The preliminary results of the crystallization and room-temperature diffraction of an inositol dehydrogenase are reported.
Keywords: inositol dehydrogenase, Lactobacillus casei BL23
Abstract
Lactobacillus casei BL23 contains two genes, iolG1 and iolG2, homologous with inositol dehydrogenase encoding genes from many bacteria. Inositol dehydrogenase catalyzes the oxidation of inositol with concomitant reduction of NAD+. The protein encoded by iolG2, LcIDH2, has been purified to homogeneity, crystallized and cryoprotected for diffraction at 77 K. The crystals had a high mosaicity and poor processing statistics. Subsequent diffraction measurements were performed without cryoprotectant at room temperature. These crystals were radiation-resistant and a full diffraction data set was collected at room temperature to 1.6 Å resolution.
1. Introduction
Inositol dehydrogenase (IDH) catalyzes the NAD+-dependent conversion of myo-inositol into scyllo-inosose (Ramaley et al., 1979 ▶; Daniellou et al., 2007 ▶). Many bacterial species contain multiple apparent IDH-encoding genes, although it is unknown whether these genes represent redundancies or whether they possess different substrate specificities. In our previous work, we proposed that the genes that are annotated as IDH could be divided into four subclasses based on specific protein sequence motifs (van Straaten et al., 2010 ▶). One species that possesses multiple IDH genes is Lactobacillus casei BL23, a probiotic lactic acid bacterium that possesses two inositol dehydrogenase genes annotated iolG1 and iolG2 (Yebra et al., 2007 ▶). We have crystallized the protein encoded by the latter, LcIDH2.
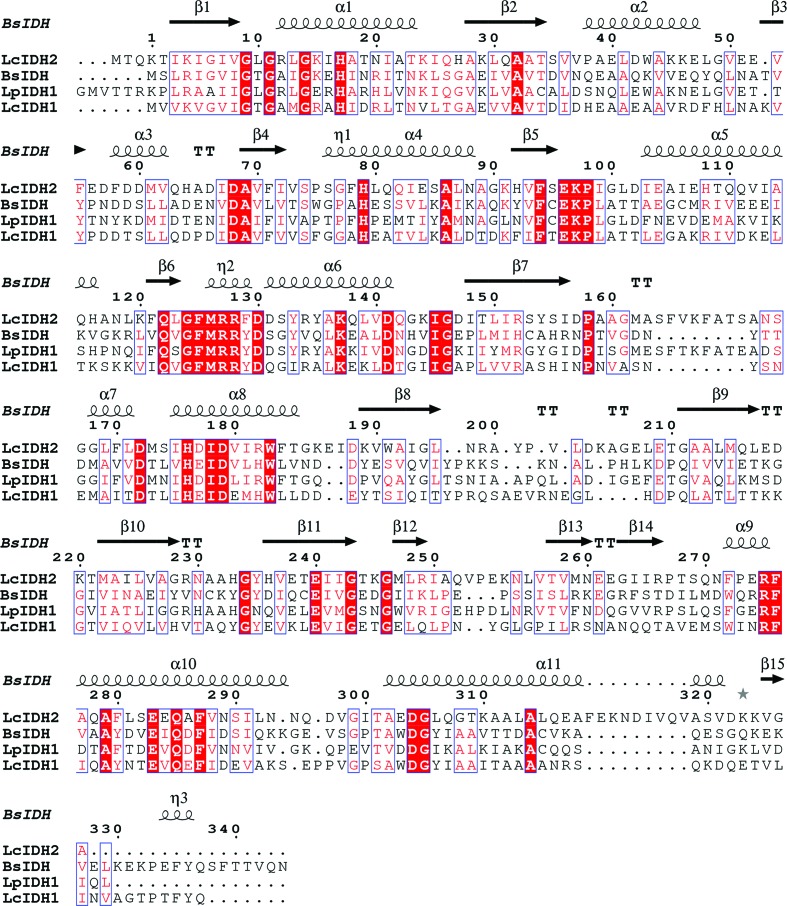
LcIDH2 has a molecular mass of 38 330 Da, a theoretical isoelectric point of 5.33, a theoretical extinction coefficient of 23950 M −1 cm−1, and similar to other IDH enzymes studied (van Straaten et al., 2010 ▶) it crystallizes as a tetramer. Sequence alignment of LcIDH2 with related IDHs, including Bacillus subtilis IDH (BsIDH), as shown in Fig. 1 ▶ (Notredame et al., 2000 ▶; Gouet et al., 2003 ▶), suggests that LcIDH2 has an extended α-helix between β7 and β8 near the putative active site whereas LcIDH1 does not. LcIDH1 and BsIDH belong to IDH subgroup 1, which lacks the extended α-helix, while IDHs from L. planatarum (LpIDH1), Thermotoga maritima (TmIDH) and Corynebacterium glutamicum (CgIDH) each possess the additional α-helix. In the aforementioned structures it is proposed that the helix alters the conformation of the active site by covering the binding pocket (van Straaten et al., 2010 ▶). LcIDH2 is a member of IDH subgroup 2, while TmIDH and CgIDH both belong to subgroup 3. Sequence alignment of BsIDH, LcIDH1, LpIDH1 and LcIDH2 shows the putative catalytic residues are conserved. The pairwise sequence identities between LcIDH2 and BsIDH, LcIDH1 and LpIDH1 are 37, 41 and 59%, respectively.
