte nuclear extracts analyzed by EMSA. Nuclear extracts from KLF4-overexpressing podocytes were pretreated with or without unlabeled competitor DNA or KLF4 antibody before incubation with labeled probes for EMSA to confirm specificity of the complexes. KRE (–) and (+) refer to probes with or without a deletion of the KRE sequence, respectively. **P < 0.01 vs. controls. †P < 0.05, ††P < 0.01 vs. the respective groups.
Discussion
In this study, we demonstrate that KLF4 is expressed in kidney podocytes and that its expression is decreased in proteinuric states. Concerning the localization of KLF4 in the podocyte, the data from both immunofluorescence double staining and immunoelectron microscopy suggest that KLF4 is expressed in both the nucleus and cytoplasm. Similar results were found with 2 commercial antibodies. A similar expression pattern of KLF4 in both the nucleus and cytoplasm has been reported in other tissues (22, 23). It has been suggested that KLF4 is stored in the cytoplasm and translocates to the nucleus after stimulation (23), which would explain why both nuclear and cytoplasmic staining were observed.
We have shown that the transient restoration of KLF4 in diseased glomeruli in vivo, by gene transfer and by transgenic overexpression, results in the restoration of podocyte phenotypes and the attenuation of proteinuria. We performed these experiments on multiple animal models to confirm the reproducibility of our findings. In particular, the ADM nephropathy model was performed using 2 different strains of mice (BALB/c and C57BL/6J). Although studies that characterize the ADM nephropathy model first used the BALB/c strain (14), several laboratories have reported that a higher dose of ADM also results in proteinuria in the C57BL/6J and mixed strains (13, 15, 16, 24), and we found that good concordance was obtained in the results from both strains.
We next investigated whether reduced KLF4 expression in itself could cause proteinuria directly, using engineered mice with a podocyte-specific targeted deletion of Klf4. The reduction of KLF4 caused minimal phenotypical changes at a baseline, but the reduced expression of KLF4 aggravated podocyte dysfunction in ADM nephropathy. Therefore, we examined the possibility that other KLF family members have overlapped roles with respect to KLF4 in the basal state. Interestingly, a recent study revealed that KLF15 is a key regulator of the podocyte phenotype (19). We observed the changes in KLF15 expression with KLF4 knockdown in podocytes, which suggests that the complementary effects of KLF4 are also found in the actions of KLF15, which may protect against the development of proteinuria at baseline. In the ADM nephropathy model in Klf4 KO mice, a more profound decrease in KLF4 without increased KLF15 could lead to increased albuminuria compared with WT controls. Further studies are required to examine how KLF4 acts synergistically with KLF15 and/or other members of the KLF family to regulate podocyte function.
Concerning the mechanism of regulation in podocyte phenotype by KLF4, we focused on epigenetic change in the genes regulating podocyte phenotype and function because of our observation that transient KLF4 overexpression in podocytes had sustained effects in vitro and in vivo. We found that KLF4 overexpression resulted in increased expression of epithelial markers; these changes were accompanied by a decrease in promoter methylation in several epithelial genes, which contrasted with an increase in promoter methylation in mesenchymal marker genes, suggesting gene-specific epigenetic control of the podocyte phenotype by KLF4.
Because nephrin is an important component of the slit membrane and downregulation of nephrin is known to cause proteinuria (25), we examined the molecular mechanisms of the KLF4-induced demethylation of the nephrin promoter. We confirmed that KLF4 expression resulted in decreased methylation of the nephrin promoter using 3 different assays (DNA methylation microarray, MSP, and BGS). Close examination of the changes in methylation of different CpG binucleotides revealed that CpG sites that were immediately adjacent to a putative KRE sequence were clearly demethylated by KLF4 overexpression. Moreover, in the knockdown experiments of KLF4 and/or KLF15 in cultured podocytes, the result of DNA methylation status in the nephrin promoter was compatible with the in vivo phenotype and the relationship between KLF4 and KLF15.
