Abstract
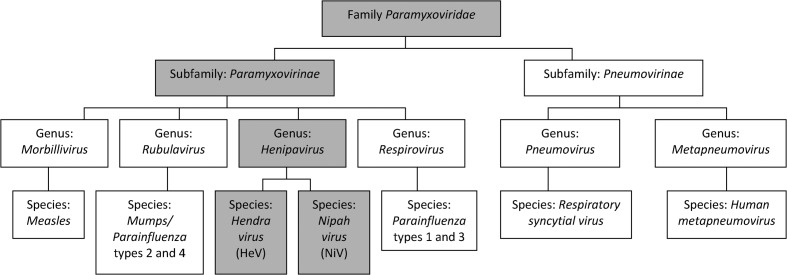
Hendra virus (HeV) was first isolated in 1994, from a disease outbreak involving at least 21 horses and two humans in the Brisbane suburb of Hendra, Australia. The affected horses and humans all developed a severe but unidentified respiratory disease that resulted in the deaths of one of the human cases and the deaths or putting down of 14 of the horses. The virus, isolated by culture from a horse and the kidney of the fatal human case, was initially characterised as a new member of the genus Morbillivirus in the family Paramyxoviridae. Comparative sequence analysis of part of the matrix protein gene of the virus and the discovery that the virus had an exceptionally large genome subsequently led to HeV being assigned to a new genus, Henipavirus, along with Nipah virus (a newly emergent virus in pigs).
The regular outbreaks of HeV-related disease that have occurred in Australia since 1994 have all been characterised by acute respiratory and neurological manifestations, with high levels of morbidity and mortality in the affected horses and humans. The modes of transmission of HeV remain largely unknown. Although fruit bats have been identified as natural hosts of the virus, direct bat–horse, bat–human or human–human transmission has not been reported. Human infection can occur via exposure to infectious urine, saliva or nasopharyngeal fluid from horses. The treatment options and efficacy are very limited and no vaccine exists.
Reports on the outbreaks of HeV in Australia are collated in this review and the available data on the biology, transmission and detection of the pathogen are summarized and discussed.
HISTORY AND CASES
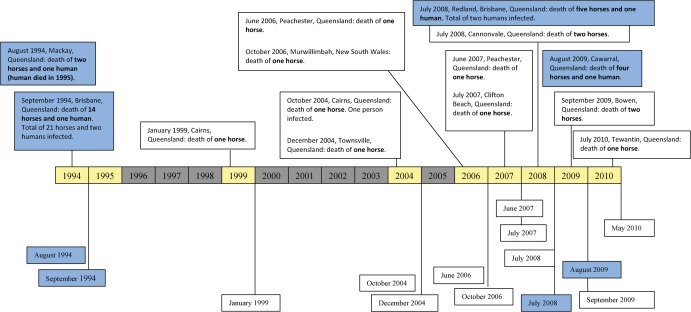
In September 1994, an acute respiratory disease was reported in 21 horses on a property in the Brisbane suburb of Hendra (in Queensland, Australia). The equine illness was characterised by high rectal temperatures, acute respiratory difficulty and influenza-like symptoms, including nasal discharge (Murray et al., 1995). This disease resulted in the death or putting down of 14 of the horses within 2 weeks (Fig. 1). A trainer and a stable-hand who had had close contact with an infected, pregnant mare, that had died of similar symptoms 2 weeks prior to the main outbreak, developed symptoms of acute respiratory disease (Murray et al., 1995). Although the stable-hand made a full recovery, the trainer died after 6 days of interstitial pneumonia. Virus was isolated, by culture, from lung tissue of one of the affected horses post-mortem and inoculated into two healthy horses to examine the aetiology of the infecting agent. Both inoculated horses became ill, with high fever and respiratory disease, after an incubation period of 6 or 10 days (Murray et al., 1995). The virus involved was identified, by comparative sequence analysis of part of the matrix protein gene, as a member of the genus Morbillivirus (Murray et al., 1995; St Georgiev, 2009). Although initially named equine morbillivirus, the pathogen was subsequently renamed as Hendra virus (HeV) after the suburb of Hendra, where the virus was first isolated (Field et al., 2010). As the result of phylogenetic analysis and the observation of the structure and large size of the virus, Wang et al. (2000) placed HeV in a new genus, Henipavirus, within the family Paramyxoviridae (Fig. 2).