Figure 1.
Multiple sequence alignment of BsIDH, LcIDH1, LcIDH2 and LpIDH1. Secondary-structure prediction for LcIDH2 suggests an α-helix between β7 and β8 lacking in BsIDH and LcIDH1.
In the course of structure solution full diffraction data sets of LcIDH2 crystals were obtained at cryo-temperature as well as at room temperature. The cryocooled crystal diffracted to higher resolution than the room-temperature crystal but also suffered from a higher mosaic spread. This prompted a molecular-replacement search using the room-temperature data set as a target. Initially, the room-temperature data set was misassigned to the lower symmetry space group P2, a monoclinic lattice. Subsequent molecular-replacement trials suggested space group P222, a primitive orthorhombic lattice, but with a strong noncrystallographic translation symmetry operator that closely approximated the body-centered orthorhombic space group I222. It was found that both the cryocooled crystal and the room-temperature data sets could be solved in I222 but not P222.
We have initiated studies utilizing both crystallography and enzymology to examine the IDH subgroups to determine whether these subgroups represent different metabolic functions.
This paper describes the crystallization and initial X-ray analysis of the crystal structure of LcIDH2. Moreover, we discuss our initial observations of the fortuitous room-temperature radiation tolerance of the protein crystals.
2. Materials and methods
2.1. Expression and purification
The gene iolG2 was amplified from genomic DNA using the following primers: forward, 5′-GGAGGAGGTAATTGGATCCATGACTCAAAAAACG-3′; reverse, 5′-CCCAATTCAATCCTGCAGTTAAGCACCTAC-3′. It was subsequently ligated into the BamHI and PstI sites of pQE-80L vector (Qiagen). This inserts a His-tag sequence MRGSHHHHHHGS before the start codon of LcIDH2.
Following ligation, the vector was transformed into competent XL1-Blue cells. The cells were cultured in 1 l Terrific Broth medium at 310 K to an OD600 of 0.6 and then moved to a 288 K shaker. Overexpression was induced by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside. The cultures were pelleted by centrifugation and resuspended in lysis buffer [25 mM Tris–HCl, 20 mM imidizole, 0.5 M NaCl, 10%(v/v) glycerol pH 8.0]. The resuspended cells were lysed by the addition of 100 µl 20 mg ml−1 lysozyme, 10 µl 20 mg ml−1 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride and 25 µl 10 mg ml−1 DNase. The suspension was placed on ice and the cell walls were lysed by sonication. Cell debris was separated from the protein supernatant by centrifugation and then filtered through a 25 µm filter. The filtrate was brought to 10% ammonium sulfate with slow stirring at 277 K to precipitate high-molecular-mass contaminants and the precipitate was removed by centrifugation. The supernatant was filtered a second time through a 25 µm filter.
