Abstract
The importance of diet in health and disease has been well characterized in the past decades. Although the earlier focus of diet research was in the context of undernutrition and the importance of adequate nutrient intake to prevent malnutrition, in the current era of epidemic obesity the focus of our efforts has evolved toward understanding the effects of excess caloric intake. The current surge in childhood obesity rates suggests a correlation of maternal metabolic syndrome and obesity with programming of the fetal epigenome for metabolic diseases later in life. Alterations of the fetal genome, epigenome and metabolome have been well documented in cases of maternal malnutrition, including both overnutrition and undernutrition. It is of great interest and importance to understand how these divergent maternal factors regulate/program the fetus for metabolic diseases, and we and others have observed that epigenetic modifications to the fetal and placental epigenome accompany these reprogramming events. The following review summarizes recent studies on the effects of maternal diet and obesity on fetal epigenetics contributing to adult diseases later in life by taking advantage of state-of-the-art genomic, epigenomic and metagenomic techniques in nonhuman primate model systems.
Keywords: high-fat diet, DOHaD, nonhuman primate, epigenetics
Introduction
According to the World Health Organization reports, obesity rates have doubled since 1980, with 500 million people suffering from obesity in 2008.1 The current global health epidemic of obesity is considered a major contributor to the increasing prevalence of type 2 diabetes, as well as atherosclerotic, cardiovascular and hypertensive morbidity and mortality. In association with this increase in adult obesity throughout the developed and developing world, there is an occurrence of disproportionate earlier onset of obesity among children (infants to adolescents). Given such anticipation (for example, tendency of the disease to appear at an earlier age of onset and with increasing severity in successive generations), the increased prevalence of childhood obesity cannot be attributed to either environment or genetics alone.
According to Barker's fetal origins of adult disease hypothesis, perturbations in the gestational milieu influence the development of diseases later in life.2, 3 This supposition gains support from several epidemiologic studies,4, 5, 6, 7, 8, 9, 10 as well as animal models of nutritional constraint11, 12 and uteroplacental insufficiency-induced intrauterine growth restriction (IUGR).13, 14 These outcomes have been suggested to occur through the static reprogramming of gene expression via alterations in chromatin structure (epigenetic regulation).
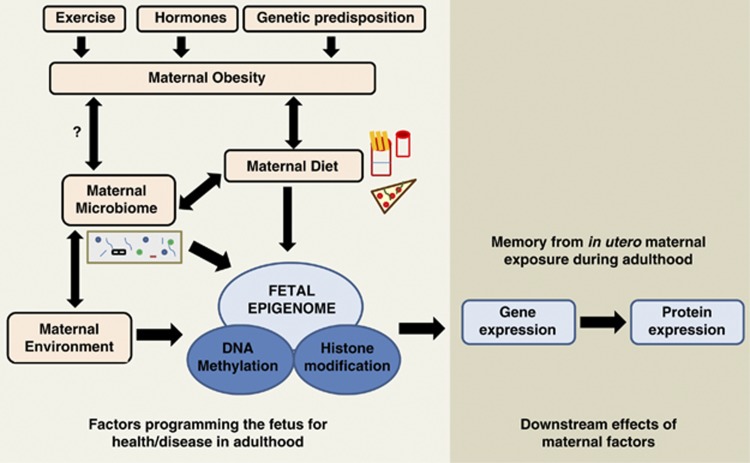
In the past decades, epigenomic studies have provided great insights into gene regulation and expression at various developmental stages. Epigenetic changes can be considered as a command to the genomic program that is species and tissue specific. Although the genetic makeup of an individual can predispose them toward these diseases, it is the epigenetic state of an individual that can be programmed by events that occur during development. The effects of maternal diet, microbiome and environment on the fetal epigenome (Figure 1 and Table 1) are currently being highly studied to elucidate molecular mechanisms and pathways affected through these factors and to dissect the exact role of obesity and/or diet in metabolic diseases. The following review summarizes the recent advances in our understanding of human and nonhuman primate (NHP) epigenetics and how the memory/signature imprinted by various maternal factors such as diet program the fetal susceptibility to adult metabolic diseases that occur later in life.