Figure 1.
The recorded Hendra-virus infections in horses and humans in Australia, the blue-shaded boxes indicating outbreaks with human fatalities.
Figure 2.
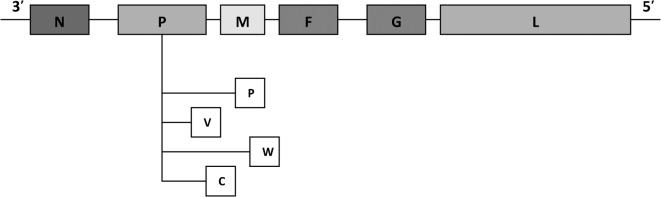
The six transcription-unit genome of Hendra virus, showing the positions of the nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), glycoprotein (G) and large-polymerase (L) genes. The components, P, V and W, are P-gene products encoding the C protein.
Although a retrospective study was performed, on laboratory records and stored specimens, including samples from rodents, marsupials, birds, amphibians and insects, no evidence of HeV infection prior to 1994 was found (Field et al., 2001; Halpin and Mungall, 2007). The 1994 outbreak therefore probably signalled the emergence of a novel virus.
Since the initial outbreak in Hendra, there have been numerous other reports of HeV-related disease in Australian horses, with occasional transmission to humans. In 1995, a second, fatal, human case of HeV was reported from the Queensland city of Mackay (Rogers et al., 1996). This case was linked to two horses that had died, on the case’s property, in 1994 — from what was initially believed to be avocado poisoning or snake-bite but retrospectively confirmed to be HeV infection. HeV activity was reported again in 1999 in Cairns, Queensland, when a thoroughbred mare suddenly died of respiratory illness (Field et al., 2000). At death, the horse displayed a yellow, frothy, nasal discharge from which HeV was isolated. After no recorded activity for another 5 years, HeV caused morbidity in horses twice in 2004, once in Cairns (again) and once in Townsville, Queensland (Hanna et al., 2006). Each of the incidents resulted in the death of a single horse. Although the veterinarian attending the Cairns case developed clinical illness, she recovered and, at the time the report of her illness was published, remained clinically well. Since 2006, HeV-related morbidity has been reported each year in Queensland, resulting in the deaths of two more humans and 15 more horses (www.cdc.gov/ncidod/dvrd/spb/mnpages/dispages/nipah.htm) and HeV-related disease becoming notifiable in Australia (Field, 2009; R. Simmons, unpubl. obs.).
Of particular relevance to the emergence of HeV was an outbreak of an acute respiratory illness in pigs on a farm in peninsular Malaysia in 1998, which resulted in the death of 105 people and the culling of over a million pigs (Chua et al., 2000). Like HeV, the aetiological agent in this outbreak — named Nipah virus (NiV) — was characterised as a member of the family Paramyxoviridae. Several outbreaks of NiV-related disease have occurred in Central and South–east Asia since 1998 (St Georgiev, 2009). Since HeV and NiV display similar genomic and physical characteristics, including having fruit bats as natural hosts and causing a similar disease syndrome in humans, NiV was included as a member of the new genus Henipavirus by Chua et al. (2000).
CHARACTERISTICS AND TAXONOMY OF THE VIRUS
HeV belongs to the family Paramyxoviridae, the members of which are characterised by having a non-segmented, negative-strand RNA (NNS) genome (Wang et al., 2000). Both members of the genus Henipavirus (i.e. HeV and NiV) have a linear ribonucleoprotein core (RNP) that is characteristic of other paramyxoviruses. Structurally, the RNP core consists of a single-stranded genomic RNA molecule to which nucleocapsid proteins (N) are bound. The genomic RNA strand exhibits negative polarity, with one N bound to the genome for every six nucleotides (Calain and Roux, 1993). Each henipavirus is spherical or filamentous, measuring 150–200 nm in diameter and up to 10,040 nm in length, and has surface projections (St Georgiev, 2009). The Henipavirus genome is markedly longer than that of other members of the Paramyxoviridae (typically 15·1–15·9 kb; Wang et al., 2000). The genome of HeV is 18,234 nucleotides long and arranged as 3′-N–P/V/C–M–F–G–L-5′ (Fig. 3; Wang et al., 2000). Nucleoproteins (N), smaller phosphoproteins (P) and a large polymerase protein (L) constitute the RNP core, which is enveloped by a receptor-binding glycoprotein (G) and a fusion protein (F) (Bishop et al., 2007). The G and F proteins appear as the characteristic spikes on the surface of the RNP core and, combined with the RNP core, are responsible for the transcription of genomic RNA to messenger RNA (St Georgiev, 2009). The matrix protein (M), which lies beneath the viral envelope and is known to be released into host cells from the RNP core, completes the architecture of HeV.