Concerning the mechanism of DNA demethylation in the nephrin promoter, we examined binding of DNMTs to the nephrin promoter. We found that DNMT1 binding was reduced at the nephrin promoter region that was demethylated by KLF4 overexpression. We next examined histone acetylation at this region and found that KLF4 expression was associated with increased acetylation of histone H3K9 at the nephrin promoter, whereas acetylation at the vimentin promoter was conversely decreased. Recent studies have shown that epigenetic mechanisms play an important role in the control of cell differentiation and function (26, 27) and both DNA methylation/demethylation and histone posttranslational modifications may be involved in the initiation of nuclear reprogramming toward pluripotency in human somatic cells (28). Although the exact mechanisms are unclear, increased acetylation of histones associated with the promoter region of a particular gene is thought to cause decreased promoter methylation and increased expression of that gene (27, 29).
These results support the hypothesis that KLF4 induces gene-specific methylation/demethylation of different target genes, which involves differential association of acetylated histones and DNMTs to the promoter regions. This produces changes in the promoter methylation, promoter activity, and ultimately cellular form and function. Although our studies were performed on kidney podocytes, these results may also be important for understanding the potency of KLF4 in cell reprogramming for other cell types, including iPS cells.
To our knowledge, this is the first report describing the effects of the pluripotent reprogramming factor KLF4 on epigenetic remodeling in the kidney, and this may be relevant for understanding the mechanisms of reprogramming in other tissues. Furthermore, we found that KLF4 is not only expressed in podocytes, but can also cause widespread changes in promoter methylation, resulting in changes in the podocyte phenotype. This raises the possibility that in situ transfer of KLF4 (achieved in this study by gene transfer) or other methods to transiently alter podocyte KLF4 activity could induce epigenetic changes sufficient to cause reversal of podocyte injury and long-lasting reduction of proteinuria. Since conventional therapies have not always been successful in producing sustained suppression of proteinuria, treatment with reprogramming factors may provide a basis for new and innovative strategies, with the final aim of reversing podocyte injury and achieving sustained suppression or even remission of proteinuria.
These results may have more important clinical implications, since proteinuria is not only an important component of chronic kidney diseases, but also a risk factor for cardiovascular diseases (30, 31). Recent reports have shown that KLF4 is implicated in vascular (32, 33) and cardiac diseases (34), so our findings suggest the possibility that a reduction in KLF4 expression in multiple tissues may provide a common mechanism linking cardiovascular and renal diseases.
In conclusion, our data indicate that KLF4 can modulate podocyte phenotype and function epigenetically both in vivo and in vitro. The results of this study suggest that the podocyte epigenome can be the target of direct intervention for the reduction of proteinuria.
Methods
Animal treatment protocols.
Studies were performed on 8-week-old male BALB/c mice or C57BL/6J mice obtained from Sankyo Laboratories. Glomerular injury was induced by treatment with ADM or puromycin. The doses and routes of treatment are described in detail in Supplemental Methods. Studies on diabetic mice were performed using C57BLKS/J db/db mice (Jackson Laboratories), which have been reported to develop detectable DN (35, 36).
Generation of podocyte-specific inducible Klf4 transgenic mice.
Tetracycline-regulated podocyte-specific inducible Klf4 transgenic mice (podocin-rtTA TetO Klf4) were generated by cloning mouse Klf4 cDNA into the plasmid pTRE-Tight-BI-DsRed-Express (Clontech/Takara Bio) to obtain pTRE-Klf4. After digestion with PvuI, the linear DNA fragment was microinjected into mouse ova for the generation of pTRE-Klf4 transgenic mice on a background of C57BL/6. pTRE-Klf4 transgenic mice were intercrossed with heterozygous podocin-rtTA (Pod/–) mice (Jackson Laboratories) to obtain podocin-rtTA TetO Klf4 on a mixed background of C57BL/6 and FVB/N. Expression of KLF4 in podocytes was induced in mice by administration of Dox (2 mg/ml) in the drinking water.
Generation of podocyte-specific Klf4 KO mice.
