An ultrahigh-resolution structure of the Z-DNA dodecamer, solved from the anomalous signal of P atoms, reveals substantial flexibility of the backbone phosphate groups.
Keywords: Z-DNA structure, Z-DNA dodecamer, phosphorus SAD phasing, ultrahigh resolution, flexibility of phosphate groups
Abstract
A large number of Z-DNA hexamer duplex structures and a few oligomers of different lengths are available, but here the first crystal structure of the d(CGCGCGCGCGCG)2 dodecameric duplex is presented. Two synchrotron data sets were collected; one was used to solve the structure by the single-wavelength anomalous dispersion (SAD) approach based on the anomalous signal of P atoms, the other set, extending to an ultrahigh resolution of 0.75 Å, served to refine the atomic model to an R factor of 12.2% and an R free of 13.4%. The structure consists of parallel duplexes arranged into practically infinitely long helices packed in a hexagonal fashion, analogous to all other known structures of Z-DNA oligomers. However, the dodecamer molecule shows a high level of flexibility, especially of the backbone phosphate groups, with six out of 11 phosphates modeled in double orientations corresponding to the two previously observed Z-DNA conformations: ZI, with the phosphate groups inclined towards the inside of the helix, and ZII, with the phosphate groups rotated towards the outside of the helix.
1. Introduction
The first crystal structures of left-handed DNA were solved over three decades ago (Wang et al., 1979 ▶; Drew et al., 1980 ▶). Currently, a number of crystal structures of the left-handed Z-form of DNA are available in the Protein Data Bank (PDB; Berman et al., 2000 ▶) and the Nucleic Acid Database (NDB; Berman et al., 1992 ▶). Most of them are palindromic hexamer duplexes, but several have different sequences, and some contain shorter or longer nucleic acid chains. A few structures of DNA–protein complexes are also known in which at least part of the nucleic acid chain adopts a Z-DNA conformation. All available crystal structures have an alternating sequence of purines and pyrimidines, most often cytosines (Cyt) and guanines (Gua), which sometimes includes modified or additionally substituted bases. Characteristically, the guanines exist in the syn conformation, with a torsion angle around the glycosidic bond of about 60°, instead of the typical 120° for the usual anti conformation, and this causes the DNA backbone to adopt a left-handed zigzag conformation in both strands of the duplex. In several Z-DNA structures, two different phosphate-group conformations are observed: the ZI form, in which the phosphate groups are shifted deeper inside the helix towards the groove, and the ZII form, in which the phosphate groups are rotated away from the groove (Saenger, 1983 ▶). Sometimes, the phosphate groups exist in double conformations.
In the crystal structures of relatively short Z-DNA oligonucleotides, the double-stranded duplexes are straight and are arranged in practically infinite helices, with the Watson–Crick nucleotide pairs stacked throughout the whole crystal. These helices are usually packed side-by-side in a hexagonal fashion, although they may be shifted along their length in various ways, since the interactions between neighboring helices are weak and are executed mostly through solvent water molecules or various ions. The helical rise of Z-DNA is approximately 3.7 Å and its helical twist is 60° per two consecutive nucleotides, so a full turn of a Z-DNA helix with a length of about 44 Å corresponds to 12 nucleotides. Supplementary Table S11 presents crystal data characteristic of various Z-DNA crystal structures available in the PDB and NDB. A great majority of the Z-DNA structures are hexamer duplexes crystallized in space group P212121, with two duplexes positioned one after another on the 21 axis parallel to the crystal c cell edge of about 44 Å in length, forming a practically infinite helix that lacks a phosphate group every six nucleotides and extends throughout the whole crystal. Since the ratio of unit-cell parameters b:a in this crystal form is close to 31/18 ≃ 31/2, all parallel helices are packed in a hexagonal fashion. Several structures are presented with P32, P65 or P6522 symmetry, with cell dimensions and Z-DNA packing related to the previous orthorhombic form but with molecules disordered in their exact positioning along the helical axis. Apart from these, a small number of Z-DNA hexamer duplex structures crystallize with a different, unrelated, cell and symmetry. A few tetrameric and decameric Z-DNA duplexes crystallize in the ‘typical’ orthorhombic cell, with apparent disorder in their positioning along the helical axis. No crystal structure of a Z-DNA dodecamer has been available in the PDB until now, but idealized theoretical models of a dodecamer with alternative phosphate-group conformations (ZI and ZII) were formulated long ago (Wang et al., 1981 ▶), and one such model has been optimized using molecular-dynamics calculations (Laaksonen et al., 1989 ▶; Eriksson & Laaksonen, 1992 ▶).
Here, we present the ultrahigh-resolution crystal structure of the Z-DNA dodecamer d(CGCGCGCGCGCG)2, solved from the anomalous signal of its P atoms, and discuss its similarities and differences with respect to the known structures of Z-DNA.
2. Materials and methods
2.1. Crystallization and diffraction data collection
The oligonucleotide d(CGCGCGCGCGCG)2 was purchased from Eurofins MWG Operon (Huntsville, USA) and used without further purification. The DNA was incubated as a 1.66 mM solution in water at 37° C for 20 min. The crystals were obtained at 20°C by the hanging-drop method after mixing a 1.66 mM solution of the oligonucleotide with a precipitant solution consisting of 40 mM sodium cacodylate pH 7.0, 12 mM spermine tetrachloride, 80 mM NaCl, 10%(v/v) 2-methyl-2,4-pentanediol (MPD) in a 2:1 ratio. The well contained a 35% MPD solution. Large crystals grew in about 3 d in the form of irregular blobs without well developed faces or edges, as illustrated in Fig. 1 ▶. For cryoprotection during data collection, the crystal was dipped for a few seconds into the same precipitant solution containing in addition 30% MPD.
Figure 1.
The crystal of d(CGCGCGCGCGCG)2 employed to collect both of the diffraction data sets used in this paper.
Two data sets were measured from the same specimen using two different X-ray wavelengths at the Advanced Photon Source (Argonne National Laboratory, Argonne, USA). The first set, referred to as ‘cg12ano’, was collected on the SER-CAT beamline 22-ID using a wavelength of 1.54 Å with the intention of utilizing the anomalous signal from the P atoms. The second set, termed ‘cg12high’, was measured on the NE-CAT beamline 24-ID-C using a short wavelength of 0.62 Å to achieve very high data resolution. The cg12high data were collected in three passes and the cg12ano data were collected in two passes, with different effective exposures per image and crystal-to-detector distances, and were eventually scaled and merged together to adequately measure the weak high-resolution reflections, as well as the strongest low-resolution reflections that were overloaded on the highly exposed diffraction images. The data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The statistics of both sets are presented in Table 1 ▶. The X-ray beam was collimated to a 50 µm diameter, much smaller than the size of the crystal used to collect data, as shown in Fig. 1 ▶, so that the two data sets originated from two separate locations on the same crystal. This crystal did not suffer significant radiation damage, as judged from the scaling B-factor values, which varied between 0.0 and 1.7 Å2 for the cg12high data and between 0.0 and 2.7 Å2 for the cg12ano data.
Table 1. Diffraction data statistics.
Values in parentheses are for the highest resolution shell.
| Data set | cg12high | cg12ano |
|---|---|---|
| Diffraction data | ||
| Beamline | 24-ID-C, APS | 22-ID, APS |
| Detector | Pilatus 6M-F | MAR300 CCD |
| Wavelength (Å) | 0.6199 | 1.5418 |
| Space group | C2 | C2 |
| Unit-cell parameters | ||
| a (Å) | 48.48 | 48.54 |
| b (Å) | 19.55 | 19.57 |
| c (Å) | 31.22 | 31.25 |
| β (°) | 116.4 | 116.4 |
| Resolution (Å) | 0.75 (0.76–0.75) | 1.64 (1.70–1.64) |
| Reflections, total | 434398 | 32149 |
| Reflections, unique | 32210 (1525)† | 2978 (108)†/5634 (208)‡ |
| Completeness (%) | 95.1 (92.2)† | 93.4 (81.2)†/94.0 (80.9)‡ |
| Multiplicity | 13.5 (7.6)† | 10.8 (7.1)†/5.7 (3.7)‡ |
| R merge (%) | 3.1 (73.4)† | 4.2 (5.5)†/3.9 (4.9)‡ |
| 〈I/σ(I)〉 | 84.7 (2.8)† | 72.0 (55.5)†/54.4 (40.6)‡ |
| Mosaicity range (°) | 0.17–0.41 | 0.15–0.38 |
| Wilson B factor (Å2) | 6.9 | 11.4 |
| PDB code | 4ocb | |
| Refinement | ||
| Resolution (Å) | 30–0.75 | |
| No. of parameters | 3612 | |
| wR2 (%) | 32.2 | |
| R factor, F o > 4σ(F o) (%) | 11.8 | |
| No. of reflections, F o > 4σ(F o) | 27517 | |
| R factor, all reflections (%) | 12.2 | |
| Total No. of reflections | 32194 | |
| R free, F o > 4σ(F o) (%) | 12.9 | |
| No. of free reflections, F o > 4σ(F o) | 1364 | |
| R free, all reflections (%) | 13.4 | |
| Total No. of free reflections | 1601 | |
| R.m.s.d. from library targets | ||
| Bond 1–2 distances (Å) | 0.021 | |
| Angle 1–3 distances (Å) | 0.050 | |
| Asymmetric unit content | ||
| DNA nucleotides§ | 12 | |
| DNA atoms | 243 | |
| DNA atom sites, disordered | 2 × 15 | |
| Average B factor for DNA (Å2) | 9.9 | |
| Fully occupied water sites | 15 | |
| Partially occupied water sites | 65 | |
| Total occupancy of all water sites | 53.7 | |
| Average B factor of all water sites (Å2) | 19.6 | |
Friedel mates treated as equivalent reflections.