Figure 3.
The phylogenetic position of Hendra virus within the family Paramyxoviridae and genus Henipavirus (Wang et al., 2000).
Although the genome of HeV is in many ways similar to those of other paramyxoviruses, several features distinguish HeV. The most obvious of these is the large genome size of HeV, which is primarily attributable to the P and L proteins (Wang et al., 1998, 2000). Sequencing of the L protein of HeV has revealed a conserved intergenic trinucleotide sequence, 3′-GAA-5′, and a stop signal that is similar to those of members of the genera Respirovirus and Morbillivirus (Wang et al., 2000). It was on the basis of the phylogenetic analysis of RNA polymerase (L) genes that a new genus, Henipavirus, was created within the sub-family Paramyxovirinae to accommodate the two ‘new’ viruses, HeV and NiV (Gould, 1996; Yu et al., 1998; Wang et al., 2000).
The complete genome of NiV has still to be reported but this virus appears to be fairly closely related to HeV, showing antigenic similarities, similar amino-acid sequences in its N (92% identity), P (71%,) and L (87%) proteins (Halpin et al., 2007) and sequence homology in its N, P, M, F and G regions (Harcourt et al., 2000). There are, however, considerable differences between the genomes of HeV and NiV. Among 1301 bp in a region from the M gene, for example, the HeV originally isolated in 1994 was found to differ by only five base pairs from an HeV isolated, from another horse, in 1999 but by 408 bp from an NiV isolate (Chua et al., 2000). In addition, an open reading frame (ORF) believed to encode a short basic protein in the P gene of HeV is absent from NiV (Wang et al., 1998).
INFECTION AND DISEASE SYNDROMES
So far, there have been only 14 independent incidents of equine and/or human HeV infection detected in Australia (Fig. 1). Horses infected with HeV display severe respiratory and/or neurological symptoms in most cases and, as such, the HeV-related disease is colloquially known as equine respiratory syndrome or equine Morbillivirus pneumonia (St Georgiev, 2009). The incubation period of HeV in horses seems to range between 48 h and 16 days (Baldock et al., 1996; St Georgiev, 2009; Field et al., 2010). Although some variation in the clinical manifestations of HeV infection — which may be attributable to differences in the virus strain circulating in each outbreak (Field et al., 2001) — has been observed, respiratory and/or neurological distress can be ascribed as the primary manifestations of HeV infection in both horses and humans. In the original outbreak of 1994, acute respiratory disease with high fevers was reported in the affected horses while influenza-like symptoms were reported in the affected humans (Murray et al., 1995). In the outbreak that occurred in 2008, however, the main manifestations in the affected horses and humans were neurological (Field et al., 2010). In parallel with the respiratory and neurological illness, HeV infection in horses is associated with facial swelling, a frothy nasal discharge and ataxia (Hooper et al., 1997). Meningitis has occurred in at least one human infected with HeV (O’Sullivan et al., 1997) and late-onset encephalitis, which may develop up to 8 months post-exposure, has been reported in humans infected with NiV (Tan et al., 2002).
TRANSMISSION DYNAMICS
Direct infection of humans and horses with HeV appears to occur as a result of contact with infectious material from horses. HeV has been isolated from the nasopharyngeal secretions, saliva, urine, foetal material and organs of horses (Daniels et al., 2001) and humans or horses coming into contact with such (presumably) infectious materials are at risk of infection (Field et al., 2007). Transmission from fruit bats of the genus Pteropus — which are believed to be a natural host of HeV (Halpin et al., 2000) — to horses probably occurs but has not been confirmed. Direct human–human transmission has also not been reported. In challenge trials in animal models, HeV has been found in the urine and saliva of experimentally infected cats, and a horse held near the experimentally infected cats showed signs of HeV infection (Williamson et al., 1998). In the same trials, attempts to show horse–cat and fruit-bat–horse transmission were, however, unsuccessful.