LcIDH2 was purified by Ni2+-affinity chromatography using an MC20 POROS column (Applied Biosystems). The column was loaded to capacity and eluted into 10 ml fractions using a linear gradient of imidazole (20–500 mM over ten column volumes). The column flowthrough was measured using a UV–Vis 280 nm detector and a peak was observed at 100 mM imidazole. The highest purity peaks were pooled together, dialyzed against four 2 l exchanges of 25 mM Tris–HCl pH 8.0 and concentrated to 20 mg ml−1 (OD280) by centrifugation at 3000 rev min−1 in a FIBERLite F500 rotor. The purified protein was divided into 50 µl aliquots and flash-cooled in liquid nitrogen for long-term storage at 253 K.
2.2. Crystallization
Crystallization trials were set up using the microbatch technique and the LcIDH2 protein was screened against commercial crystallization suites from Qiagen. The protein solution was diluted to 10 mg ml−1 and was mixed in a 1:1 ratio with crystallization buffer to give a final volume of 2 µl. The wells were covered with a layer of paraffin oil and stored at room temperature. A well with a crystallization component consisting of 2.8 M ammonium sulfate, 0.2 M citric acid pH 5.0 produced an initial hit with crystals having an edge length of >100 µm. The condition was optimized to 2.8 M ammonium sulfate, 0.18 M citric acid pH 5.0 to produce crystals with a well defined morphology (Fig. 2 ▶).
Figure 2.

Crystals of LcIDH2. The crystals were grown in a 2 µl drop of 1 µl 10 mg ml−1 protein buffered in 25 mM Tris–HCl pH 8.0, 1 µl 2.8 M ammonium sulfate, 0.8 M citric acid at pH 5.0.
2.3. Data collection and processing
Initially, crystals were prepared for data collection by harvesting them with loops from MiTeGen and dipping them in cryoprotectant. A variety of cryoprotectants were attempted (sodium malonate, ethylene glycol, mineral oil and Paratone-N) but upon visual inspection 20% glycerol had no apparent effect on the integrity of the crystal. The cryoprotected crystals were then flash-cooled in liquid nitrogen in preparation for data collection.
Crystal diffraction and data collection were performed on the Canadian Light Source insertion-device beamline 08ID-1. Crystals were mounted 200 mm from an MX300 Rayonix detector and kept cool by a dry nitrogen cryostream. Diffraction images were collected at an X-ray wavelength of 0.9795 Å over an angular range of 180° with an oscillation angle of 0.2° and an exposure time of 0.5 s per image.
Visual inspection of the images showed reflections with significant streaking and apparent overlapping neighbors. Indexing and cell refinement of the collected images in iMosflm (Battye et al., 2011 ▶) yielded space group I222. An initial unit-cell refinement calculated a mosaic spread of ∼0.7° and unit-cell parameters a = 43.45, b = 121.65, c = 130.94 Å. The data set was integrated in I222 and scaled using SCALA (Evans, 2006 ▶) with a cutoff of 1.6 Å. An initial pass at molecular replacement was performed using a previously solved structure of LcIDH1 (unpublished work) from our laboratory. The structure was pruned in CHAINSAW (Stein, 2008 ▶) prior to use as a molecular-replacement model. The molecular-replacement solution was inspected in Coot (Emsley & Cowtan, 2004 ▶) and, while the packing appeared to be reasonable, it did not produce the expected tetramer arrangement that has been observed for previous IDH structures (van Straaten et al., 2010 ▶). A more rigorous application of automated molecular replacement using MrBUMP (Keegan & Winn, 2007 ▶) resulted in a tetramer in the unit cell. The model was used for rigid-body and reciprocal refinement in PHENIX (Adams et al., 2010 ▶) but the resulting electron-density maps were too poor for model building and the R work and R free remained high at 51 and 54%, respectively.