Friedel mates treated as independent reflections.
Cyt1 lacks the 5′-terminal phosphate group.
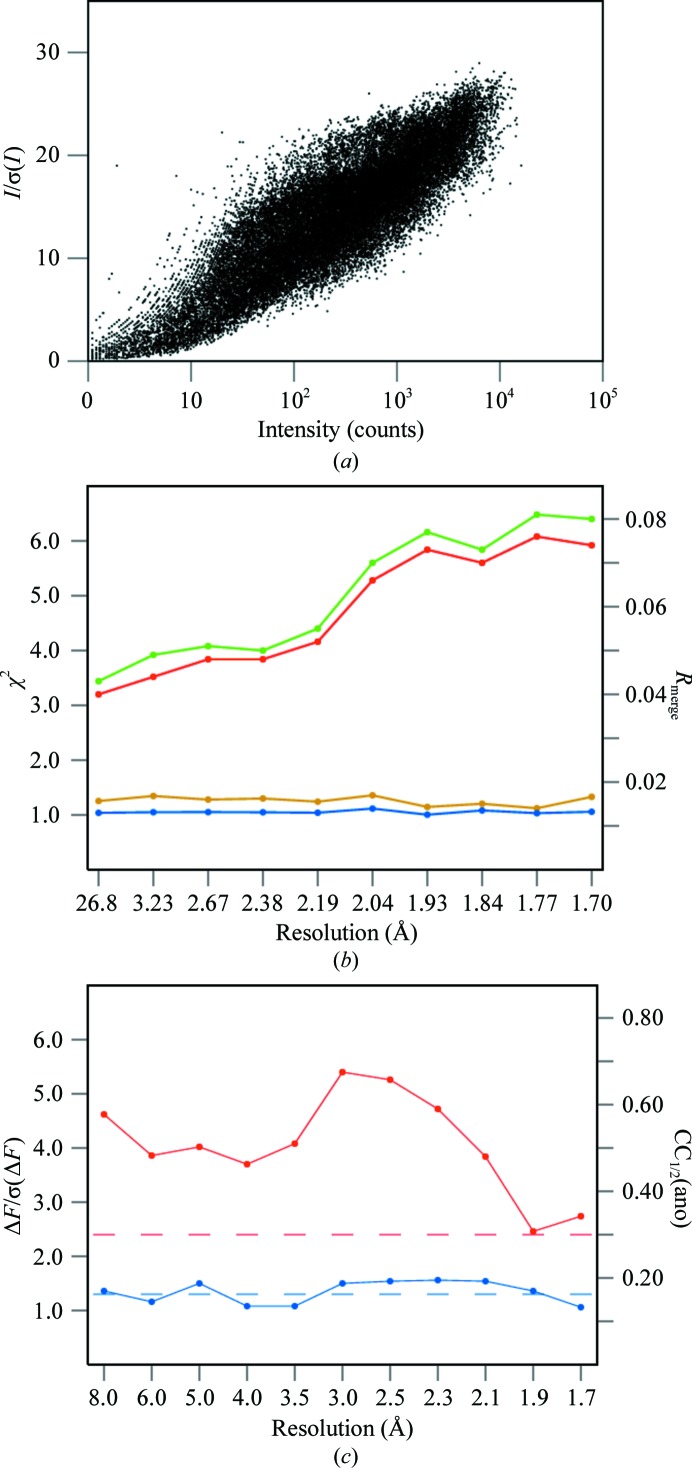
Several properties of the cg12ano data set are illustrated in Fig. 2 ▶. The overall strength of the intensities is presented in Fig. 2 ▶(a) as the relation of the signal-to-noise ratio I/σ(I) and the intensity of all reflections (Diederichs, 2010 ▶), and the asymptote of this dependence is higher than 30. The graph does not have the typical sigmoidal shape, as a result of merging data from two data-collection passes with different effective exposures. The SCALEPACK values of R merge and χ2 as a function of resolution are shown in Fig. 2 ▶(b) for merging with Friedel mates treated as equivalent or independent reflections. The differences between these two cases depend on the amount of anomalous signal in the data. Fig. 2 ▶(c) shows values of the anomalous signal-to-noise, ΔF/σ(ΔF), and the correlation coefficient, CC1/2(ano), between the signed anomalous differences in two random halves of all data obtained from SHELXC. The anomalous signal is significant if ΔF/σ(ΔF) is higher than 1.3 and CC1/2(ano) is higher than about 30% (Schneider & Sheldrick, 2002 ▶).
Figure 2.
Various quality criteria of the cg12ano data set. (a) Dependence between signal-to-noise ratio, I/σ(I), and intensity, I, of all reflections. (b) Values of R merge (red and green) and χ2 (blue and yellow) as a function of resolution resulting from treating Friedel mates as equivalent (green and yellow) or independent (red and blue) reflections obtained from SCALEPACK. (c) Anomalous signal to noise, ΔF/σ(ΔF), (blue) and CC1/2(ano) (red) values in resolution bins. The dashed lines show the significance levels of these parameters.
2.2. Structure solution
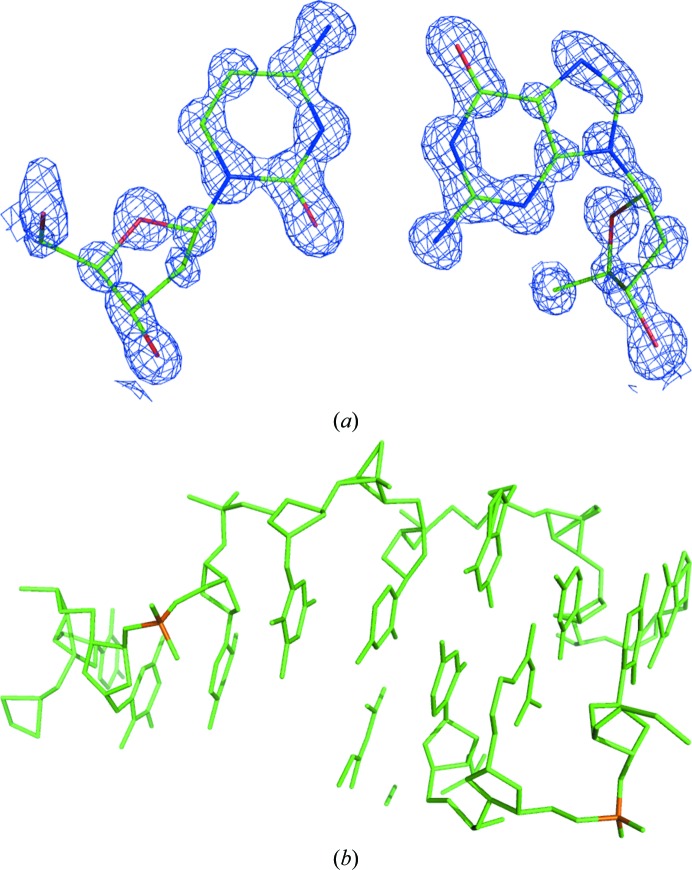
The anomalous data in the cg12ano set were analyzed with the XPREP program (Sheldrick, 2003 ▶) and the extracted anomalous differences were submitted to SHELXD (Sheldrick, 2008 ▶) with instructions to find 11 anomalous sites. The resulting constellation of anomalous sites and the native data prepared by XPREP from the cg12high set were then submitted to SHELXE (Sheldrick, 2008 ▶) for density modification, with the solvent content estimated as 32%. The resulting phase set had a pseudo-free correlation coefficient between E obs and E calc of 76%, and the corresponding map clearly revealed all of the atoms in the structure (Fig. 3 ▶ a). However, when the same procedure was performed with the lower resolution native data obtained by XPREP from the cg12ano set, the E obs/E calc pseudo-free correlation coefficient was 44% and the phases obtained from SHELXE did not lead to an interpretable map. This is the first use of the P-SAD approach to solve a novel structure; the only previous use of P-SAD was its application to the already known d(CGCGCG)2 duplex (Dauter & Adamiak, 2001 ▶).
Figure 3.