Floxed Klf4 mice (Klf4flox/flox) on a background of C57BL/6, originally developed in the laboratory of Klaus H. Kaestner et al. (2), were obtained from the Mutant Mouse Regional Resource Centers (University of Missouri-MMRRC, Columbia, Missouri, USA), supported by the NIH. In Klf4flox/flox mice, recombination of the Klf4 loxP allele deletes exons 2 and 3 and causes a frameshift mutation in exon, which abolishes KLF4 function completely. Klf4flox/flox mice were bred with transgenic mice expressing Cre recombinase under the control of the podocin promoter (podocin-Cre mice) on a background of C57BL/6, originally developed in the laboratory of Larence Holzman et al. (37) and obtained from Jackson Laboratory, to generate podocyte-specific Klf4-deficient mice. Female, heterozygous podocin-Cre mice were mated with homozygous, male Klf4flox/flox mice. Female offspring (F1) heterozygous for both podocin-Cre and Klf4flox/flox were bred back to the homozygous Klf4flox/flox males. The offspring (F2 generation) that were homozygous for Klf4flox/flox and heterozygous for podocin-Cre were considered KO mice, and their littermates that did not carry the podocin-driven Cre-recombinase insert were used as WT controls.
Gene transfer experiments.
For the Klf4 gene transfer experiments, KLF4 expression plasmid (pCMV-SPORT6-KLF4, 1 mg/kg) or empty vector was administered systemically by rapid tail-vein injection for hydrodynamic-based in vivo gene transfer, as described previously (16). For the Klf4 antisense cDNA experiments, Klf4 cDNA was subcloned in an antisense orientation into the plasmid pCMV-SPORT6, then administered as described above. Protocols for siRNA experiments are described in Supplemental Methods.
Biochemical studies.
Urine collections were performed in metabolic cages; urine albumin and Cr concentrations were determined using a direct competitive ELISA kit (Albuwell; Exocell), and a Cr assay kit (BioVision), respectively.
Histological studies and immunofluorescence staining.
The kidneys were removed and fixed in 4% paraformaldehyde and then embedded in paraffin blocks. Histological sections were stained with PAS solution, and a quantitative assessment of the PAS-positive area was performed as described below. Other kidney samples were fresh frozen in OCT compound. Normal kidney samples were obtained from patients undergoing nephrectomy for renal cell carcinoma. Immunofluorescence staining of cryostat sections was performed as described by us previously (13, 38). In brief, sections were fixed in ice-cold acetone at 4°C prior to blocking with PBS containing 1% BSA, then incubated overnight at 4°C with primary antibody. After washing, the samples were incubated with a FITC- or rhodamine-labeled secondary antibody and detected using primary and secondary antibodies listed in Supplemental Table 3. F-actin staining was performed using phalloidin red (Invitrogen). Quantification of PAS-positive areas and immunofluorescent staining were performed as described previously (13). Images of 15 to 20 glomeruli per section or of all the glomeruli in the biopsy sample (for human samples) were acquired with a laser confocal microscope (LSM710; Zeiss). The glomerular area stained purple (for the PAS stain) or stained green (for the immunofluorescence experiments) was quantified by color channel analysis and pixel counting using Photoshop CS4 software (Adobe) and divided by the area of the glomerular tuft. The mean value of the control samples was assigned the arbitrary unit of 1.
Electron microscopy.
Transmission electron microscopy was performed on glutaraldehyde-fixed, epoxy-embedded kidney samples. Immunoelectron microscopy was performed as described previously (39). For the quantification of foot process effacement, the glomerular basement membrane (GBM) was traced and measured using an image processing and analysis program (Scion Corp.). The number of podocyte foot processes present was divided by the total length of the GBM and expressed as the number per micrometer of GBM length (40, 41).
Laser microdissection.
Laser microdissection was performed according to previous reports. Freshly frozen kidney tissue was cut into 7-μm–thick sections. Cryosections were excised using a P.A.L.M. MicroBeam IP 230V Z microscope for laser pressure catapulting (P.A.L.M. Microlaser Technologies) as described previously (42, 43).
Cell culture and in vitro assays.