Figure 1.
Overview of the maternal factors shaping the fetal epigenome and effects on fetal gene expression in adulthood.
Table 1. Epigenomic studies associated with high-fat diet.
| Finding | Reference |
|---|---|
| In rats, maternal high-fat diet during gestation has been associated with histone modification of Pck1 gene in offsprings. | 17 |
| Alteration in methylation levels of leptin promoter due to high-fat diet induced obesity. | 23 |
| In nonhuman primate model of maternal high-fat diet, alterations in hepatic H3K14 acetylation levels have been observed in the offspring. | 34 |
| Differential Npas2 promoter occupancy of fetal histone H3K14 acetylation has been observed in response to maternal high-fat diet. | 35 |
Overnutrition (high-fat/calorie-dense diet)
In light of the underlying importance of maternal diet, the prospective-longitudinal Southampton Women's survey was begun in 1998 to understand the effects of maternal diet, body mass index and environment/social factors on the offspring. Although the study is still ongoing, preliminary results indicate the presence of two cohorts namely ‘prudent diet' and ‘high-fat diet' (HFD).15, 16 For example, a recent study demonstrated that maternal HFD during gestation led to increased mRNA expression of gluconeogenic genes, elevated plasma glucose levels and histone modification of the Pck1 gene.17 The methylation status of several genes involved in metabolic, vascular and endocrine function are altered because of changes in diet during prenatal and/or postnatal life (reviewed in Lillycrop and Burdge18). Overall, the consensus has been that maternal diet alters the epigenomic regulation and signatures of certain genes during gestation, which program the offspring to a higher risk of acquiring metabolic syndrome-related diseases.19 In summary, maternal consumption of HFD begins the cycle of ‘susceptibility to obesity and metabolic diseases' for the future generations through the epigenomic signatures left behind by the altered consumption of HFD diet. The only solution being ‘adequate/optimal intake of calories and fat' as confirmed by the NHP model of obesity.20, 21
Maternal metabolic disease states
In the past decades, ‘obesity begets obesity' has been commonly stated by researchers. Obesity is a result of imbalance between energy intake (diet) and energy expenditure (exercise) along with genetic predisposition and alteration of maternal hormones such as leptin, insulin, ghrelin and growth hormone. An European-based project ‘OBELIX' was initiated in 2009 to understand the association between risk for obesity and metabolic syndrome later in life due to prenatal and postnatal exposure to endocrine-disrupting chemicals.22 Methylation of the leptin promoter is altered because of HFD-induced obesity.23 A recent retrospective epidemiological study demonstrated that the offspring exposed to maternal diabetes have an increased cardiovascular risk profile.24 The possible effect of gestational diabetes in altering fetal epigenetic mechanisms has been discussed in further detail elsewhere.25 Thus, the altered metabolic maternal state has an evident effect on altering the fetal epigenome and transcriptome; resolving these observations with our findings in the Japanese macaque will be of fundamental importance in the coming years.