To explore the possibility of transmission by vectors, blood-fed mosquitoes (from the properties affected by HeV outbreaks in Brisbane and Mackay) and Cyclopodia albertisii (obligate ectoparasites of fruit bats) have been surveyed for HeV (Field et al., 2001). Although one pooled sample of mosquitoes was found PCR-positive for the virus, all subsequent investigations of potential vectors have given negative results.
HeV has been detected in all four species of fruit bat found on mainland Australia (Pteropus conspicillatus, P. alecto, P. poliocephalus and P. scapulatus) and isolated from three of the species (Mackenzie, 1999; Halpin et al., 2000; Field et al., 2001, 2007). Remarkably, when Field et al. (2001) checked >1000 fruit bats for anti-HeV antibodies, almost one in every two of the bats (47%) was found seropositive. Caution when handling fruit bats is therefore warranted, as is the adherence to strict universal precautions when in contact with an infected human.
Given the high seroprevalence of HeV in fruit bats and the presence of spatial and temporal clustering in the known outbreaks of HeV in Australian horses and humans, viral spill-over from fruit bats to horses seems a plausible explanation of the equine infections (Field et al., 2007). All known outbreaks of HeV in horses and humans have occurred in tropical or sub-tropical regions of Australia where fruit bats are common, either in Queensland or northern New South Wales. There appears to be some evidence of seasonality in the known outbreaks, all but four of which have occurred in June, July, August or September (Fig. 1), and this may be indicative of a seasonal cycle in viral excretion by infected fruit bats (Field et al., 2007).
PATHOGENESIS AND TREATMENT
The molecular mechanisms that lie behind the pathogenicity and virulence of HeV have been investigated in some detail. Generally, pathogenicity in the Henipavirus genus is dependant on the ability of the viruses to circumvent the host’s cellular interferon response (Guillaume et al., 2004), which is believed to be controlled by the P-gene products: P, V, W and C (Eaton et al., 2005; Shaw et al., 2005). As indicated by their names, the cell-attachment glycoprotein (G) and the fusion protein (F) play a significant role in the attachment and fusion of the viruses to host-cell receptors (Bossart et al., 2005). Interestingly, both HeV and NiV display attachment properties akin to those of the Pneumovirinae, with a type-II membrane G glycoprotein that does not haemagglutinate or show neuraminidase activity (Yu et al., 1998). Unlike the Pneumovirinae, however, henipaviruses can use cell-surface protein receptors to bind in the absence of the N-acetyl neuraminic acid that is usually found on the outer membrane of mammalian cells (Eaton et al., 2004).
The features of HeV and NiV pathogenicity, along with the interferon system, have offered significant targets in the development of vaccines against the viruses. Despite a focus on NiV vaccines (Guillaume et al., 2004; Tamin et al., 2009), an HeV vaccine has been trialled in laboratory ferrets and is expected to be released within the decade (www.csiro.au/science/Hendra-Virus.html). An antiviral drug, ribavirin, has recently been demonstrated to reduce viral load in HeV infection (Williamson and Torres-Velez, 2010) although the drug’s use to treat patients infected with HeV has met with varied success (Wright et al., 2004; Playford et al., 2010). Prophylactic immunoglobulins (Ig) are now available in Queensland and are soon to be offered to individuals considered to be at-risk of HeV infection (R. Simmons, unpubl. obs.). In the absence of reliable drug therapy, rapid detection and careful adherence to disease-control procedures are currently the most efficient strategies in the management of HeV outbreaks.
DIAGNOSIS AND DETECTION
The isolation of the virus from a fluid or tissue sample is the definitive method of confirming HeV infection (Hyatt et al., 2001). Clinical samples can be inoculated onto Vero or RK13 cells (Halpin et al., 2000; Rockx et al., 2010), with the subsequent formation of syncytia and punctate holes in the surface of the cell monolayer indicative of the presence of HeV (or NiV). Each syncytium contains at least 20 nuclei and the arrangement of the nuclei within a syncytium can be used to differentiate between HeV and NiV (Hyatt et al., 2001). HeV has been isolated from several different tissues but attempts at viral isolation are often supported by serological, enzymatic and genetic procedures (Daniels et al., 2001). In fatal infections, the virus can be cultured from tissue samples collected post-mortem, including liver, spleen and kidney necropsies. In non-fatal cases, HeV may be isolated from throat swabs, urinary-tract swabs and/or serum (Williamson et al., 1998; Field et al., 2010).