The conditionally immortalized human podocyte cell line was provided by Moin A. Saleem (University of Bristol, Bristol, United Kingdom) (44). Podocytes overexpressing KLF4 were obtained by stable transfection of the KLF4 expression vector pCMV-SPORT6-KLF4 or empty vector. Podocytes with tetracycline-inducible KLF4 expression were obtained by stable transfection of pTet-On-Advanced (Clontech) and pTRE-KLF4 into podocytes, and KLF4 expression was induced by administration of Dox (1 μg/ml). Experiments on stable cell lines were performed on at least 2 clonal lines to rule out a clonal effect. For the in vitro siRNA transfection studies, commercially synthesized siRNA oligonucleotides (listed in Supplemental Table 2) and control siRNA (HiGC; 12935-400) were purchased from Invitrogen and transfected into cultured podocytes according to the manufacturer’s instructions. Albumin permeability was assessed by quantification of the passage of FITC-labeled BSA across the podocyte monolayer in differentiated podocytes (45–47).
Western blot analysis.
Podocytes were homogenized in RIPA lysis buffer supplemented with protease cocktail inhibitor (Invitrogen). Proteins were resolved by SDS-PAGE and transferred onto PVDF) membranes. Western blotting was performed as described by us previously (48), using antibodies listed in Supplemental Table 3.
Real-time quantitative RT-PCR.
Gene expression was quantitatively analyzed by real-time RT-PCR (38), using primers and probes described in Supplemental Table 2.
Cloning and methylation of the human nephrin gene promoter.
DNA fragments derived from the human nephrin gene promoter were obtained by PCR and subcloned into pGL3-basic plasmid (Promega) or pCpGfree-promoter (InVivogen) for assays of methylated and unmethylated promoter activity, as described in Supplemental Methods.
MSP and BGS.
Bisulfite conversion of fragmented purified DNA was performed using the EpiTect Bisulfite Kit (QIAGEN). For MSP, bisulfite-modified DNA was amplified with methylation-specific or unmethylation-specific primer pairs. Methylated (Meth) and unmethylated (Unmeth) control DNA were produced by treatment of purified PCR product with or without bacterial methylase SssI, respectively. For BGS, bisulfite-modified DNA was amplified in a nested PCR with BGS1 and BGS2 primers. Amplified PCR products were cloned into TA cloning vector pT7-Blue and sequenced. Primer sequences used for the analysis of nephrin and vimentin promoter methylation are described in Supplemental Table 2.
ChIP assay and EMSA.
Nuclear extracts were harvested, and ChIP assay was performed with anti-KLF4, anti–acetyl-H3K9, anti-DNMT1, anti-DNMT3a, or anti-DNMT3b antibodies, using standard techniques. The primers used for analysis of different regions of the nephrin and vimentin promoters are described in Supplemental Table 2. EMSA was performed as described previously (49), using oligonucleotide sequences shown in Supplemental Table 2.
Microarray-based genome-wide DNA methylation profiling and gene-expression profiling.
DNA samples from KLF4-expressing podocytes and their controls were subjected to DNA methylation profiling using the HumanMethylation450 DNA Analysis BeadChip (Illumina). RNA samples from KLF4-expressing podocytes and their controls were subjected to the expression analysis on SurePrint G3 Human GE 8x60K v2 Microarray (Agilent) according to Agilent standard protocols. Protocols are described in Supplemental Methods. The DNA methylation profile data was uploaded to the NCBI GEO database (GSE41689). A gene expression and the integrated promoter methylation level belonging to the same GenBank accession number was also shown in the GEO database (GSE54395).
Statistics.
Results were expressed as the mean ± SEM. Statistical comparisons were made by ANOVA followed by Scheffe’s post-hoc test between more than 2 groups and by a 2-tailed Student’s t test between 2 groups. Statistical significance was defined as P < 0.05.
Study approval.
All animal experiments were performed in accordance with the Animal Experimentation Guidelines of the Keio University School of Medicine and were approved by Koichi Matsuo, Laboratory Animal Center, Keio University School of Medicine. For the human studies, biopsy samples were obtained from renal biopsies at Keio University Hospital and Tokyo Electric Power Company Hospital with informed consent of the patients, in accordance with the Declaration of Helsinki.