NHP models of HFD exposure in utero
Biomedical animal models are a boon for human medical research and clinical applications. Researchers have been striving to use the most appropriate model system to study human diseases, depending upon the question to be addressed. For example, mutation studies can be easily performed in rodent model systems owing to the fast generation time and litter size, whereas a porcine model has been traditionally used for neonatal medical research. However, the ultimate goal has been to establish an animal model that adequately mimics the human structure, function and physiology. A NHP model could serve as a solution owing to the results from fossil records and comparative genomics that indicate that chimpanzees are the closest human relative followed by gorilla, orangutan and the macaque, with the latter sharing ∼93% identity even with a divergence time from humans of 23 to 30 million years ago.26, 27
Several studies using macaque models have been important for our understanding of HFD, obesity, metabolic syndrome, diabetes and cardiovascular disease.28, 29, 30, 31 For example, a caloric restriction study performed in the rhesus macaque demonstrated that a reduced risk of atherosclerosis could be observed in association with increased HDL2 levels and decreased triglyceride levels.32 Similarly, a Japanese macaque33 model has been successfully developed by our collaborative group in efforts aimed at understanding the impact on NHP fetuses exposed in utero to a maternal HFD.21 With respect to the maternal phenotypic profile, we have previously described that over time (for example, 3–5 years) two distinct maternal phenotypes become apparent among our dams. These dams can be considered sensitive (HFD-S) or resistant (HFD-R) to our HFD. HFD-S dams display a unique metabolic phenotype in both the nonpregnant and pregnant state. By the 4th year on the HFD, the HFD-S group demonstrates a >35% increase in body weight, a >3-fold increase in insulin AUC (area under the curve) in response to a glucose tolerance test, a >5-fold increase in leptin levels (leptin/body weight ratio) and a near doubling in body fat. However, fasting glucose levels do not significantly differ in either the HFD-S or HFD-R groups despite a significant alteration (control, 2.53±0.50; HFD-R, 2.81±0.70; HFD-S, 7.87±1.50; P<0.05 for HFD-S versus control) in the homeostasis model assessment of insulin resistance (or HOMAIR, which serves as a general measure of insulin resistance in reference to basal glucose and insulin). By comparison, HFD-R animals retained similar body weight and metabolic phenotype as the controls throughout the entire 4 years.20, 21
In our initial studies establishing our NHP model of maternal HFD exposure in utero, we demonstrated that a maternal high-fat/calorie-dense diet altered fetal hepatic chromatin structure and led to alterations in fetal and postnatal gene expression.20, 34, 35 These observations supported our hypothesis that a high-fat maternal diet leading to obesity modifies the gestational milieu and profoundly influences the postnatal and fetal phenotype in association with fetal histone (H3) covalent modifications. Specifically, we observed that the fetal chromatin structure was not globally altered but rather modified via site-specific alterations in H3 acetylation with lysine-specific punctuate modifications (that is, significant alterations in H3K14 but not H3K9 nor and H3 methylation demarcation); these occurred with appropriate and anticipated alterations in their associative epigenetic machinery.34 We have observed that, although the maternal phenotype is relatively mild, regardless of maternal HFD-R or HFD-S status the fetus develops nonalcoholic fatty liver and displays an altered fetal hepatic epigenomic profile. As the offspring are delivered in the early final trimester of gestation (term being 167 days), appreciable differences in overall fetal weight are avoided.
Of interest, we have shown that reversing the maternal diet in year 5 from high fat to control (‘HFD reversal') bears appreciable differences on the fetal phenotype, with partial reversion trending toward control values.21 Given that we observed significant alterations in the maternal and fetal phenotype following in utero exposure to a high-fat maternal diet, we aimed at characterizing the full spectrum of epigenomic differences in our model system. Specifically and consistent with those of other investigators,36 we failed to observe a significant change in mean fetal hepatic mRNA expression over offspring of control diet animals for MeCP2. However, a significant modest increase in the expression of Dnmt1 and a decrease in the expression of HDAC1 mRNA occurred in offspring of animals exposed to a HFD. Moreover, when we similarly examined gene expression differences in a limited number of offspring, allowed to go to term and then killed at postnatal day 30 (n=3) following in utero exposure to maternal high-fat or control diet for 2 years, we observed a persistent alteration in affected gene expression. Thus, HDAC1 remained downregulated in its expression in HFD-exposed offspring at postnatal day 30. Consistent with our mRNA expression profile, fetal nuclear extracts from offspring of HFD animals were observed to be significantly relatively depleted of HDAC1 protein and in vitro HDAC functional activity. Taken together, these initial data illustrated the capacity of a calorie-dense high-fat maternal diet to alter the fetal primate epigenome in an acetyl site-specific manner on the H3 tail. Such site-specific ‘permissive' or ‘restrictive' modifications of the histone code were felt to likely associate with the reprogrammed expression of fetal genes. These in turn potentially favor the presumptive obese postnatal phenotype in an anticipatory manner. Collectively, our observations lent a potential molecular basis to the developmental origins of adult disease in primates.