Given the high mortality associated with HeV infection in humans and horses and the lack of an effective vaccine or treatment, samples suspected of containing the virus should only be cultured in laboratories with the highest biosafety classification (i.e. level 4; Williamson and Torres-Velez, 2010). In outbreak situations in Australia, serological testing, such as ELISA, can be performed at level-3 facilities or even, if the serum has been pre-treated to inactivate the virus, at level-2 facilities (Daniels et al., 2001). Once the virus is isolated, characterisation and confirmation tests may be performed by the inoculation of uninfected animals such as golden hamsters (Williamson and Torres-Velez, 2010) as well as by immunohistopathology (Guillame et al., 2009), serology (Black et al., 2001) and using PCR (Smith et al., 2001).
Immunohistochemistry (IHC) and Electron Microscopy
One of the most successful and basic tests for the detection of HeV post-mortem involves the use of a Hendra-specific monoclonal or polyclonal antiserum on formalin-embedded tissue, to detect HeV-specific antigen (Daniels et al., 2001). Many different tissue samples can be used for such IHC, including spleen, lung, brain, mediastinal lymph nodes, kidney, uterus, placenta, and foetal matter. Although primary infection occurs in the vascular endothelium, HeV antigens may be cleared from the lung relatively soon after infection and tissue sampling should therefore not be confined to the lung (Hooper et al., 2001).
Following the post-mortem isolation of virus, electron microscopy can be used to demonstrate HeV in viral cultures. At a minimum density of 108 plaque-forming units/ml, viral cultures can be viewed either directly after negative staining or after specific labelling using immuno-electronmicroscopy techniques (Hyatt et al., 2001).
Serology
The serum neutralization test is the ‘gold standard’ reference test for HeV. The neutralization test works on the principle that the usual cytopathogenic effect (CPE) of the virus, on a cell monolayer, will be blocked by the HeV-specific antibodies present in serum from an animal that, or patient who, is seropositive for HeV. In the standard assay, a test serum, which may be diluted 1∶2 or even 1∶5 if the serum sample is small (Halpin et al., 2000), is incubated with HeV before the virus is added to a Vero-cell monolayer in culture. The cultures are checked for a CPE 3 days later. In the last decade, the standard serum neutralization test has been modified to produce a rapid, immune plaque assay that takes advantage of the development of syncytia in HeV-infected Vero-cell cultures within 24 h of culture inoculation and allows viral load to be quantified (Crameri et al., 2002). This plaque assay can be used to test sera after any viruses present have been inactivated with methanol and gives a result after just 1 day of incubation. In situations requiring urgent, rapid detection, such as an outbreak, ELISA and real-time PCR are, however, more commonly used. Although an indirect ELISA for the detection of anti-HeV antibodies is the test that is most routinely employed, antigen-capture ELISA are rapidly gaining credence and Luminex-based tests are also being used (Daniels et al., 2001; Hayman et al., 2008; Chiang et al., 2010).
Real-time PCR
A real-time assay based on a TaqMan PCR has been used for the detection of HeV in Australia (Halpin et al., 2000; Smith et al., 2001). For this test, RNA is extracted, either using a commercial, viral-RNA extraction kit or manually (Smith et al., 2001). Complementary DNA (cDNA) is generated from the viral RNA template before the cDNA is amplified using primers and probes targeting the M gene of HeV (Smith et al., 2001). The sensitivity of this assay, which Smith et al. (2001) described as 1000 times better than that of conventional PCR, can be improved by the use of additional primers targeting different regions of the HeV genome (such as the P gene), to the point where direct culture becomes unnecessary (Feldman et al., 2009). Several other, modified PCR-based assays, including some based on TaqMan or SYBR Green, are in use at diagnostic facilities in Australia (R. Simmons, unpubl. obs.). Of these modified assays, one targeted at the N gene and based on SYBR-Green chemistry appears the most sensitive, with a detection limit of just 200 HeV virions (Feldman et al., 2009).