(a) A fragment of the electron-density map at the 1.5σ contour level calculated after P-SAD phasing and density modification by the programs SHELXD and SHELXE, with the final model of a Cyt–Gua pair shown. (b) The model encompassing a total of 243 atoms of the dodecamer obtained from direct-methods phasing by SHELXD (some fragments belong to symmetry-equivalent molecules).
The structure solution was also attempted by direct methods applied to the high-resolution native data set. The SHELXD program was run against the cg12high data at 0.75 Å resolution with default input parameters. Among 10 000 phase sets, 17 had an E calc/E obs correlation coefficient on all and weak reflections, CC(all) and CC(weak), of above 52 and 36%, respectively, whereas all other phase trials had a CC(all) lower than 25% and a CC(weak) lower than 12%. The best set had a CC(all) of 57.8% and a CC(weak) of 43.23%, and the obtained atomic model encompassed the whole molecule, correctly showing all 243 atoms expected in the dodecamer (Fig. 3 ▶ b). In addition, this structure could also be solved by molecular replacement using the d(CGCGCG)2 hexamer as a search model.
2.3. Structure refinement
The model built into the map obtained from SHELXE was submitted to REFMAC for isotropic refinement (Murshudov et al., 2011 ▶), accompanied by an automatic search for solvent sites by ARP/wARP (Perrakis et al., 1999 ▶), then to SHELXL (Sheldrick, 2008 ▶) for isotropic, and later anisotropic, refinement. The geometry restraints based on the standard target library (Parkinson et al., 1996 ▶) were applied to bond lengths and angles and planar nucleotide bases. The default SHELXL restraints ISOR, SIMU and DELU were applied to the anisotropic atomic displacement parameters (ADPs). The H-atom positions were recalculated at every refinement cycle in idealized positions and their isotropic ADPs were fixed at values 20% higher than the ADPs of their parent atoms. The occupancies of fragments (phosphate groups) adopting alternative conformations were refined with their sum constrained to unity. Eventually, the occupancies of solvent water O atoms were also refined, and when the occupancy parameter was refined to a value exceeding 0.95 it was fixed at unity. Cycles of refinement were interspersed with visual inspection sessions using Coot (Emsley & Cowtan, 2004 ▶) and, if necessary, the model was corrected manually, for example, by introducing alternative conformations of several phosphate groups. After applying the conjugate-gradient least-squares (CGLS) minimization method, the last round of refinement was performed using the full-matrix least-squares (FMLS) option, with the parameter shifts damped to zero, to obtain reliable estimations of all refined and derived parameters of the model. The final statistics of the refined model of d(CGCGCGCGCGCG)2 are presented in Table 1 ▶. The refined model and the corresponding structure factors have been deposited in the PDB with identification code 4ocb.
3. Results and discussion
3.1. Anomalous signal of P atoms
As stated above, the structure was solved using the anomalous signal of the P atoms present in the cg12ano data set collected using a wavelength of 1.54 Å. The initial SHELXD runs revealed potentially successful solutions with high E obs/E calc correlation coefficient values. However, there was no sharp contrast in the heights of the anomalous scatterer peaks identified by SHELXD, suggesting that some phosphorus sites may be disordered. In the final SHELXD run the program was asked to find 15 sites, and the results are presented in Table 2 ▶, in which the anomalous sites identified by SHELXD are compared with the positions of P atoms in the eventually refined structure of the dodecamer.
Table 2. Comparison of the positions of anomalous sites found by SHELXD and the refined positions of P atoms.
The occupancies and B factors (in Å2) are given for the refined P-atom sites and the relative occupancies are given for anomalous scatterer sites estimated by SHELXD. The distances (in Å) between the positions of the P atoms and the anomalous sites are also listed, as well as the distances between two P-atom sites in disordered, alternative conformations. SHELXD peaks Q12, Q15 and those following Q15 correspond to noise.
| Atom | Occupancy | B factor | Anomalous peak | Peak height | Distance Q—P | Distance Pa—Pb |
|---|---|---|---|---|---|---|
| P2 | 1.00 | 8.14 | Q1 | 1.00 | 0.21 | |
| P3a | 0.77 | 8.51 | Q6 | 0.52 | 0.39 | 1.57 |
| P3b | 0.23 | 9.80 | Q6 | 0.52 | 1.28 | 1.57 |
| P4 | 1.00 | 12.63 | Q4 | 0.74 | 0.18 | |
| P5a | 0.65 | 13.63 | Q11 | 0.38 | 0.40 | 2.49 |
| P5b | 0.35 | 15.71 | Q14 | 0.33 | 0.38 | 2.49 |
| P6 | 1.00 | 15.88 | Q9 | 0.49 | 0.08 | |
| P7 | 1.00 | 8.89 | Q2 | 0.81 | 0.16 | |
| P8a | 0.65 | 8.39 | Q3 | 0.78 | 0.23 | 0.57 |
| P8b | 0.35 | 9.81 | Q3 | 0.78 | 0.35 | 0.57 |
| P9a | 0.26 | 8.51 | Q7 | 0.51 | 1.20 | 1.46 |
| P9b | 0.74 | 11.39 | Q7 | 0.51 | 0.38 | 1.46 |
| P10 | 1.00 | 15.10 | Q8 | 0.49 | 0.29 | |
| P11a | 0.39 | 8.41 | Q13 | 0.34 | 0.30 | 1.57 |
| P11b | 0.61 | 12.15 | Q10 | 0.41 | 0.65 | 1.57 |
| P12a | 0.62 | 9.26 | Q5 | 0.53 | 0.50 | 0.84 |
| P12b | 0.38 | 8.86 | Q5 | 0.53 | 0.39 | 0.84 |
In the final structure, five phosphates are presented in a single conformation and six phosphates are modeled in double conformations. Among the first 14 anomalous sites identified by SHELXD, 13 corresponded to phosphate atoms, and only one peak (number 12 in the list) constituted noise. Four anomalous peaks are located between four pairs of disordered P atoms positioned relatively close to each other, separated by 1.57 Å or less. For the two pairs of most distant alternative phosphorus sites (2.49 and 1.57 Å), each individual site corresponds to a separate anomalous peak. This result is in keeping with the resolution limit of the cg12ano data set, nominally equal to 1.64 Å.
3.2. Refined model of the dodecamer
The chain of 12 alternating Cyt and Gua nucleotides forms the regular left-handed Z-type helix, together with the identical chain related by the crystallographic dyad perpendicular to the helix axis in the middle of its length, analogous to the helices observed in duplexes of hexameric Z-DNA structures.
In agreement with the typical features of Z-DNA, the cytosine nucleotides exist in the syn conformation of the base with respect to the deoxyribofuranose ring and the guanosine nucleotides adopt the anti conformation. In spite of the presence of spermine in the crystallization medium, no features that could correspond to the polyamine were identified in the electron-density map. All solvent sites in the crystal structure are interpreted as water molecules, although some of them may be partially occupied by ions such as Na+, counterbalancing the negative charge of the DNA phosphate groups. However, no solvent sites could convincingly be ascribed to metal ions on the basis of the coordination geometry, and the electron density alone cannot differentiate the isoelectronic Na+ and H2O moieties.