The underlying cause for the failure of molecular-replacement remains unknown but may have been due to a combination of high mosaic spread in the reflection data, resulting in inaccurate calculation of structure factors, as well as low sequence similarity between LcIDH1 and LcIDH2. In an effort to improve the quality of the diffraction data, and possibly improve the molecular-replacement solution, it was decided that protein crystal diffraction would be tested at room temperature in the absence of glycerol.
A crystal was mounted in a MiTeGen loop and covered by a plastic capillary that included a few microlitres of mother liquor to prevent evaporation. Crystals were mounted at a crystal-to-detector distance of 200 mm and diffraction images were collected at 0.9795 Å over an angular range of 180° with an oscillation angle of 0.5° and an exposure time of 0.5 s per image. The diffraction images were auto-indexed in iMosflm and the results again indicated an orthorhombic lattice with P222 as the highest symmetry and an initial refinement unit cell with parameters a = 43.9, b = 123.3, c = 132.1 Å. The second possible lattice for indexing was primitive monoclinic with a lower symmetry space group, P2, and unit-cell parameters a = 43.9, b = 123.2, c = 132.1 Å, β = 90.01°.
The room-temperature data set was integrated in P1, with unit-cell parameters a = 49.8, b = 85.3, c = 193.85 Å, α = 98.9, β = 87.8, γ = 90.1°, and a molecular-replacement solution was attempted with MrBUMP and CHAINSAW. The LpIDH1 structure (PDB entry 3cea; Structural Genomics Consortium, unpublished work) solved to a resolution of 2.4 Å was used as a search model. This protein shares 46% sequence identity with LcIDH2. A solution was found with eight monomers in the unit cell. A series of alternating cycles of simulated annealing, rigid-body refinement and model building were applied in PHENIX (Adams et al., 2010 ▶). The resulting model was refined to an R work and R free of 36 and 44%, respectively. The improvements to the model permitted solution of the structure in P2 and further model building and refinement decreased the R work and R free to 15 and 17%, respectively.
The unit-cell parameters of the primitive monoclinic lattice are very close to orthorhombic and this motivated a search for the true lattice and space group in the orthorhombic system. The room-temperature data set was indexed in P222 and scaled in SCALA. It was found that a resolution cutoff of 1.6 Å resulted in an 〈I/σ(I)〉 of 3.7 in the outer shell and at higher resolutions the 〈I/σ(I)〉 fell below 1.5.
The calculated Matthews coefficient (Matthews, 1968 ▶) for the P222 unit cell was 2.52 Å3 Da−1, with a solvent content of 49%, and predicts two monomers in the unit cell. MOLREP (Vagin & Teplyakov, 2010 ▶) found two monomers in the unit cell and a pseudo-translation vector of 〈0.5, 0.5, 0.5〉 in fractional coordinates. The integrated data set was run through phenix.xtriage, which also calculated a pseudo-translation vector of 〈0.5, 0.5, 0.5〉 with an off-origin peak that was 80% of the origin peak. The presence of the pseudo-translation vector suggested that the room-temperature LcIDH2 data set is approximating a lower symmetry space group and may fit better in I222 instead of P222. The room-temperature data set was successfully integrated and refined in the highest symmetry space group available, I222. The refinement statistics are summarized in Table 1 ▶.
Table 1. Summary of data-collection statistics for the cryoprotected and room-temperature data sets.