Supplementary Material
Acknowledgments
This study was supported by grants for scientific research (20590984, 20680105, 2155542, and 23890205) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan; the Nateglinide Memorial Toyoshima Research and Education Fund; the Salt Science Foundation, Tokyo, Japan; and Daiwa Securities Health Foundation, Tokyo, Japan. K. Hayashi was supported by a Grant-in-Aid for JSPS Fellows (2155542). We are grateful to Takeshi Marumo (Research Center for Advanced Science and Technology, Tokyo University) for helpful discussions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(6):2523–2537. doi:10.1172/JCI69557.
References
- 1.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32(11–12):1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129(11):2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22(4):356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 4.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29(6):549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4, and CBP: implications for terminal nephron differentiation. Am J Physiol. 2005;288(5):F899–F909. doi: 10.1152/ajprenal.00370.2004. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3(3):340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321(5889):699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 11.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2(1):10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K, Sasamura H, Ishiguro K, Sakamaki Y, Azegami T, Itoh H. Regression of glomerulosclerosis in response to transient treatment with angiotensin II blockers is attenuated by blockade of matrix metalloproteinase-2. Kidney Int. 2010;78(1):69–78. doi: 10.1038/ki.2010.81. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wang YP, Tay YC, Harris DC. Progressive adriamycin nephropathy in mice: sequence of histologic and immunohistochemical events. Kidney Int. 2000;58(4):1797–1804. doi: 10.1046/j.1523-1755.2000.00342.x. [DOI] [PubMed] [Google Scholar]
- 15.Dai C, Stolz DB, Kiss LP, Monga SP, Holzman LB, Liu Y. Wnt/β-catenin signaling promotes podocyte dysfunction and albuminuria. J Am Soc Nephrol. 2009;20(9):1997–2008. doi: 10.1681/ASN.2009010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai C, Saleem MA, Holzman LB, Mathieson P, Liu Y. Hepatocyte growth factor signaling ameliorates podocyte injury and proteinuria. Kidney Int. 2010;77(11):962–973. doi: 10.1038/ki.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 18.Bu X, et al. Systemic administration of naked plasmid encoding HGF attenuates puromycin aminonucleoside-induced damage of murine glomerular podocytes. Am J Physiol. 2011;301(4):F784–F792. doi: 10.1152/ajprenal.00210.2011. [DOI] [PubMed] [Google Scholar]
- 19.Mallipattu SK, et al. Kruppel-like factor 15 (KLF15) is a key regulator of podocyte differentiation. J Biol Chem. 2012;287(23):19122–19135. doi: 10.1074/jbc.M112.345983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26(3):796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Beneden K, et al. Valproic acid attenuates proteinuria and kidney injury. J Am Soc Nephrol. 2011;22(10):1863–1875. doi: 10.1681/ASN.2010111196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godmann M, Kosan C, Behr R. Kruppel-like factor 4 is widely expressed in the mouse male and female reproductive tract and responds as an immediate early gene to activation of the protein kinase A in TM4 Sertoli cells. Reproduction. 2010;139(4):771–782. doi: 10.1530/REP-09-0531. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik DK, Gupta M, Das S, Basu A. Kruppel-like factor 4, a novel transcription factor regulates microglial activation and subsequent neuroinflammation. J Neuroinflammation. 2010;7:68. doi: 10.1186/1742-2094-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeansson M, Bjorck K, Tenstad O, Haraldsson B. Adriamycin alters glomerular endothelium to induce proteinuria. J Am Soc Nephrol. 2009;20(1):114–122. doi: 10.1681/ASN.2007111205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haraldsson B, Nystrom J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev. 2008;88(2):451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 26.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 27.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 28.Mali P, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou JN, et al. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem Pharmacol. 2007;73(9):1297–1307. doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012;379(9811):165–180. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 31.Sarnak MJ, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation. 2003;108(17):2154–2169. doi: 10.1161/01.CIR.0000095676.90936.80. [DOI] [PubMed] [Google Scholar]