CONCLUSIONS
The mechanisms of HeV emergence are largely unknown, as are the modes of the pathogen’s transmission between fruit bats, horses and humans. Some evidence exists for spill-over from fruit bats to horses during outbreaks (Turmelle and Olivial, 2009) but such transmission remains to be demonstrated (Williamson et al., 1998). From an epidemiological perspective, an understanding of the role of possible secondary or intermediate hosts, such as cats, must also be considered (Hyatt et al., 2004). There is currently no vaccine for this highly pathogenic virus and treatment is restricted to general antiviral drugs or prophylactic immunoglobulin therapy, with limited success. The handling of infectious HeV specimens must therefore be restricted to laboratories with high-level biosafety containment and outbreak situations must be treated with great concern by the relevant public-health and veterinary authorities. Although HeV is only known to have caused sporadic and relatively minor outbreaks in the past, it is a virus of significant veterinary, medical and biosecurity importance, especially given the high mortality associated with equine and human infections. It must be acknowledged that HeV is a newly emergent virus, with a known history of only 16 years. The transmission routes of the virus need to be identified and the tests for the diagnosis of HeV infection and the strategies for the management of such infection need to be improved, ideally before further, possibly major, outbreaks occur.
REFERENCES
- 1.Baldock FC, Douglas IC, Halpin K, Field HE, Young PL, Black PF.(1996)Epidemiological investigations into the 1994 equine morbillivirus outbreaks in Queensland, Australia. Singapore Veterinary Journal 2057–61. [Google Scholar]
- 2.Bishop KA, Stantchev TS, Hickey AC, Khetawat D, Bossart KN, Krasnoperov V, Gill P, Feng, YR, Wang L, Eaton BT, Wang LF, Broder CC.(2007)Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. Journal of Virology 815893–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black PF, Cronin JP, Morrissy CJ, Westbury HA.(2001)Serological examination for evidence of infection with Hendra and Nipah viruses in Queensland piggeries. Australian Veterinary Journal 79424–426. [DOI] [PubMed] [Google Scholar]
- 4.Bossart KN, Crameri GS, Dimitrov AS, Mungall BA, Feng YR, Patch JR, Choudhary A, Wang LF, Eaton BT, Broder CC.(2005)Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. Journal of Virology 796690–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calain P, Roux L.(1993)The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. Journal of Virology 644822–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang CF, Lo MK, Rota PA, Spiropoulou CF, Rollin PE.(2010)Use of monoclonal antibodies against Hendra and Nipah viruses in antigen capture ELISA. Virology Journal 7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh WJ, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field HE, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BWJ.(2000)Nipah virus: a recently emergent deadly paramyxovirus. Science 2881432–1435. [DOI] [PubMed] [Google Scholar]
- 8.Crameri G, Wang LF, Morrissy C, White J, Eaton BT.(2002)A rapid immune plaque assay for the detection of Hendra and Nipah viruses and anti-virus antibodies. Journal of Virological Methods 9941–51. [DOI] [PubMed] [Google Scholar]
- 9.Daniels P, Ksiazek TG, Eaton BT.(2001)Laboratory diagnosis of Nipah and Hendra virus infections. Microbes and Infection 3289–295. [DOI] [PubMed] [Google Scholar]
- 10.Eaton BT, Wright PJ, Wang, LF, Sergeyev O, Michalski WP, Bossart KN, Broder CC.(2004)Henipaviruses: recent observations on regulation of transcription and the nature of the cell receptor. Archives of Virology. Supplementum Unnumbered122–131. [PubMed] [Google Scholar]
- 11.Eaton BT, Broder CC, Wang LF.(2005)Hendra and Nipah viruses: pathogenesis and therapeutics. Current Molecular Medicine 5805–816. [DOI] [PubMed] [Google Scholar]