Leveraging these collective observations, we then set out to study the serum metabolome of HFD-R and HFD-S dams, and to test whether these observed phenotypes were transferred in utero to the developing fetus. We then characterized the fetal metabolome from dams fed control, HFD or HFD reversal. We hypothesized that this approach would not only decipher the fetal and maternal serum metabolome under maternal HFD conditions leading to maternal obesity but also would potentially identify candidate biomarker(s) associated with the development of obesity. Together, in these initial studies,20 approximately 1300 chromatographic features were detected. Through MVDA, this number was reduced to 60 possible metabolites, and using comparative t-tests 22 metabolites were found to have statistical significance (P<0.05) over the entire study period. By virtue of maternal HFD alone, fetal phenotypic differences are accompanied by altered metabolite concentrations of seven metabolites (P<0.05). Altered serum concentrations of cholesterol, members of the vitamin E tocopherol complex, glycine, aminomalonic acid, citric acid and 2-hydroxybutyrate, ascorbic acid and α-tocopherol were observed. Taken together, these studies provided the first evidence that in primate fetal life maternal obesity and in utero HFD exposure associate with elevations in 2-hydroxybutyrate, which may serve as a marker for a correlation between fetal lipid oxidation, hypertriglyceridemia and NAFLD.20
Armed with these intriguing differences across the epigenome and metabolome, we thereafter sought to identify specific reprogrammed pathways of interest in the developing fetus. By using mRNA copy number analysis, we found that the components of the peripheral circadian machinery were transcribed in the NHP fetal liver in an intact phase–antiphase manner and that Npas2, a paralog of the Clock transcription factor, serves as the rate-limiting transcript by virtue of its relative low abundance (10- to 1000-fold lower). We further demonstrated that exposure to a maternal HFD in utero significantly altered the expression of fetal hepatic Npas2 (up to 7.1-fold, P<0.001) compared with that in control diet-exposed animals and is reversible in fetal offspring from obese dams reversed to a control diet (1.3-fold, P>0.05(ref. 35)). Although the Npas2 promoter remained largely unmethylated, differential Npas2 promoter occupancy of acetylation of fetal histone H3 at lysine 14 (H3K14ac) occurred in response to maternal HFD exposure compared with control diet-exposed animals. Furthermore, we found that disruption of Npas2 is consistent with HFD exposure in juvenile animals, regardless of in utero diet exposure.35 In summary, these data suggest that fetal and juvenile peripheral Npas2 expression is uniquely vulnerable to diet exposure and notably HFD exposure in critical gestational intervals.
Thus, comparison between maternal HFD-S (obese) and resistant dams (lean) (alongside dietary reversal in obese gravidae) reinforced this notion to support our summary findings that the maternal HFD—and not maternal obesity—is probably the significant and meaningful factor in altering the fetal epigenome and metabolome.20, 34, 35
Further human cohort studies, using similar diet groups and phenotypes, would confirm the maternal HFD effect versus the effect of obesity. In short, maternal HFD consumption alters gene expression and protein translation via DNA methylation and histone and chromatin modifications. These fine-tuning changes in genes lead to offspring susceptibility toward a wide array of diseases/conditions.