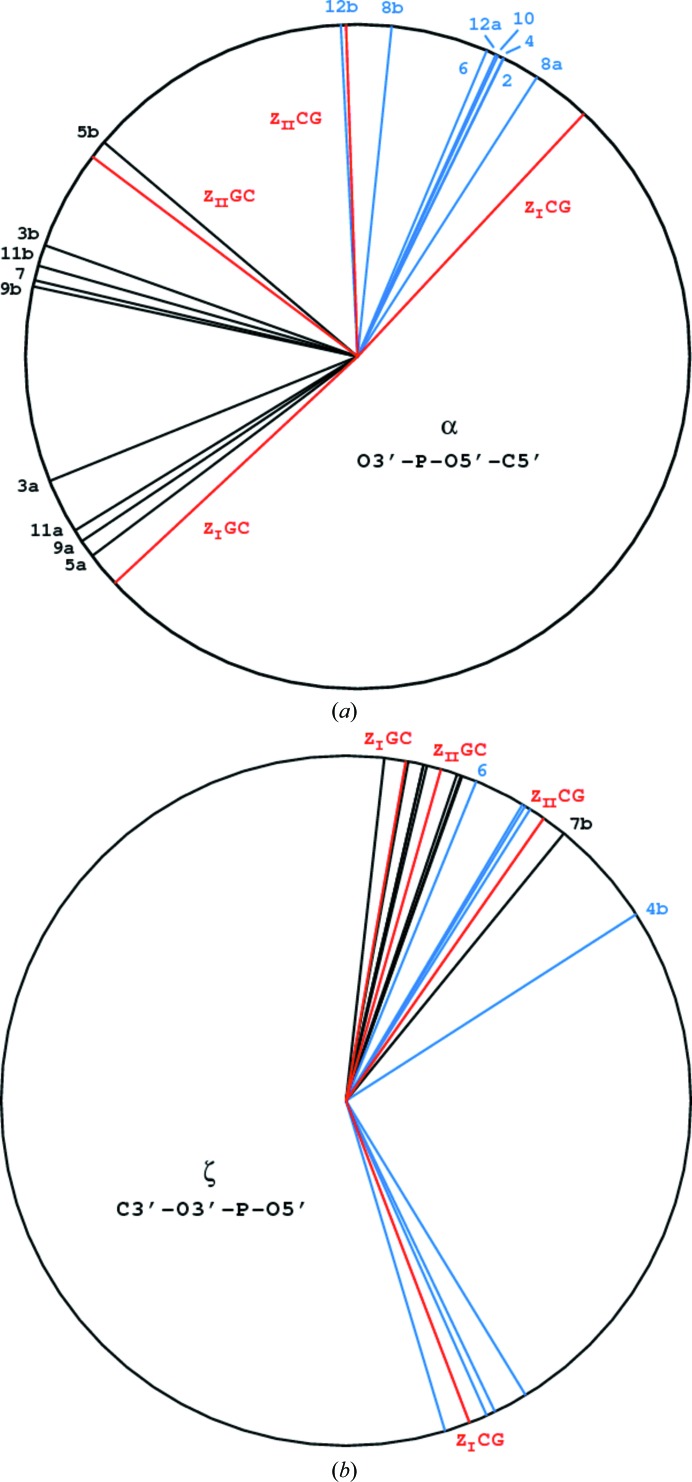
Most atoms of the nucleotide bases and sugars are well defined in the electron density, but significant disorder exists in the conformations of a number of phosphate groups. Six of them are modeled in double conformations, marked A and B, approximately corresponding to the ZI and ZII types. Four of them (P3, P5, P9 and P11) are in the GC stage of the oligomer and two (P8 and P12) in the CG stage, as illustrated in Fig. 4 ▶. The torsion angles of the sugar-phosphate backbone of the dodecamer are presented in Table 3 ▶ and illustrated in Fig. 5 ▶. The exact conformational torsion angles in the phosphate groups differ somewhat from each other and from the values of the idealized ZI and ZII structures presented in PDB models 2zna and 3zna, respectively (Wang et al., 1981 ▶) and listed by Saenger (1983 ▶). As shown in Fig. 5 ▶, the α (O3′—P—O5′—C5′) angles of the A and B conformations are closer to each other than the idealized ZI and ZII values in both the CG and GC stages of the dodecamer. The ζ (C3′—O3′—P—O5′) angles of the A and B conformers in the CG stages are generally similar to the idealized values, with two outliers, phosphates 4b and 6. The backbone, particularly the phosphate groups in the dodecamer, is much more flexible than in the hexamer model (PDB entry 3p4j; Brzezinski et al., 2011 ▶), in which all phosphate groups are well defined in single but different (ZI or ZII) conformations. However, double conformations of the phosphate groups have been observed in several hexamer duplex structures. For example, in the d(CGCGCA)–d(TGCGCG)–[Ru(NH3)6]3+ structure (PDB entry 2hto; Bharanidharan et al., 2007 ▶) four phosphate groups exist in double conformations, all at the GC stage of the backbone, and in the recently published d(CGCGCG)2 structure (PDB entry 3wbo; Chatake, 2013 ▶) seven phosphates are modeled in double conformations, three in the GC steps and four in the CG steps.
Figure 4.
Representations of the individual phosphate groups in either single or double conformations for the CG stage (a) and the GC stage (b) of the dodecamer backbone.
Table 3. Torsion angles (°) in the backbone chain and sugars of the dodecamer.
| Angle name | Angle definition | Cyt1 | Cyt3a | Cyt3b | Cyt5a | Cyt5b | Cyt7a | Cyt7b | Cyt9a | Cyt9b | Cyt11a | Cyt11b | ZI | ZII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | O3′—P—O5′—C5′ | −158.0 | 160.4 | −143.1 | 139.8 | 166.7 | −146.2 | 167.8 | −148.4 | 164.1 | −137 | 143 | ||
| β | P—O5′—C5′—C4′ | −119.6 | 171.3 | −129.6 | 172.9 | 155.5 | −124.5 | 170.7 | −121.8 | 167.8 | −139 | 164 | ||
| γ | O5′—C5′—C4′—C3′ | 54.7 | 52.9 | 52.4 | 66.0 | 49.2 | 49.3 | 55.7 | 56 | 66 | ||||
| δ | C5′—C4′—C3′—O3′ | 143.4 | 139.6 | 141.4 | 152.1 | 144.4 | 145.5 | 133.5 | 151.2 | 138 | 147 | |||
| ∊ | C4′—C3′—O3′—P | −91.7 | −94.6 | −102.5 | −100.5 | −89.7 | −95.3 | −84.3 | −106.5 | −94 | −100 | |||
| ζ | C3′—O3′—P—O5′ | 77.0 | 79.6 | 70.3 | 83.6 | 50.8 | 76.4 | 70.5 | 71.2 | 80 | 74 | |||
| χ | O4′—C1′—N1—C2 | −147.5 | −154.1 | −158.5 | −153.9 | −151.6 | −150.3 | −159 | −148 | |||||
| ν0 | C4′—O4′—C1′—C2′ | −25.0 | −28.6 | −29.8 | −24.7 | −26.7 | −25.8 | |||||||
| ν1 | O4′—C1′—C2′—C3′ | 36.6 | 38.1 | 41.6 | 36.4 | 38.2 | 37.4 | |||||||
| ν2 | C1′—C2′—C3′—C4′ | −34.1 | −32.3 | −36.8 | −34.1 | −34.0 | −34.2 | |||||||
| ν3 | C2′—C3′—C4′—O4′ | 20.6 | 16.5 | 20.5 | 20.1 | 19.1 | 20.2 | |||||||
| ν4 | C3′—C4′—O4′—C1′ | 2.7 | 7.3 | 4.8 | 2.8 | 4.9 | 3.2 | |||||||
| Saenger type | ZI | ZII | ZI | ZII | ZII | ZI | ZII | ZI | ZII | |||||
| Phosphate occupancy | 0.78 | 0.22 | 0.65 | 0.35 | 0.26 | 0.74 | 0.72 | 0.28 | ||||||
| Pseudorotation P | 157.4 | 149.9 | 153.9 | 157.4 | 154.2 | 156.4 | ||||||||
| Pseudorotation τ | 36.9 | 37.3 | 41.0 | 36.9 | 37.8 | 37.3 | ||||||||
| Sugar pucker | C2′-endo | C2′-endo | C2′-endo | C2′-endo | C2′-endo | C2′-endo | ||||||||
| Angle name | Angle definition | Gua2a | Gua2b | Gua4a | Gua4b | Gua6 | Gua8a | Gua8b | Gua10a | Gua10b | Gua12a | Gua12b | ZI | ZII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | O3′—P–O5′—C5′ | 63z.9 | 64.0 | 67.1 | 57.3 | 84.1 | 65.0 | 65.4 | 92.9 | 47 | 92 | |||
| β | P—O5′—C5′—C4′ | −173.1 | −174.1 | −170.7 | −174.3 | −174.4 | −172.6 | 179.4 | −165.1 | 179 | −167 | |||
| γ | O5′—C5′—C4′—C3′ | 176.5 | 178.8 | −176.4 | 177.7 | 179.5 | −168.9 | 155.4 | −169 | 157 | ||||
| δ | C5′—C4′—C3′—O3′ | 94.3 | 85.4 | 122.9 | 92.9 | 94.6 | 94.0 | 150.6 | 84.9 | 99 | 94 | |||
| ∊ | C4′—C3′—O3′—P | −129.9 | 159.2 | −124.7 | 169.8 | −170.9 | −112.6 | 179.7 | −109.0 | 179.4 | −104 | −179 | ||
| ζ | C3′—O3′—P-O5′ | −64.4 | 58.7 | −58.6 | 32.7 | 67.8 | −65.9 | 57.7 | −73.3 | 59.2 | −69 | 55 | ||
| χ | O4′—C1′—N9—C4 | 67.1 | 61.1 | 56.8 | 58.8 | 58.0 | 78.8 | 64.7 | 68 | 62 | ||||
| ν0 | C4′—O4′—C1′—C2′ | −6.4 | −10.6 | −4.2 | −2.9 | −6.5 | −11.7 | −11.6 | ||||||
| ν1 | O4′—C1′—C2′—C3′ | −9.5 | −5.2 | −12.8 | −15.4 | −10.8 | 30.6 | −17.7 | ||||||
| ν2 | C1′—C2′—C3′—C4′ | 20.5 | 17.9 | 23.4 | 26.2 | 22.8 | −36.0 | 37.4 | ||||||
| ν3 | C2′—C3′—C4′—O4′ | −24.5 | −23.7 | −26.5 | −28.5 | −27.0 | 30.8 | −44.8 | ||||||
| ν4 | C3′−C4′−O4′−C1′ | 19.7 | 22.8 | 19.4 | 20.2 | 21.1 | −12.4 | 35.2 | ||||||
| Saenger type | ZI | ZI | ZI | ZI | ZII | ZI | ZI | ZII | ||||||
| Phosphate occupancy | 0.65 | 0.35 | 0.62 | 0.38 | ||||||||||
| Pseudorotation P | 33.3 | 43.2 | 27.4 | 24.2 | 32.0 | 180.5 | 32.8 | |||||||
| Pseudorotation τ | 24.2 | 24.6 | 26.4 | 28.7 | 26.9 | 36.0 | 44.4 | |||||||
| Sugar pucker | C3′-endo | C4′-exo | C3′-endo | C3′-endo | C3′-endo | C3′-exo | C3′-endo | |||||||
Figure 5.