In order to make a comparison of the scaling statistics, we chose to set the resolution ranges to equal values. Numbers in parentheses refer to the highest resolution shell.
| Cryoprotected | Room-temperature | |
|---|---|---|
| Beamline | 08ID-1, CLS | 08ID-1, CLS |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Temperature (K) | 77 | 293 |
| Detector | Rayonix MX300HE CCD X-ray detector | Rayonix MX300HE CCD X-ray detector |
| Crystal-to-detector distance (mm) | 200 | 200 |
| Rotation range per image (°) | 0.2 | 0.5 |
| Total rotation angle (°) | 180 | 180 |
| Exposure time per image (s) | 0.5 | 0.5 |
| Space group | I222 | I222 |
| a, b, c (Å) | 43.45, 121.65, 130.94 | 43.92, 123.33, 132.12 |
| Resolution range (Å) | 32.74–1.60 (1.69–1.60) | 32.74–1.60 (1.69–1.60) |
| Total No. of reflections | 331587 (46827) | 349808 (50153) |
| No. of unique reflections | 46240 (6655) | 47854 (6922) |
| Completeness (%) | 100 (96.4) | 99 (100) |
| Multiplicity | 7.2 (7.0) | 7.3 (7.2) |
| 〈I/σ(I)〉 | 18.2 (5.9) | 12.3 (3.6) |
| R merge | 0.055 (0.245) | 0.076 (0.563) |
| R r.i.m | 0.059 (0.264) | 0.082 (0.607) |
| R p.i.m | 0.022 | 0.030 |
| Overall B factor (Å2) | 19.580 | 19.230 |
3. Results and discussion
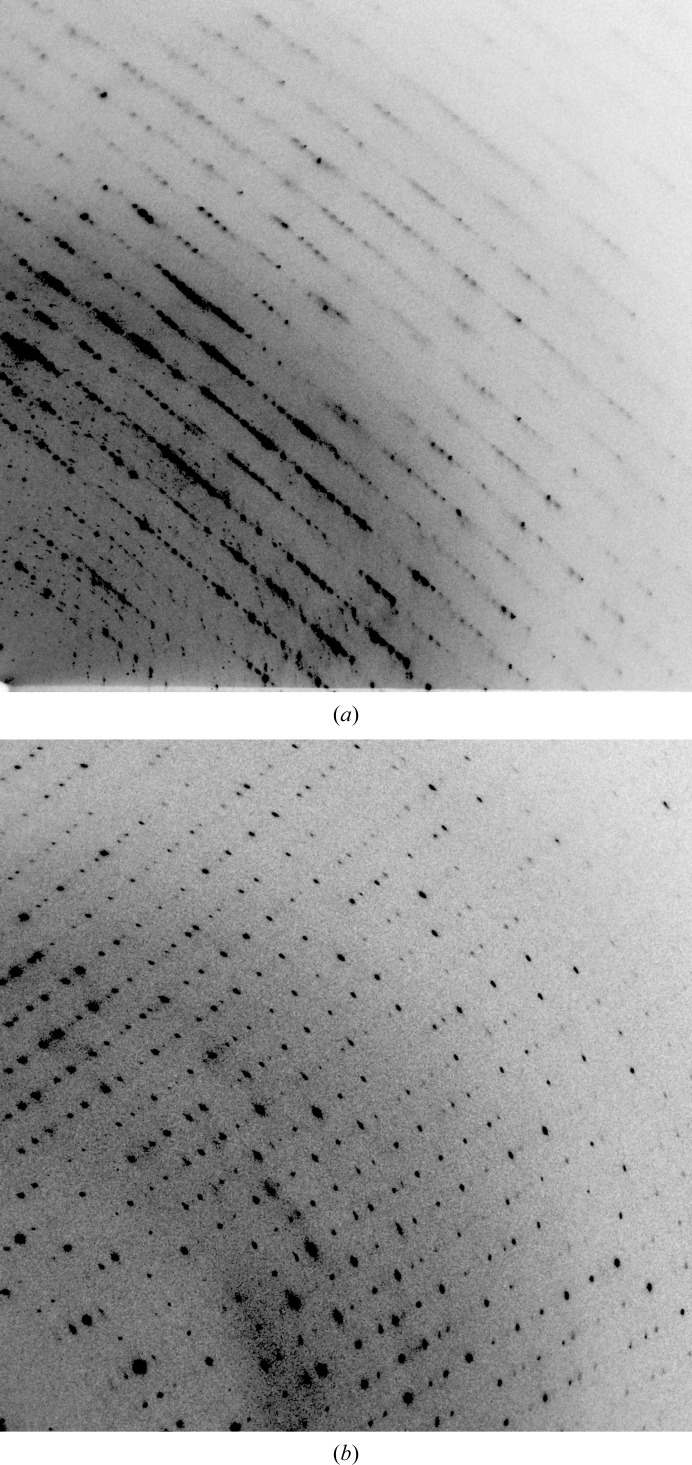
The pursuit of a crystallographic structure for LcIDH2 resulted in two interesting artifacts: the room-temperature radiation tolerance of a protein crystal and its pseudo-translation symmetry. Microbatch crystallization yielded crystals that diffracted to high resolution at 77 K as well as 293 K but with potentially different space groups. For either possible space group the unit-cell parameters of the crystals appear to contract as a result of flash-cooling in liquid nitrogen. Comparison of diffraction images from a cryocooled crystal and a room-temperature crystal is shown in Fig. 3 ▶. Comparison of the integrated data sets in iMosflm showed that the room-temperature crystal had an average signal-to-noise ratio 〈I/σ(I)〉 per image that began at 14 and decreased to 7 (decreasing from 2 to 0.4 in the highest resolution bin) over 360 images, but the 〈I/σ(I)〉 of the cryocooled crystal remained constant at 14 (0.7 in the highest