Modifiable maternal factors altering the fetal epigenome in humans
Undernutrition (calorie and protein restriction)
Undernutrition has been of great concern for the developing world. Seminal follow-up studies based on the Dutch Hunger Winter and Nigerian famine during civil war have established that children born during these periods acquired a thrifty phenotype associated later in life with elevated cholesterol and triglyceride levels,5 impaired glucose tolerance4, 5, 6, 7, 8, 9, 10 and higher body mass index, resulting in a higher risk for cardiovascular disease.7, 8, 9, 10, 37 Through several previous studies, it has been shown that calorie restriction during pregnancy hinders the fetal development and programs the offspring for the thrifty phenotype.
For decades, the importance of maternal intake of folate, vitamin B6 and B12 in preventing neural tube malformations and protein restriction in avoiding the adverse effects of IUGR has been widely understood (reviewed in Brown et al.38 and Simpson et al.39). The amount of protein intake in developing countries is lower owing to poverty, resulting in a carbohydrate-rich diet. Protein provides the essential amino acids that the body cannot synthesize, including methionine that is important in one-carbon metabolism. One-carbon metabolism has an important role in methyl transfer reactions, for example, methylation performed by DNA methyltransferases. Maternal methyl donor supplementation has been shown to prevent inheritance of obesity in the offspring.40 A recent study in an IUGR rat model demonstrated that maternal protein restriction resulted in elevated cholesterol levels in adult offspring mediated via histone modification at cholesterol 7alpha-hydrolase.41 Uteroplacental insufficiency and IUGR in the rat results in covalent modifications of chromatin structure or changes in DNA methylation patterns, which leads to persistent changes in postnatal gene expression.42 Protein restriction leads to inadequate levels of essential amino acids, that is, amino acids derived from the diet and not synthesized de novo by the body. Thus, maternal protein restriction interferes with proper functioning of one-carbon metabolism, which is tightly coupled with the methyltransferases, leading to altered epigenetic features.
The microbiome and metabolism
Previously, microbes were always mentioned in the context of ‘pathogens' that need to be eliminated from the human body, as they were viewed only in the context of disease burden. However, we have come to appreciate that humans serve as hosts to coevolving microbes residing in highly plethoric communities. Indeed, microbiota are present from the time of birth, with up to 10-fold the number of microorganisms to adult human cells and a collective genome (the ‘metagenome') that exceeds our human genome in terms of gene content by more than 100-fold.43 Moreover, we appreciate that the human microbiota are a metabolically and antigenically vibrant and diverse community that may function as mutualists (symbiotically beneficial), commensals (of neither harm nor benefit) or pathogens (of host detriment).
The widely accepted notion of ‘obesity begets obesity' is part of the story, as we have seen a profound effect of diet in shaping the microbiome community in the host.44 Several studies of interest to this monograph are worth commenting on. First, Turnbaugh et al.45 analyzed the microbiota of 154 individuals (monozygotic and dizygotic twin pairs) comparing for leanness and obesity. The results demonstrated shared microbiota composition between family members, but each individual's core microbiota varied in specific lineages present. The gut microbiome was found to be altered due to obesity when compared to the lean counterpart at the phylum level, reduced bacterial diversity (in obese) and altered metabolic pathways.45 Second, and just recently, Wu et al.44 demonstrated the long-term dietary role in shaping the human gut enterotypes. Detectable alterations in microbiome composition could be observed because of diet within 24 h (high fat-low fiber versus low fat-high fiber), but the enterotype identity remained stable, thus concluding that enterotype identity is associated with long-term dietary consumption. Thus, diet could be viewed as a modulator (therapeutic intervention) to modify the microbiome community.44
Obesity has also been associated with alterations in human microbiome composition in turn affecting short-chain fatty acid metabolism.46 Diabetes and diet have also been shown to alter the gut microbiome, but there has been no causal relationship described between altered microbiome and risk of obesity and diabetes owing to confounding human factors.47 The importance of symbiotic microbial community is undeniable. The coevolution of humans and microbiota has led to a mutually beneficial state. Hence, it is intuitive to think for example the presence of organisms that use sugars/fat as substrates and metabolize them to derive energy, thereby protecting the host against obesity.