Torsion angles around the backbone bonds involving a P atom (a) for the α bond (O3′—P—O5′—C5′) and (b) for the ζ bond (C3′—O3′—P—O5′). The canonical values for the ZI and ZII phosphate conformations (Saenger, 1983 ▶) are shown in red, the bonds in the CG stage in black and those in the GC stage in blue.
In spite of the disorder of the phosphates, the location of water molecules inside the helix groove is analogous to the typical situation found in other Z-DNA structures. Two atoms, O2-Cyt and N2-Gua, which form the ‘internal’ Watson–Crick hydrogen bond, are also hydrogen-bonded to one water molecule each. The water molecules connected to N2-Gua are also hydrogen-bonded to a phosphate oxygen OP2 of the next residue (except in residue Gua6), but the water molecules connected to O2-Cyt are bonded to the next water molecule, which forms a hydrogen bond to the OP2 phosphate O atom, also belonging formally to the next residue. However, this arrangement is not highly regular and many water sites are disordered, not fully occupied and characterized by relatively weak electron densities.
Within the atoms forming the Watson–Crick pairs at the external side of the helix, all O6-Gua atoms are engaged in hydrogen bonds to water molecules, but two of the N4-Cyt atoms do not have hydrogen-bond partners. Only one N7-Gua8 atom is hydrogen-bonded; the other five have no partners at all. The water molecules again display a substantial degree of disorder and many sites are partially occupied.
3.3. Accuracy of the structure
The ultrahigh resolution of the diffraction data permitted the classic, small-molecule-style estimation of the uncertainties of all coordinate and ADP parameters of all non-H atoms in the atomic model from the inversion of the least-squares matrix. In fact, the resolution of 0.75 Å is higher than the maximum limit of about 0.8 Å achievable with Cu Kα radiation and four-circle diffractometers, the approach traditionally used in the crystallography of small organic molecules. The number of refined parameters for the Z-DNA dodecamer is 3612 and the number of reflections used in refinement is 32 194, yielding an N refl/N par ratio of 8.9, sufficient for meaningful anisotropic refinement of all (non-H) atoms by the FMLS approach.
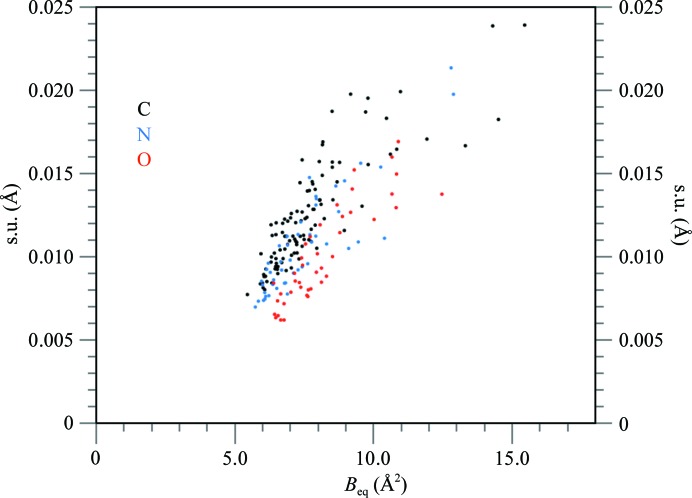
Fig. 6 ▶ shows the positional accuracy (standard uncertainty, s.u.) of all non-disordered dodecamer atoms estimated from FMLS refinement as a function of their B factors. In general, the uncertainties are proportional to the B factors and depend on the atomic number Z at, but the differences between the accuracies of O, N and C atoms are not as clearly pronounced as in the case of the Z-DNA hexamer duplex d(CGCGCG)2 refined at 0.55 Å resolution (PDB entry 3p4j; see Fig. 3 of Brzezinski et al., 2011 ▶). The proportionality ratio s.u. × Z at/B is about 0.02 for the dodecamer and 0.0065 for the hexamer. Similarly, the Wilson B factor is 6.9 Å2 for the dodecamer and only 2.5 Å2 for the hexamer. This results in a lower resolution limit for the diffraction data of the dodecamer (0.75 Å) than for the hexamer (0.55 Å), in spite of the very similar size and content of both crystal unit cells.
Figure 6.
The standard uncertainties estimated from the FMLS refinement for all non-disordered atoms in the dodecamer as a function of their B factors. C atoms are shown in black, N atoms in blue and O atoms in red.
In the 3p4j structure of the Z-DNA hexamer, the variability of bonds and angles of the same type found in cytidines and guanosines was comparable to the level of uncertainty of these parameters obtained from the FMLS procedure. Table 4 ▶ shows the analogous values obtained for the dodecamer compared with those of the hexamer. The accuracy of the geometrical parameters is approximately three times lower and the spread of equivalent bonds and angles is about three times larger in the structure of the dodecamer than in the hexamer 3p4j, reflecting the differences in data resolution and final R factors, but also suggesting a higher flexibility for the dodecamer structure. However, the variability of the geometrical parameters in the dodecamer is comparable to that observed for the structure of the hexamer (PDB entry 1i0t; Tereshko et al., 2001 ▶) refined at 0.6 Å resolution.
Table 4. Deviation of bond lengths (Å) and angles (°) from the mean values of their individual types and uncertainty ranges.
Only fully occupied atoms in single conformations are included in the statistics.
| Dodecamer | Hexamer 3p4j | Hexamer 1i0t | ||||
|---|---|---|---|---|---|---|
| Moiety | N | R.m.s.d.† | Range of s.u.‡ | R.m.s.d.† | Range of s.u.‡ | R.m.s.d.† |
| Bonds | ||||||
| Cytidines | 54 | 0.0107 | 0.0051–0.0103 | 0.0033 | 0.0017–0.0035 | 0.0137 |
| Guanosines | 77 | 0.0127 | 0.0049–0.0171 | 0.0038 | 0.0018–0.0034 | 0.0139 |
| Sugars | 95 | 0.0132 | 0.0041–0.0172 | 0.0070 | 0.0017–0.0035 | 0.0137 |
| Angles | ||||||
| Cytidines | 72 | 0.89 | 0.36–0.67 | 0.41 | 0.10–0.19 | 1.17 |
| Guanosines | 112 | 0.84 | 0.35–1.28 | 0.38 | 0.10–0.19 | 1.07 |
| Sugars | 125 | 1.26 | 0.30–1.05 | 1.24 | 0.09–0.18 | 1.38 |
Root-mean-square deviations (r.m.s.d.s) of bonds and angles from the mean values of their individual types
Range of standard uncertainties (s.u.) of bond lengths and angles estimated from the FMLS refinement. Standard uncertainties are not available for 1i0t.