resolution shell). The cryocooled crystal presented a higher average signal-to-noise ratio in the highest resolution shell and a lower R merge after scaling when compared with the room-temperature crystal. The decrease in 〈I/σ(I)〉 for the room-temperature crystal relative to the cryocooled crystal over the exposure of a full data-set collection suggests it still succumbs to primary radiation damage but to a lesser extent than a typical protein crystal (Stern et al., 2009 ▶; Garman, 2010 ▶; Kauffmann et al., 2006 ▶; Howells et al., 2009 ▶).
Figure 3.
Diffraction images from cryocooled and room-temperature diffraction experiments. (a) Diffraction image from a cryocooled crystal. (b) Diffraction image from room-temperature data collection. The deleterious effects of the cryoprotectant are evident in the diffuse scattering and increase in the mosaic spread (left). Reflections in the room-temperature diffraction data set are well defined, separated and appear to have a low mosaic spread.
Molecules related by noncrystallographic symmetry (NCS) operators frequently occur in crystal structures owing in part to the oligomeric structure of their constituent protein molecules, and may aid in real-space averaging and improvement of phases (Rossmann & Blow, 1962 ▶). Vexing problems with NCS may arise when pseudo-translation closely places a monomer in a position that mimics a different Bravais lattice (Zwart et al., 2008 ▶; Guelker et al., 2009 ▶; Chook et al., 1998 ▶; Sauter & Zwart, 2009 ▶). Analysis of the LcIDH2 room-temperature crystal suggested it had a pseudo-translation symmetry operator. P222 and I222 belong to the same orthorhombic crystal system and point group but possess different Bravais lattices, the former being primitive and the latter body-centered. A P222 molecular-replacement solution places the second monomer in the unit cell at 〈x + 1/2, y + 1/2, z + 1/2〉 by pseudo-translation. This position is identical to a symmetry-equivalent position of I222. Moreover, the symmetry-related equivalent positions in P222 are a subset of I222. The final test in determining the true space group was the successful refinement of the room-temperature crystal in I222.