Primate fetal development is thought to occur within an intrauterine microbiota-free environment, and yet within a short interval following birth the human microbiome colonizes and ‘differentiates' until the adult complement of 90 trillion or so microbiota is achieved.48 On the basis of a relative paucity of data, it is proposed that the naive neonatal microbiome is first established with rupture of the amniotic membranes, with further microbiota being introduced as the fetus traverses the vaginal birth canal. By the time of delivery, the neonate has been successively exposed to the maternal vaginal microbial ecosystem.43 Passage through the vaginal canal is an integral part of this process, as mode of delivery alters the neonatal microbiome.43 We recently demonstrated that the vaginal microbiome is altered during pregnancy and also varies depending on vaginal collection site.49 However, as a comprehensive characterization of how the maternal diet potentially affects the developing fetal and neonatal microbiome has not yet been undertaken, ascribing dominant roles to maternal habitus or mode of delivery is premature. With the emergence of state-of-the-art NextGen sequencing methodology, great strides will be made in better defining the beneficial effects and the factors altering the human microbiome. The importance of understanding the link between the human microbiome, the immune system and the host diet has been extensively reviewed by other authors.50
In summary, studies validate the altered expression/presence of microbial communities owing to altered physiological/phenotypic state, but the cause–effect relationship is yet to be established. Nevertheless, future studies could give us better insights into maintaining optimal human health through microbial alteration.
Conclusions
Our current understandings underscore the importance of balanced diet and maternal health. Undernutrition and overnutrition both result in an altered fetal phenotype, which in adulthood is susceptible to metabolic and cardiovascular diseases. The susceptibility is based upon several maternal factors such as obesity, nutrition, microbiome and environment, which alter the fetal epigenomic regulation resulting in altered gene and protein expression. Currently, NHP are crucial models to improve our understandings of developmental origins of adult disease. The interdisciplinary research has begun and would certainly help us further dissect the interrelated mechanisms controlling our epigenome.
Acknowledgments
This work was supported by the NIH Director New Innovator Pioneer Award DP21DP2OD001500-01 (KAT) and Burroughs Welcome Fund Preterm Birth Initiative 1008819.01. We thank the members of the Aagaard Lab for their helpful discussions regarding the manuscript, including Dr Melissa Suter, Dr Min Hu, Ms Lori Showalter, Ms Cynthia Shope and Dr Aishe Chen.
The authors declare no conflict of interest.
Footnotes
This article was published as part of a supplement funded with an unrestricted educational contribution from Desjardins Sécurité Financière.
References
- World Health Organization. Obesity and Overweight [Online], Fact sheet no. 311, updated March 2011, http://www.who.int/mediacentre/factsheets/fs311/en/index.html.