- 12.Eaton BT, Broder CC, Middleton D, Wang LF.(2006)Hendra and Nipah viruses: different and dangerous. Nature Reviews, Microbiology 423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman K. S, Foord A, Heine HG, Smith IL, Boyd V, Marsh GA, Wood JLN, Cunningham AA, Wang LF.(2009)Design and evaluation of consensus PCR assays for henipaviruses. Journal of Virological Methods 16152–57. [DOI] [PubMed] [Google Scholar]
- 14.Field HE.(2009)Hendra virus revisited. Virologica Sinica 24105–109. [Google Scholar]
- 15.Field HE, Barratt PC, Hughes RJ, Shield J, Sullivan ND.(2000)A fatal case of Hendra virus infection in a horse in north Queensland: clinical and epidemiological features. Australian Veterinary Journal 78279–280. [DOI] [PubMed] [Google Scholar]
- 16.Field HE, Young P, Yob JM, Mills J, Hall L, Mackenzie JS.(2001)The natural history of Hendra and Nipah viruses. Microbes and Infection 3307–314. [DOI] [PubMed] [Google Scholar]
- 17.Field HE, Breed AC, Shield J, Hedlefs RM, Pittard K, Pott B, Summers PM.(2007)Epidemiological perspectives on Hendra virus infection in horses and flying foxes. Australian Veterinary Journal 85268–270. [DOI] [PubMed] [Google Scholar]
- 18.Field HE, Schaaf K, Kung N, Simon C, Waltisbuhl D, Hobert H, Moore F, Middleton D, Crook A, Smith G, Daniels P, Glanville RJ, Lovell D.(2010)Hendra virus outbreak with novel clinical features, Australia. Emerging Infectious Diseases 16338–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould AR.(1996)Comparison of the deduced matrix and fusion protein sequences of equine morbillivirus with cognate genes of the Paramyxoviridae. Virus Research 4317–31. [DOI] [PubMed] [Google Scholar]
- 20.Guillaume V, Contamin H, Loth P, Georges-Courbot M-C, Lefeuvre A, Marrianeau P, Chua KB, Lam SK, Buckland R, Deubel V, Wild TF.(2004)Nipah virus: vaccination and passive protection studies in a hamster model. Journal of Virology 78834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillaume V, Thong Wong K, Looi RY, Georges-Courbot M-C, Barrot L, Buckland R, Wild TF, Horva B.(2009)Acute Hendra virus infection: analysis of the pathogenesis and passive antibody protection in the hamster model. Virology 387459–465. [DOI] [PubMed] [Google Scholar]
- 22.Halpin K, Mungall BA.(2007)Recent progress in Henipavirus research. Comparative Immunology, Microbiology and Infectious Diseases 30287–307. [DOI] [PubMed] [Google Scholar]
- 23.Halpin K, Young PL, Field HE, Mackenzie JS.(2000)Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. Journal of General Virology 811927–1932. [DOI] [PubMed] [Google Scholar]
- 24.Halpin K, Hyatt AD, Plowright RK, Epstein JH, Daszak P, Field HE, Wang LF, Daniels PW.(2007)Emerging viruses: coming in on a wrinkled wing and a prayer. Clinical Infectious Diseases 44711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna JN, McBride WJ, Brookes DL, Shield J, Taylor CA, Smith I, Craig SB, Smith GA.(2006)Hendra virus infection in a veterinarian. Medical Journal of Australia 185562–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcourt BH, Tannin A, Ksiazek TG, Rollin PE, Anderson LJ, Bellini WJ, Rota PA.(2000)Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271334–349. [DOI] [PubMed] [Google Scholar]
- 27.Hayman DT, Suu-Ire R, Breed AC, McEachern JA, Wang L, Wood JL, Cunningham AA.(2008)Evidence of henipavirus infection in West African fruit bats. PLoS One 3e2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper PT, Ketterer PJ, Hyatt AD, Russell GM.(1997)Lesions of experimental equine morbillivirus pneumonia in horses. Veterinary Pathology 34312–322. [DOI] [PubMed] [Google Scholar]
- 29.Hooper PT, Zaki SR, Middleton D, Daniels PW.(2001)Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes and Infection 3315–322. [DOI] [PubMed] [Google Scholar]
- 30.Hyatt AD, Zaki SR, Goldsmith CS, Wise TG, Hengstberger SG.(2001)Ultrastructure of Hendra virus and Nipah virus within cultured cells and host animals. Microbes and Infection 3297–306. [DOI] [PubMed] [Google Scholar]
- 31.Hyatt AD, Daszak, P, Cunningham AA, Field HE, Gould AR.(2004)Henipaviruses: gaps in the knowledge of emergence. Ecohealth 125–38. [Google Scholar]