- 32.Zhou G, et al. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest. 2012;122(12):4727–4731. doi: 10.1172/JCI66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, et al. Kruppel-like factor 15 is critical for vascular inflammation. J Clin Invest. 2013;123(10):4232–4241. doi: 10.1172/JCI68552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao X, et al. Kruppel-like factor 4 regulates pressure-induced cardiac hypertrophy. J Mol Cell Cardiol. 2010;49(2):334–338. doi: 10.1016/j.yjmcc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma K, McCue P, Dunn SR. Diabetic kidney disease in the db/db mouse. Am J Physiol. 2003;284(6):F1138–F1144. doi: 10.1152/ajprenal.00315.2002. [DOI] [PubMed] [Google Scholar]
- 36.Deb DK, et al. Combined vitamin D analog and AT1 receptor antagonist synergistically block the development of kidney disease in a model of type 2 diabetes. Kidney Int. 2010;77(11):1000–1009. doi: 10.1038/ki.2010.22. [DOI] [PubMed] [Google Scholar]
- 37.Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB. Podocyte-specific expression of cre recombinase in transgenic mice. Genesis. 2003;35(1):39–42. doi: 10.1002/gene.10164. [DOI] [PubMed] [Google Scholar]
- 38.Ishiguro K, Hayashi K, Sasamura H, Sakamaki Y, Itoh H. “Pulse” treatment with high-dose angiotensin blocker reverses renal arteriolar hypertrophy and regresses hypertension. Hypertension. 2009;53(1):83–89. doi: 10.1161/HYPERTENSIONAHA.108.122721. [DOI] [PubMed] [Google Scholar]
- 39.Sohara E, et al. Pathogenesis and treatment of autosomal-dominant nephrogenic diabetes insipidus caused by an aquaporin 2 mutation. Proc Natl Acad Sci U S A. 2006;103(38):14217–14222. doi: 10.1073/pnas.0602331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H, et al. Inhibition of podocyte FAK protects against proteinuria and foot process effacement. J Am Soc Nephrol. 2010;21(7):1145–1156. doi: 10.1681/ASN.2009090991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heikkila E, et al. beta-Catenin mediates adriamycin-induced albuminuria and podocyte injury in adult mouse kidneys. Nephrol Dial Transplant. 2010;25(8):2437–2446. doi: 10.1093/ndt/gfq076. [DOI] [PubMed] [Google Scholar]
- 42.Kohda Y, Murakami H, Moe OW, Star RA. Analysis of segmental renal gene expression by laser capture microdissection. Kidney Int. 2000;57(1):321–331. doi: 10.1046/j.1523-1755.2000.00824.x. [DOI] [PubMed] [Google Scholar]
- 43.Cohen CD, Grone HJ, Grone EF, Nelson PJ, Schlondorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61(1):125–132. doi: 10.1046/j.1523-1755.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- 44.Saleem MA, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 45.Satchell SC, Anderson KL, Mathieson PW. Angiopoietin 1 and vascular endothelial growth factor modulate human glomerular endothelial cell barrier properties. J Am Soc Nephrol. 2004;15(3):566–574. doi: 10.1097/01.ASN.0000115397.22519.03. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Kang YS, Dai C, Kiss LP, Wen X, Liu Y. Epithelial-to-mesenchymal transition is a potential pathway leading to podocyte dysfunction and proteinuria. Am J Pathol. 2008;172(2):299–308. doi: 10.2353/ajpath.2008.070057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rico M, et al. WT1-interacting protein and ZO-1 translocate into podocyte nuclei after puromycin aminonucleoside treatment. Am J Physiol. 2005;289(2):F431–F441. doi: 10.1152/ajprenal.00389.2004. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu-Hirota R, Sasamura H, Kuroda M, Kobayashi E, Hayashi M, Saruta T. Extracellular matrix glycoprotein biglycan enhances vascular smooth muscle cell proliferation and migration. Circ Res. 2004;94(8):1067–1074. doi: 10.1161/01.RES.0000126049.79800.CA. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Liu Y, Zhang H, Chen G, Wang K, Xiao X. KLF4 promotes the expression, translocation, and releas eof HMGB1 in RAW264.7 macrophages in response to LPS. Shock. 2008;30(3):260–266. doi: 10.1097/shk.0b013e318162bef7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.