- Paneth N, Susser M. Early origin of coronary heart disease (the ‘Barker hypothesis'). BMJ 1995; 310: 411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP. The malnourished baby and infant. Br Med Bull 2001; 60: 69–88. [DOI] [PubMed] [Google Scholar]
- de Rooij S, Painter R, Roseboom T, Phillips D, Osmond C, Barker D et al. Glucose tolerance at age 58 and the decline of glucose tolerance in comparison with age 50 in people prenatally exposed to the Dutch famine. Diabetologia 2006; 49: 637–643. [DOI] [PubMed] [Google Scholar]
- Lumey L, Stein AD, Kahn HS, Romijn J. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am J Clin Nutr 2009; 89: 1737–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet 1998; 351: 173–177. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, Van der Meulen JHP, Osmond C, Barker DJP, Ravelli ACJ, Bleker OP. Plasma lipid profiles in adults after prenatal exposure to the Dutch famine. Am J Clin Nutr 2000; 72: 1101–1106. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, Van der Meulen JHP, Osmond C, Barker DJP, Ravelli ACJ, Schroeder-Tanka JM et al. Coronary heart disease after prenatal exposure to the Dutch famine, 1944-45. Heart 2000; 84: 595–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom TJ, Van Der Meulen JHP, Ravelli ACJ, Osmond C, Barker DJP, Bleker OP. Plasma fibrinogen and factor VII concentrations in adults after prenatal exposure to famine. Br J Haematol 2000; 111: 112–117. [DOI] [PubMed] [Google Scholar]
- Hult M, Tornhammar P, Ueda P, Chima C, Edstedt Bonamy A-K, Ozumba B et al. Hypertension, diabetes and overweight: looming legacies of the Biafran famine. PLoS One 2010; 5: e13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley SC, Jackson AA. Increased systolic blood pressure in adult rats induced by fetal exposure to maternal low protein diets. Clin Sci 1994; 86: 217–222; discussion 121. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC, Phillips GJ, Benediktsson R, Gardner DS, Edwards CR, Jackson AA et al. Protein intake in pregnancy, placental glucocorticoid metabolism and the programming of hypertension in the rat. Placenta 1996; 17: 169–172. [DOI] [PubMed] [Google Scholar]
- Joss-Moore LA, Wang Y, Campbell MS, Moore B, Yu X, Callaway CW et al. Uteroplacental insufficiency increases visceral adiposity and visceral adipose PPARγ2 expression in male rat offspring prior to the onset of obesity. Early Hum Dev 2010; 86: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, Hsu JL et al. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics 2004; 18: 43–50. [DOI] [PubMed] [Google Scholar]
- Crozier SR, Inskip HM, Godfrey KM, Cooper C, Harvey NC, Cole ZA et al. Weight gain in pregnancy and childhood body composition: findings from the Southampton Women's Survey. Am J Clin Nutr 2010; 91: 1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier SR, Robinson SM, Borland SE, Inskip HM. Dietary patterns in the Southampton Women's Survey. Eur J Clin Nutr 2006; 60: 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakovsky RS, Zhang X, Zhou D, Pan YX. Gestational high fat diet programs hepatic phosphoenolpyruvate carboxykinase gene expression and histone modification in neonatal offspring rats. J Physiol 2011; 589: 2707–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Burdge GC. Epigenetic changes in early life and future risk of obesity. Int J Obes (Lond) 2011; 35: 72–83. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA. Effect of maternal diet on the epigenome: implications for human metabolic disease. Proc Nutr Soc 2011; 70: 64–72. [DOI] [PubMed] [Google Scholar]
- Cox J, Williams S, Grove K, Lane RH, Aagaard-Tillery KM. A maternal high-fat diet is accompanied by alterations in the fetal primate metabolome. Am J Obstet Gynecol 2009; 201: 281.e1–281.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 2009; 119: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legler J, Hamers T, van Eck van der Sluijs-van de Bor M, Schoeters G, van der Ven L, Eggesbo M et al. The OBELIX project: early life exposure to endocrine disruptors and obesity. Am J Clin Nutr 2011; 94: 1933S–1938S. [DOI] [PubMed] [Google Scholar]
- Milagro FI, Campion J, Garcia-Diaz DF, Goyenechea E, Paternain L, Martinez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem 2009; 65: 1–9. [DOI] [PubMed] [Google Scholar]
- West NA, Crume TL, Maligie MA, Dabelea D. Cardiovascular risk factors in children exposed to maternal diabetes in utero. Diabetologia 2011; 54: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Morera JL, Rodriguez-Rodero S, Menendez-Torre E, Fraga MF. The possible role of epigenetics in gestational diabetes: cause, consequence, or both. Obstet Gynecol Int 2010; 2010: 605163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J Hum Evol 2005; 48: 237–257. [DOI] [PubMed] [Google Scholar]