3.4. Arrangement of helices and comparison with other Z-DNA structures
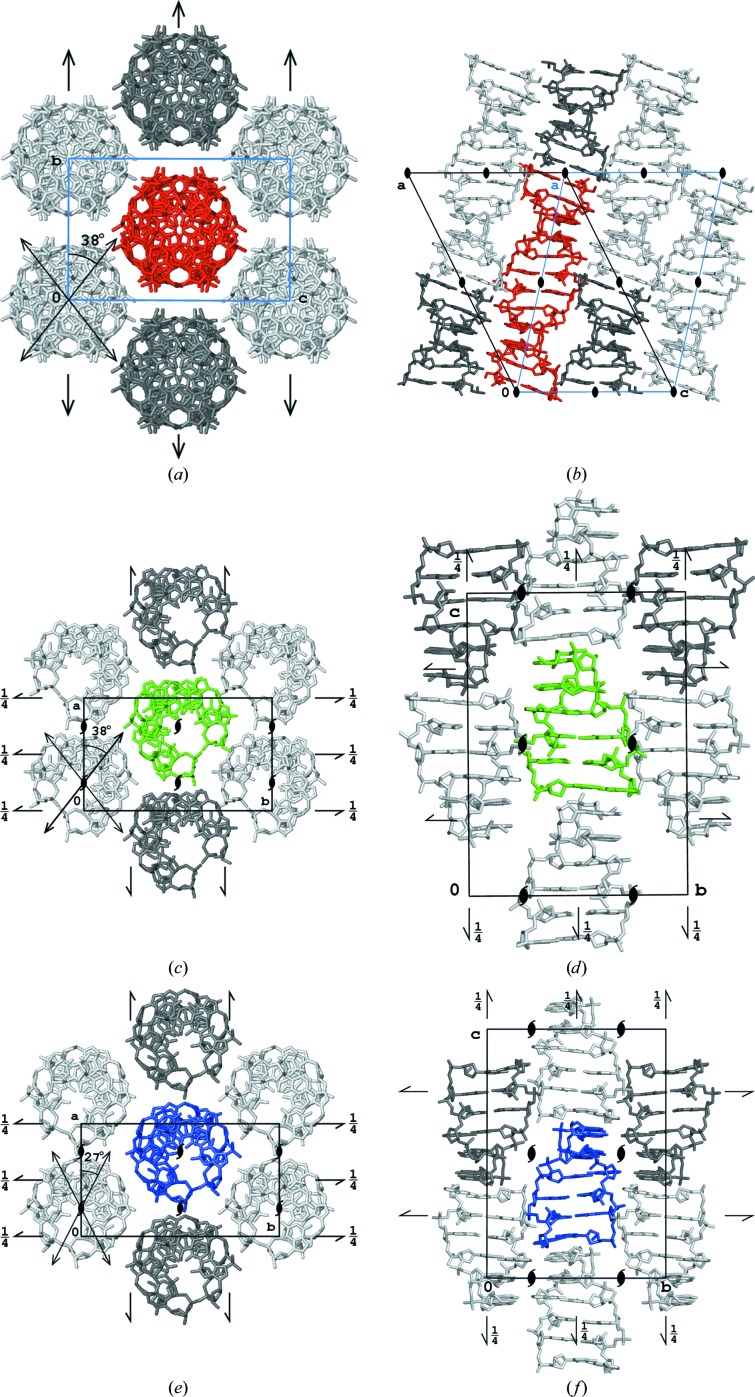
The d(CGCGCGCGCGCG)2 helix is straight and elongated along the diagonal (1, 0, 1) direction of the standard C-centered monoclinic unit cell, as schematically illustrated in Figs. 7 ▶(a) and 7 ▶(b). This cell can be expressed as a nonstandard, I-centered monoclinic cell in which the helices are parallel to the c cell direction with length 44.5 Å. This value is within the range 42–45 Å (Supplementary Table S1) observed for crystals of Z-DNA hexamers, in which two hexamers correspond to one full turn of the helix oriented along one of the unit-cell edges.
Figure 7.
Packing of molecules in the structures of (a, b) the dodecamer, (c, d) hexamer 3p4j and (e, f) hexamer 1i0t viewed along the helices and along the perpendicular direction parallel to the shortest edge of the corresponding unit cell. For the dodecamer, the standard monoclinic C-centered cell is shown in black and the nonstandard monoclinic I-centered cell is shown in blue.
There are no structures of straight Z-DNA duplexes crystallized in space group C2 in the PDB, but the packing of the dodecamer duplexes described here in the monoclinic C2 cell is, in fact, analogous to the arrangement of hexamer duplexes in the orthorhombic P212121 crystal forms. As pointed out previously (Egli et al., 1991 ▶; Brzezinski et al., 2011 ▶), there are two types, A and B, of orthorhombic Z-DNA hexamer crystals which differ by the rotation of the helix along its length and a slight translation with respect to the symmetry axes. This is illustrated in Figs. 7 ▶(c) and 7 ▶(d) and in Figs. 7 ▶(e) and 7 ▶(f). The arrangement of helices in the plane perpendicular to their length is hexagonal, and is identical in all three compared crystal types. The dodecamers form infinite helices throughout the crystal, but are different from the hexameric crystals in that these helices lack one phosphate group every 12 base pairs instead of every six base pairs.
The individual hexamer duplexes have twofold symmetry axes perpendicular to their helices at half length, which are strict in palindromic sequences and approximate otherwise. In the orthorhombic crystal forms, these symmetry elements are noncrystallographic. Their orientation is different in the two orthorhombic crystal types, forming an angle of about 27° with the shortest a edge of the cell in the A-form and an angle of about 38° in the B-form, as shown in Figs. 7 ▶(c) and 7 ▶(e) and listed in Supplementary Table S1. The d(CGCGCGCGCGCG)2 dodecamer duplex also possesses a twofold symmetry axis perpendicular to its length, and it is identical to the crystallographic twofold axis of the monoclinic system. Moreover, analogous to the hexameric forms, it is possible to identify the approximate twofold axes at every quarter of the length of the dodecamer duplex, which are perturbed only by the presence of the phosphate groups between residues 6 and 7. These approximate, noncrystallographic dyads are oriented 38° from the direction of the shortest b edge of the unit cell. The mutual orientation of the helices in the crystal of the dodecamer is the same as in the B form of the crystals of hexamers.
The similarity of these two structures is apparent after superposition of a single dodecamer duplex on a pair of hexamer duplexes from the 3p4j structure, with the root-mean-square deviation (r.m.s.d) of all corresponding atoms being 0.58 Å. If only the bases from both structures are overlapped, the r.m.s.d is 0.47 Å and the result is illustrated in Fig. 8 ▶. As mentioned above, the periodicity along the practically infinite Z-DNA helix in the dodecamer is 44.5 Å, which corresponds to an average distance between the planes of the Watson–Crick base pairs of 3.71 Å. In the crystals of hexamers this distance varies between 3.50 and 3.75 Å, and in the 3p4j structure it is 3.74 Å.
Figure 8.
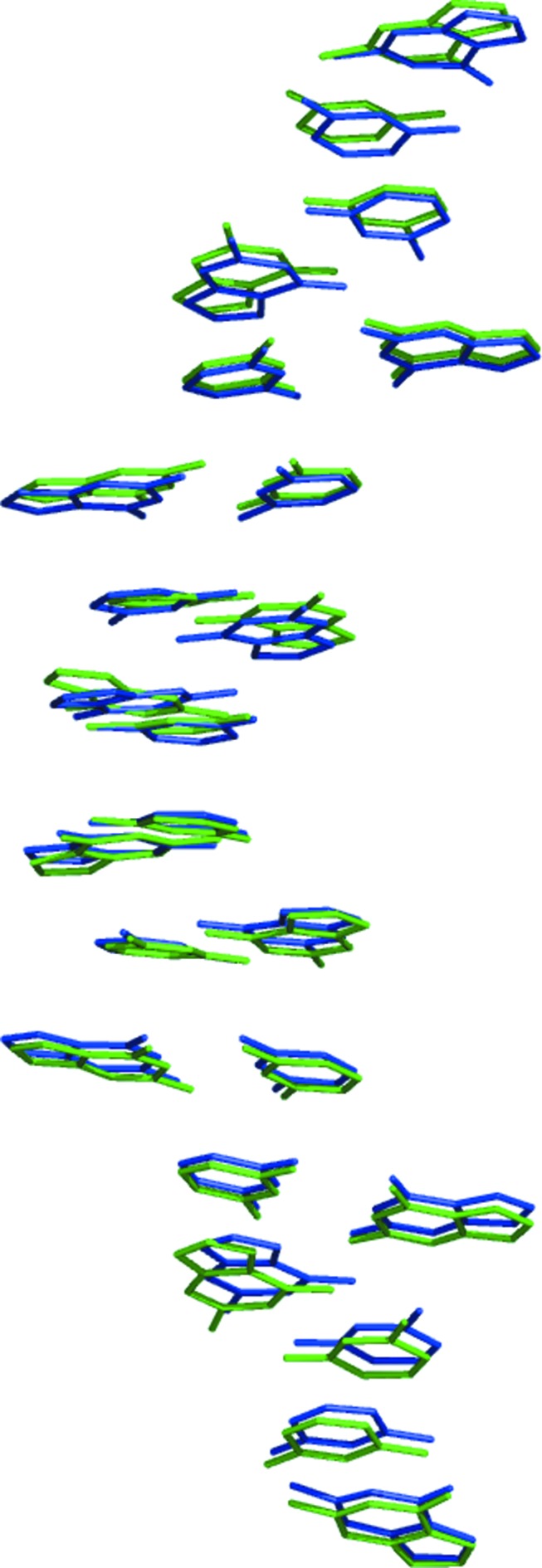
The 12 base pairs of the dodecamer (green) and from two of the 3p4j hexamer duplexes (blue) superimposed onto each other.
4. Conclusions
The crystal structure of the d(CGCGCGCGCGCG)2 dodecamer of Z-DNA is the first example of using the anomalous signal of the native P atoms to solve a novel crystal structure of a nucleotide oligomer. This approach is analogous to the use of the anomalous signal of S atoms in cysteines and methionines for phasing crystal structures of proteins. The arrangement of dodecamer duplexes is highly analogous to the organization in crystal structures of other Z-DNA oligomers, with duplexes effectively forming infinite helices packed in a parallel, hexagonal fashion. However, the mutual disposition of the neighboring helices in the crystal of the dodecamer differs from that observed in other known structures of Z-DNA and corresponds to space group C2. The dodecamer displays a significant degree of flexibility, especially in the backbone, with six out of 11 phosphate groups modeled in double conformations, positioned either closer to the inside of the helix, analogous to the conformation known as ZI, or towards the external side of the helix, similar to the ZII conformation.