4. Conclusions
The symmetry of the P222 molecular-replacement solution combined with poor refinement statistics encouraged indexing the images in the I222 space group. Scaling and molecular replacement followed the same procedure as the P222 solution with an applied resolution cutoff of 1.6 Å and resulted in similar scaling statistics when compared with I222. Initial rounds of refinement produced an R work and R free of 19 and 22%, respectively. Once the high-resolution model of the room-temperature data set was obtained, we applied it to the solution of the cryocooled crystal. The molecular-replacement solution in MOLREP found one monomer in the asymmetric unit. The molecular-replacement solution was processed in PHENIX in a similar way as the room-temperature data set. The resulting R work and R free were 23 and 24%, respectively. The data-processing statistics are summarized in Table 1 ▶. This structure of the LcIDH2 apoenzyme will be used to study the conformational changes in substrate and cofactor binding in further LcIDH2 structures.
Acknowledgments
This work was supported by NSERC Discovery Grants to DARS and DRJP. We thank Dr Barry Ziola and Dr Vanessa Pittet for assistance with genomic DNA isolation and Dr Karin van Straaten for assistance with data collection and processing. We thank the staff of macromolecular beamline 08ID-1, Canadian Light Source (CLS), Saskatoon Saskatchewan, Canada for their technical assistance during data collection. The structural studies described in this paper were performed at the CLS, which is supported by NSERC, the National Research Council of Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada and the University of Saskatchewan.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Chook, Y. M., Lipscomb, W. N. & Ke, H. (1998). Acta Cryst. D54, 822–827. [DOI] [PubMed]
- Daniellou, R., Zheng, H., Langill, D. M., Sanders, D. A. R. & Palmer, D. R. J. (2007). Biochemistry, 46, 7469–7477. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Garman, E. F. (2010). Acta Cryst. D66, 339–351. [DOI] [PMC free article] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Guelker, M., Stagg, L., Wittung-Stafshede, P. & Shamoo, Y. (2009). Acta Cryst. D65, 523–534. [DOI] [PMC free article] [PubMed]
- Howells, M., Beetz, T., Chapman, H. N., Cui, C., Holton, J. M., Jacobsen, C. J., Kirz, J., Lima, E., Marchesini, S., Miao, H., Sayre, D., Shapiro, D. A., Spence, J. C. H. & Starodub, D. (2009). J. Electron Spectrosc. Relat. Phenom. 170, 4–12. [DOI] [PMC free article] [PubMed]
- Kauffmann, B., Weiss, M. S., Lamzin, V. S. & Schmidt, A. (2006). Structure, 14, 1099–1105. [DOI] [PubMed]
- Keegan, R. M. & Winn, M. D. (2007). Acta Cryst. D63, 447–457. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Notredame, C., Higgins, D. G. & Heringa, J. (2000). J. Mol. Biol. 302, 205–217. [DOI] [PubMed]
- Ramaley, R., Fujita, Y. & Freese, E. (1979). J. Biol. Chem. 254, 7684–7690. [PubMed]
- Rossmann, M. G. & Blow, D. M. (1962). Acta Cryst. 15, 24–31.
- Sauter, N. K. & Zwart, P. H. (2009). Acta Cryst. D65, 553–559. [DOI] [PMC free article] [PubMed]
- Stein, N. (2008). J. Appl. Cryst. 41, 641–643.
- Stern, E. A., Yacoby, Y., Seidler, G. T., Nagle, K. P., Prange, M. P., Sorini, A. P., Rehr, J. J. & Joachimiak, A. (2009). Acta Cryst. D65, 366–374. [DOI] [PubMed]
- Straaten, K. E. van, Zheng, H., Palmer, D. R. J. & Sanders, D. A. R. (2010). Biochem. J. 432, 237–247. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Yebra, M. J., Zúñiga, M., Beaufils, S., Pérez-Martínez, G., Deutscher, J. & Monedero, V. (2007). Appl. Environ. Microbiol. 73, 3850–3858. [DOI] [PMC free article] [PubMed]
- Zwart, P. H., Grosse-Kunstleve, R. W., Lebedev, A. A., Murshudov, G. N. & Adams, P. D. (2008). Acta Cryst. D64, 99–107. [DOI] [PMC free article] [PubMed]