- Consortium RMGSA, Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science 2007; 316: 222–234. [DOI] [PubMed] [Google Scholar]
- Clarkson TB, Mehaffey MH. Coronary heart disease of females: lessons learned from nonhuman primates. Am J Primatol 2009; 71: 785–793. [DOI] [PubMed] [Google Scholar]
- Shively CA, Register TC, Clarkson TB. Social stress, visceral obesity, and coronary artery atherosclerosis: product of a primate adaptation. Am J Primatol 2009; 71: 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah JS, Verdery RB, Bodkin NL, Hansen BC, Le NA, Howard BV. Changes in lipoprotein concentrations during the development of noninsulin-dependent diabetes mellitus in obese rhesus monkeys (Macaca mulatta). J Clin Endocrinol Metab 1991; 72: 1067–1072. [DOI] [PubMed] [Google Scholar]
- Shamekh R, Linden EH, Newcomb JD, Tigno XT, Catherine Jen KL, Pellizzon MA et al. Endogenous and diet-induced hypercholesterolemia in nonhuman primates: effects of age, adiposity, and diabetes on lipoprotein profiles. Metabolism 2011; 60: 1165–1177. [DOI] [PubMed] [Google Scholar]
- Verdery RB, Ingram DK, Roth GS, Lane MA. Caloric restriction increases HDL2 levels in rhesus monkeys (Macaca mulatta). Am J Physiol 1997; 273 (Part 1): E714–E719. [DOI] [PubMed] [Google Scholar]
- Hayasaka K, Horai S, Gojobori T, Shotake T, Nozawa K, Matsunaga E. Phylogenetic relationships among Japanese, rhesus, Formosan, and crab-eating monkeys, inferred from restriction-enzyme analysis of mitochondrial DNAs. Mol Biol Evol 1988; 5: 270–281. [DOI] [PubMed] [Google Scholar]
- Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol 2008; 41: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R et al. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB J 2011; 25: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 2007; 97: 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli ACJ, Van Der Meulen JHP, Osmond C, Barker DJP, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am J Clin Nutr 1999; 70: 811–816. [DOI] [PubMed] [Google Scholar]
- Brown LD, Green AS, Limesand SW, Rozance PJ. Maternal amino acid supplementation for intrauterine growth restriction. Front Biosci (Schol Ed) 2011; 3: 428–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Bailey LB, Pietrzik K, Shane B, Holzgreve W. Micronutrients and women of reproductive potential: required dietary intake and consequences of dietary deficiency or excess. Part I--folate, vitamin B12, vitamin B6. J Matern Fetal Neona 2010; 23: 1323–1343. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Travisano M, Tahiliani KG, Rached MT, Mirza S. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond) 2008; 32: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohi G, Marchand K, Revesz A, Arany E, Hardy DB. Maternal protein restriction elevates cholesterol in adult rat offspring due to repressive changes in histone modifications at the cholesterol 7α-hydroxylase promoter. Mol Endocrinol 2011; 25: 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, McKnight RA, Caprau D, O'Grady S, Fu Q, Yu X et al. Intrauterine growth restriction affects hippocampal dual specificity phosphatase 5 gene expression and epigenetic characteristics. Physiol Genomics 2011; 43: 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 2011; 140: 1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. A core gut microbiome in obese and lean twins. Nature 2009; 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzielinska Z, Januszewicz A, Wiecek A, Prejbisz A, Zielinski T, Chudek J et al. Reduced kidney function estimated by cystatin C and clinical outcomes in hypertensive patients with coronary artery disease: association with homocysteine and other cardiovascular risk factors. Kidney Blood Press Res 2010; 33: 139–148. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care 2010; 33: 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. Evolution of mammals and their gut microbes. Science 2008; 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K, Versalovic J, Petrosino J, Mistretta TA, Riehle K, Coarfa C, Raza S et al. Metagenomic-based approach to a comprehensive characterization of the vaginal microbiome signature in pregnancy. Am J Obstet Gynecol 2011; 204 (Suppl 1): S42. [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 2011; 474: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]