5. Related literature
The following references are cited in the Supporting Information: Wang et al. (1981 ▶, 1984 ▶), Tereshko et al. (2001 ▶), Narayana et al. (2006 ▶), Egli et al. (1991 ▶), Bancroft et al. (1994 ▶), Chatake et al. (2005 ▶), Schuerman et al. (2003 ▶), Thiyagarajan et al. (2002 ▶, 2004 ▶), Harper et al. (1998 ▶), Van Meervelt et al. (1990 ▶), Moore et al. (1995 ▶), Brzezinski et al. (2011 ▶), Dauter & Adamiak (2001 ▶), Drozdzal et al. (2013 ▶), Ohishi et al. (1991 ▶, 2002 ▶, 2007 ▶, 2008 ▶), Gessner et al. (1989 ▶), Wilds et al. (2002 ▶), Coll et al. (1986 ▶, 1989 ▶), Sanishvili et al. (2007 ▶), Ohishi, Nakanishi et al. (1996 ▶), Ohishi, Terasoma et al. (1996 ▶), Fenn et al. (2011 ▶), Pan & Sundaralingam (2003 ▶), Ho et al. (1985 ▶), Brown et al. (1986 ▶), Schuerman et al. (1998 ▶), Bharanidharan et al. (2007 ▶), Sadasivan & Gautham (1995 ▶), Karthe & Gautham (1998 ▶), Kagawa et al. (1991 ▶), Fujii et al. (1982 ▶), Chevrier et al. (1986 ▶), Geierstanger et al. (1991 ▶), Zhou & Ho (1990 ▶), Ginell et al. (1990 ▶), Schneider et al. (1992 ▶), Schroth et al. (1993 ▶), Eichman et al. (1999 ▶), Cervi et al. (1993 ▶), Peterson et al. (1996 ▶), Karthe et al. (1996 ▶), Mooers et al. (1997 ▶), Pan et al. (1997 ▶), Mandal, Chandrasekaran et al. (2012 ▶), Brennan et al. (1986 ▶), Ban et al. (1996 ▶), Westhof et al. (1988 ▶), Crawford et al. (1980 ▶), Kumar et al. (1992 ▶), Mandal, Venkadesh et al. (2012 ▶), Malinina et al. (1994 ▶, 1998 ▶), Drew & Dickerson (1980 ▶), Venkadesh et al. (2009 ▶), Parkinson et al. (1995 ▶), Zhang et al. (1992 ▶), Doi et al. (1993 ▶), Pallan et al. (2012 ▶), Atwell et al. (2001 ▶) and Chattopadhyaya et al. (1990 ▶).
Supplementary Material
PDB reference: d(CGCGCGCGCGCG)2, 4ocb
Supplementary table of all Z-DNA structures in the PDB. DOI: 10.1107/S1399004714004684/tz5053sup1.pdf
Acknowledgments
This project was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and with Federal funds from the National Cancer Institute, National Institutes of Health (Contract No. NO1-CO-12400). Diffraction data were collected at the NE-CAT beamline 24-ID and SER-CAT beamline 22-ID at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: TZ5053).
References
- Atwell, S., Meggers, E., Spraggon, G. & Schultz, P. G. (2001). J. Am. Chem. Soc. 123, 12364–12367. [DOI] [PubMed]
- Ban, C., Ramakrishnan, B. & Sundaralingam, M. (1996). Biophys. J. 71, 1215–1221. [DOI] [PMC free article] [PubMed]
- Bancroft, D., Williams, L. D., Rich, A. & Egli, M. (1994). Biochemistry, 33, 1073–1086. [DOI] [PubMed]
- Berman, H. M., Olson, W. K., Beveridge, D. L., Westbrook, J., Gelbin, A., Demeny, T., Hsieh, S. H., Srinivasan, A. R. & Schneider, B. (1992). Biophys. J. 63, 751–759. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bharanidharan, D., Thiyagarajan, S. & Gautham, N. (2007). Acta Cryst. F63, 1008–1013. [DOI] [PMC free article] [PubMed]
- Brennan, R. G., Westhof, E. & Sundaralingam, M. (1986). J. Biomol. Struct. Dyn. 3, 649–665. [DOI] [PubMed]
- Brown, T., Kneale, G., Hunter, W. N. & Kennard, O. (1986). Nucleic Acids Res. 14, 1801–1809. [DOI] [PMC free article] [PubMed]
- Brzezinski, K., Brzuszkiewicz, A., Dauter, M., Kubicki, M., Jaskolski, M. & Dauter, Z. (2011). Nucleic Acids Res. 39, 6238–6248. [DOI] [PMC free article] [PubMed]
- Cervi, A. R., Guy, A., Leonard, G. A., Téoule, R. & Hunter, W. N. (1993). Nucleic Acids Res. 21, 5623–5629. [DOI] [PMC free article] [PubMed]
- Chatake, T. (2013). J. Synchrotron Rad. 20, 864–868. [DOI] [PMC free article] [PubMed]
- Chatake, T., Tanaka, I., Umino, H., Arai, S. & Niimura, N. (2005). Acta Cryst. D61, 1088–1098. [DOI] [PubMed]
- Chattopadhyaya, R., Grzeskowiak, K. & Dickerson, R. E. (1990). J. Mol. Biol. 211, 189–210. [DOI] [PubMed]
- Chevrier, B., Dock, A. C., Hartmann, B., Leng, M., Moras, D., Thuong, M. T. & Westhof, E. (1986). J. Mol. Biol. 188, 707–719. [DOI] [PubMed]
- Coll, M., Saal, D., Frederick, C. A., Aymami, J., Rich, A. & Wang, A. H.-J. (1989). Nucleic Acids Res. 17, 911–923. [DOI] [PMC free article] [PubMed]
- Coll, M., Wang, A. H.-J., van der Marel, G. A., van Boom, J. H. & Rich, A. (1986). J. Biomol. Struct. Dyn. 4, 157–172. [DOI] [PubMed]
- Crawford, J. L., Kolpak, F. J., Wang, A. H.-J., Quigley, G. J., van Boom, J. H., van der Marel, G. & Rich, A. (1980). Proc. Natl Acad. Sci. USA, 77, 4016–4020. [DOI] [PMC free article] [PubMed]
- Dauter, Z. & Adamiak, D. A. (2001). Acta Cryst. D57, 990–995. [DOI] [PubMed]
- Diederichs, K. (2010). Acta Cryst. D66, 733–740. [DOI] [PubMed]
- Doi, M., Inoue, M., Tomoo, K., Ishida, T., Ueda, Y., Akagi, M. & Urata, H. (1993). J. Am. Chem. Soc. 115, 10432–10433.
- Drew, H. R. & Dickerson, R. E. (1980). J. Mol. Biol. 152, 723–736. [DOI] [PubMed]
- Drew, H., Takano, T., Tanaka, S., Itakura, K. & Dickerson, R. E. (1980). Nature (London), 286, 567–573. [DOI] [PubMed]
- Drozdzal, P., Gilski, M., Kierzek, R., Lomozik, L. & Jaskolski, M. (2013). Acta Cryst. D69, 1180–1190. [DOI] [PubMed]
- Egli, M., Williams, L. D., Gao, Q. & Rich, A. (1991). Biochemistry, 30, 11388–11402. [DOI] [PubMed]
- Eichman, B. F., Schroth, G. P., Basham, B. E. & Ho, P. S. (1999). Nucleic Acids Res. 27, 543–550. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Eriksson, M. A. L. & Laaksonen, A. (1992). Biopolymers, 32, 1035–1059. [DOI] [PubMed]
- Fenn, T. D., Schnieders, M. J., Mustyakimov, M., Wu, C., Langan, P., Pande, V. S. & Brunger, A. T. (2011). Structure, 19, 523–533. [DOI] [PMC free article] [PubMed]
- Fujii, S., Wang, A. H.-J., van der Marel, G., van Boom, J. H. & Rich, A. (1982). Nucleic Acids Res. 10, 7879–7892. [DOI] [PMC free article] [PubMed]
- Geierstanger, B. H., Kagawa, T. F., Chen, S.-L., Quigley, G. J. & Ho, P. S. (1991). J. Biol. Chem. 266, 20185–20191. [DOI] [PubMed]
- Gessner, R. V., Frederick, C. A., Quigley, G. J., Rich, A. & Wang, A. H.-J. (1989). J. Biol. Chem. 264, 7921–7935. [DOI] [PubMed]
- Ginell, S. L., Kuzmich, S., Jones, R. A. & Berman, H. M. (1990). Biochemistry, 29, 10461–10465. [DOI] [PubMed]
- Harper, A., Brannigan, J. A., Buck, M., Hewitt, L., Lewis, R. J., Moore, M. H. & Schneider, B. (1998). Acta Cryst. D54, 1273–1284. [DOI] [PubMed]
- Ho, P. S., Frederick, C. A., Quigley, G. J., van der Marel, G. A., van Boom, J. H., Wang, A. H.-J. & Rich, A. (1985). EMBO J. 4, 3617–3623. [DOI] [PMC free article] [PubMed]
- Kagawa, T. F., Geierstanger, B. H., Wang, A. H.-J. & Ho, P. S. (1991). J. Biol. Chem. 266, 20175–20184. [DOI] [PubMed]
- Karthe, P. & Gautham, N. (1998). Acta Cryst. D54, 501–509. [DOI] [PubMed]
- Karthe, P., Krishnaswamy, S. & Gautham, N. (1996). Acta Cryst. A52, C149.