- 32.Mackenzie JS.(1999)Emerging viral diseases: an Australian perspective. Emerging Infectious Diseases 51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray K, Selleck P, Hooper P, Hyatt A, Gould LG, Westbury H, Hiley L, Silvey L, Rodwell B, Ketterer P.(1995)A morbillivirus that caused fatal disease in horses and humans. Science 26894–97. [DOI] [PubMed] [Google Scholar]
- 34.O’Sullivan JD, Allworth AM, Paterson DL, Snow TM, Boots R, Gleeson LJ, Gould AR, Hyatt AD, Bradfield J.(1997)Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet 34993–95. [DOI] [PubMed] [Google Scholar]
- 35.Playford EG, McCall B, Smith G, Slinko V, Allen G, Smith I, Moore F, Taylor C, Kung Y-H, Field HE.(2010)Human Hendra virus encephalitis associated with equine outbreak Australia, 2008. Emerging Infectious Diseases 16219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rockx B, Bossart KN, Feldmann F, Geisbert JB, Hickey AC, Brining D, Callison J, Safronetz D, Marzi A, Kercher L, Long D, Broder CC, Feldmann H, Geisbert TW.(2010)A novel model of lethal Hendra virus infection in African green monkeys and the effectiveness of ribavirin treatment. Journal of Virology 849831–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers RJ, Douglas IC, Baldock FC, Glanville RJ, Seppanen KT, Glesson LJ, Selleck PN, Dunn KJ.(1996)Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Australian Veterinary Journal 74243–244. [DOI] [PubMed] [Google Scholar]
- 38.Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF.(2005)Nuclear localization of Nipah virus W protein allows for inhibition of both virus and Toll-like receptor 3-triggered signalling pathways. Journal of Virology 796078–6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith IL, Halpin K, Warrilow D, Smith GA.(2001)Development of a fluorogenic RT-PCR assay (TaqMan) for the detection of Hendra virus. Journal of Virological Methods 9833–40. [DOI] [PubMed] [Google Scholar]
- 40.St Georgiev V.(2009)Paramyxoviridae: Nipah virus and Hendra virus. National Institute of Allergy and Infectious Diseases, NIH, Vol. 2: Impact on Global Healthed. St Georgiev V.Totowa, NJ: Humana; 143–150. [Google Scholar]
- 41.Tamin A, Harcourt BH, Lo MK, Roth JA, Wolf MC, Lee B, Weingartl H, Audonnet JC, Bellini WJ, Rota PA.(2009)Development of a neutralization assay for Nipah virus using pseudotype particles. Journal of Virological Methods 1601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan CT, Goh KJ, Wong KT, Sarji SA, Chua KB, Chew NK, Murugasu P, Loh YL, Chong HT, Tan KS, Thayaparan T, Kumar S, Jusoh MR.(2002)Relapsed and late-onset Nipah encephalitis. Annals of Neurology 51703–708. [DOI] [PubMed] [Google Scholar]
- 43.Turmelle AS, Olivial KJ.(2009)Correlates of viral richness in bats (order Chiroptera). Ecohealth 6522–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang LF, Michalski WP, Yu M, Pritchard LI, Crameri GS, Shiell B, Eaton BT.(1998)A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses and other animals. Journal of Virology 721482–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang LF, Yu M, Hansson E, Pritchard LI, Shiell B, Michalski WP, Eaton BT.(2000)The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. Journal of Virology 749972–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson MM, Torres-Velez FJ.(2010)Henipavirus: a review of laboratory animal pathology. Veterinary Pathology 47871–880. [DOI] [PubMed] [Google Scholar]
- 47.Williamson MM, Hooper PT, Selleck PW, Gleeson LJ, Daniels PW, Westbury, HAPK Murray.(1998)Transmission studies of Hendra virus (equine morbillivirus) in fruit bats, horses and cats. Australian Veterinary Journal 76813–818. [DOI] [PubMed] [Google Scholar]
- 48.Wright PJ, Crameri, GS, Eaton BT.(2004)RNA synthesis during infection by Hendra virus: an examination by quantitative real-time PCR of RNA accumulation, the effect of ribavirin and the attenuation of transcription. Archives of Virology 150521–532. [DOI] [PubMed] [Google Scholar]
- 49.Yu M, Hansson E, Shiell B, Michalski WP, Eaton BT, Wang LF.(1998)Sequence analysis of the Hendra virus nucleoprotein gene: comparison with other members of the subfamily Paramyxovirinae. Journal of General Virology 791775–1780. [DOI] [PubMed] [Google Scholar]