- Kumar, V. D., Harrison, R. W., Andrews, L. C. & Weber, I. T. (1992). Biochemistry, 31, 1541–1550. [DOI] [PubMed]
- Laaksonen, A., Nilsson, L. G., Jönsson, B. & Teleman, O. (1989). Chem. Phys. 129, 175–183.
- Malinina, L., Tereshko, V., Ivanova, E., Subirana, J. A., Zarytova, V. & Nekrasov, Y. (1998). Biophys. J. 74, 2482–2490. [DOI] [PMC free article] [PubMed]
- Malinina, L., Urpí, L., Salas, X., Huynh-Dinh, T. & Subirana, J. A. (1994). J. Mol. Biol. 243, 484–493. [DOI] [PubMed]
- Mandal, P. K., Chandrasekaran, A. R., Madhanagopal, B. R., Venkadesh, S. & Gautham, N. (2012). J. Cryst. Growth, 354, 20–26.
- Mandal, P. K., Venkadesh, S. & Gautham, N. (2012). Acta Cryst. F68, 1420–1426. [DOI] [PMC free article] [PubMed]
- Mooers, B. H. M., Eichman, B. F. & Ho, P. S. (1997). J. Mol. Biol. 269, 796–810. [DOI] [PubMed]
- Moore, M. H., Van Meervelt, L., Salisbury, S. A., Lin, P. K. T. & Brown, D. M. (1995). J. Mol. Biol. 251, 665–673. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Narayana, N., Shamala, N., Ganesh, K. N. & Viswamitra, M. A. (2006). Biochemistry, 45, 1200–1211. [DOI] [PubMed]
- Ohishi, H., Kunisawa, S., van der Marel, G., van Boom, J. H., Rich, A., Wang, A. H.-J., Tomita, K. & Hakoshima, T. (1991). FEBS Lett. 284, 238–244. [DOI] [PubMed]
- Ohishi, H., Nakanishi, I., Inubushi, K., van der Marel, G., van Boom, J. H., Rich, A., Wang, A. H.-J., Hakoshima, T. & Tomita, K. (1996). FEBS Lett. 391, 153–156. [DOI] [PubMed]
- Ohishi, H., Odoko, M., Grzeskowiak, K., Hiyama, Y., Tsukamoto, K., Maezaki, N., Ishida, T., Tanaka, T., Okabe, N., Fukuyama, K., Zhou, D. Y. & Nakatani, K. (2008). Biochem. Biophys. Res. Commun. 366, 275–280. [DOI] [PubMed]
- Ohishi, H., Suzuki, K., Ohtsuchi, M., Hakoshima, T. & Rich, A. (2002). FEBS Lett. 523, 29–34. [DOI] [PubMed]
- Ohishi, H., Terasoma, N., Nakanishi, I., van der Marel, G., van Boom, J. H., Rich, A., Wang, A. H.-J., Hakoshima, T. & Tomita, K. (1996). FEBS Lett. 398, 291–296. [DOI] [PubMed]
- Ohishi, H., Tozuka, Y., Da-Yang, Z., Ishida, T. & Nakatani, K. (2007). Biochem. Biophys. Res. Commun. 358, 24–28. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pallan, P. S., Marquez, V. E. & Egli, M. (2012). Biochemistry, 51, 2639–2641. [DOI] [PMC free article] [PubMed]
- Pan, B., Ban, C., Wahl, M. C. & Sundaralingam, M. (1997). Biophys. J. 73, 1553–1561. [DOI] [PMC free article] [PubMed]
- Pan, B. & Sundaralingam, M. (2003). Acta Cryst. D59, 433–437. [DOI] [PubMed]
- Parkinson, G. N., Arvanitis, G. M., Lessinger, L., Ginell, S. L., Jones, R., Gaffney, B. & Berman, H. M. (1995). Biochemistry, 34, 15487–15495. [DOI] [PubMed]
- Parkinson, G., Vojtechovsky, J., Clowney, L., Brünger, A. T. & Berman, H. M. (1996). Acta Cryst. D52, 57–64. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Peterson, M. R., Harrop, S. J., McSweeney, S. M., Leonard, G. A., Thompson, A. W., Hunter, W. N. & Helliwell, J. R. (1996). J. Synchrotron Rad. 3, 24–34. [DOI] [PubMed]
- Sadasivan, C. & Gautham, N. (1995). J. Mol. Biol. 248, 918–930. [DOI] [PubMed]
- Saenger, W. (1983). Principles of Nucleic Acid Structure. New York: Springer-Verlag.
- Sanishvili, R., Besnard, C., Camus, F., Fleurant, M., Pattison, P., Bricogne, G. & Schiltz, M. (2007). J. Appl. Cryst. 40, 552–558.
- Schneider, B., Ginell, S. L., Jones, R., Gaffney, B. & Berman, H. M. (1992). Biochemistry, 31, 9622–9628. [DOI] [PubMed]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed]
- Schroth, G. P., Kagawa, T. F. & Ho, P. S. (1993). Biochemistry, 32, 13381–13392. [DOI] [PubMed]
- Schuerman, G., Van Hecke, K. & Van Meervelt, L. (2003). Acta Cryst. D59, 1525–1528. [DOI] [PubMed]
- Schuerman, G. S., Van Meervelt, L., Loakes, D., Brown, D. M., Lin, P. K. T., Moore, M. H. & Salisbury, S. A. (1998). J. Mol. Biol. 282, 1005–1011. [DOI] [PubMed]
- Sheldrick, G. M. (2003). XPREP v. 6.14. Bruker–Nonius Inc., Madison, Wisconsin, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tereshko, V., Wilds, C. J., Minasov, G., Prakash, T. P., Maier, M. A., Howard, A., Wawrzak, Z., Manoharan, M. & Egli, M. (2001). Nucleic Acids Res. 29, 1208–1215. [DOI] [PMC free article] [PubMed]
- Thiyagarajan, S., Rajan, S. S. & Gautham, N. (2004). Nucleic Acids Res. 32, 5945–5953. [DOI] [PMC free article] [PubMed]
- Thiyagarajan, S., Satheesh Kumar, P., Rajan, S. S. & Gautham, N. (2002). Acta Cryst. D58, 1381–1384. [DOI] [PubMed]
- Van Meervelt, L., Moore, M. H., Lin, P. K., Brown, D. M. & Kennard, O. (1990). J. Mol. Biol. 216, 773–781. [DOI] [PubMed]
- Venkadesh, S., Mandal, P. K. & Gautham, N. (2009). Acta Cryst. F65, 8–13. [DOI] [PMC free article] [PubMed]
- Wang, A. H.-J., Hakoshima, T., van der Marel, G., van Boom, J. H. & Rich, A. (1984). Cell, 37, 321–331. [DOI] [PubMed]
- Wang, A. H.-J., Quigley, G. J., Kolpak, F. J., Crawford, J. L., van Boom, J. H., van der Marel, G. & Rich, A. (1979). Nature (London), 282, 680–686. [DOI] [PubMed]
- Wang, A. H.-J., Quigley, G. J., Kolpak, F. J., van der Marel, G., van Boom, J. H. & Rich, A. (1981). Science, 211, 171–176. [DOI] [PubMed]
- Westhof, E., Hosur, M. V. & Sundaralingam, M. (1988). Biochemistry, 27, 5742–5747. [DOI] [PubMed]
- Wilds, C. J., Pattanayek, R., Pan, C., Wawrzak, Z. & Egli, M. (2002). J. Am. Chem. Soc. 124, 14910–14916. [DOI] [PubMed]
- Zhang, H., van der Marel, G. A., van Boom, J. H. & Wang, A. H.-J. (1992). Biopolymers, 32, 1559–1569. [DOI] [PubMed]
- Zhou, G. & Ho, P. S. (1990). Biochemistry, 29, 7229–7236. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: d(CGCGCGCGCGCG)2, 4ocb
Supplementary table of all Z-DNA structures in the PDB. DOI: 10.1107/S1399004714004684/tz5053sup1.pdf