Abstract
In order to obtain more information about the population structure of Chilean Trypanosoma cruzi, and their genetic relationship with other Latino American counterparts, we performed the study of T. cruzi samples detected in the midgut content of Triatoma infestans insects from three endemic regions of Chile. The genetic characteristics of these samples were analysed using microsatellite markers and PCR conditions that allow the detection of predominant T. cruzi clones directly in triatomine midgut content. Population genetic analyses using the Fisher’s exact method, analysis of molecular variance (AMOVA) and the determination of FST showed that the northern T. cruzi population sample was genetically differentiated from the two southern population counterparts. Further analysis showed that the cause of this genetic differentiation was the asymmetrical distribution of TcIII T. cruzi predominant clones. Considering all triatomines from the three regions, the most frequent predominant lineages were TcIII (38%), followed by TcI (34%) and hybrid (8%). No TcII lineage was observed along the predominant T. cruzi clones. The best phylogenetic reconstruction using the shared allelic genetic distance was concordant with the population genetic analysis and tree topology previously described studying foreign samples. The correlation studies showed that the lineage TcIII from the III region was genetically differentiated from the other two, and this differentiation was correlated with geographical distance including Chilean and mainly Brazilian samples. It will be interesting to investigate whether this geographical structure may be related with different clinical manifestation of Chagas disease.
INTRODUCTION
Chagas disease is well recognized as the most serious human parasitic disease of the Americas in terms of its social and economic impact. Trypanosoma cruzi is the etiological agent of Chagas disease, transmitted by triatomine bugs; it is one of the most important vector-borne diseases in Latin America, infecting approximately 15 million people (WHO, 2007). Several reports have shown that T. cruzi is mainly diploid, with a clonal population structure with possible rare genetic exchange (Tibayrenc et al., 1986; Brisse et al., 2000; Machado and Ayala, 2001; Gaunt et al., 2003; Tibayrenc, 2003; de Freitas et al., 2006). T. cruzi has been classified into two major phylogenetic lineages, T. cruzi I (TCI) and T. cruzi II (TCII) (Souto et al., 1996; Momen, 1999). Later, based on multilocus enzyme electrophoresis and random amplified polymorphic DNA, TCII was further subdivided into five discrete phylogenetic clusters (Brisse et al., 2000). Today, the six T. cruzi lineages are named as: TcI, TcII, TcIII, TcIV, TcV and TcVI, respectively, which will be used in the present paper (Zingales et al., 2009). Several lines of evidence support the idea that TcV and TcVI lineages are hybrid, originated by a relatively recent genetic fusion between TcII and TcIII (Machado and Ayala, 2001; Westenberger et al., 2005). Later, using mitochondrial sequences and five microsatellite markers, it was shown that the two hybrids, TcV and TcVI lineages, are not separate clusters but rather make up a single hybrid lineage, which was confirmed recently by our group (de Freitas et al., 2006; Venegas et al., 2009a).
These phylogenetic analyses conducted in laboratory cultured T. cruzi strains and clones have provided some information related to the population structure of this parasite, such as the observation that the TcI lineage has been found in a vast geographical and ecological area of Latin America and has been associated both with sylvatic and domestic transmission cycles, as well as both with arboreal and terrestrial mammals (Brisse et al., 2000; Tibayrenc, 2003; Miles et al., 2009), whereas lineages TcII–TcVI exhibit a more restricted geographic and ecological distribution. Thus, the majority of T. cruzi clones of lineages TcV and TcVI were found in the southern cone of South America, while the TcIV lineage has been more frequently found in Venezuela and the Amazon Basin (Brisse et al., 2000; Miles et al., 2009). The TcIII lineage was more frequent in the Amazon Basin and in Paraguay and mainly associated with terrestrial mammals (Miles et al., 1977, 2009; Fernandes et al., 1998; Zingales et al., 1998; Yeo et al., 2005). Very interestingly, recently population genetic studies using a high-resolution microsatellite analysis, nuclear and mitochondrial genes have shown that the TcIII lineage is geographically differentiated and is strongly associated with a terrestrial ecotope (Marcili et al., et al., 2009; Llewellyn et al., 2009 a, b). However, it is worth mentioning that since the majority of these studies have been conducted in laboratory cultured parasites, stocks or isolates, they may not represent the real population structure and ecological distribution of natural populations of T. cruzi, due to selective pressure that the laboratory culture and mouse passages exert over the clones and mixtures of lineages which the T. cruzi samples contain (Deane et al., 1984; Macedo and Pena, 1998; Solari et al., 2001; Coronado et al., 2006).
Chagas disease is widespread in Chile, distributed in rural and peri-urban areas in the seven northern regions of the country. The principal and domiciliary vector of T. cruzi is Triatoma infestans, but the peridomestic and sylvatic triatomines Mepraia spinolai and Mepraia gajardoi also bear T. cruzi. The interruption of the domestic cycle of transmission of T. cruzi was completed by the Chilean Health Minister in 1999 (Lorca et al., 2001). Nevertheless, a permanent entomological vigilance and serological study of people at risk must be conducted to avoid the re-colonisation of domestic niches by new triatomines.
The main aims of the present manuscript were to determine whether Chilean T. cruzi populations are geographically structured and whether this phenomenon may be due to genetic differences among or within lineages. In addition, the genetic relationship of these natural T. cruzi populations with other Latino American counterparts, was investigated. We think that obtaining more insights about the organisation and the factors which originated the T. cruzi populations, is highly relevant since this information could be important in the knowledge about the epidemiology and pathogenesis of Chagas disease.
MATERIALS AND METHODS
Population of Triatomines
The triatomines were captured by the Program of Eradication of the Domiciliary Infestation of Triatoma infestans of the Ministry of Health during period 2005–2007. The sample size in the study corresponded to 15, 37 and 12 T. infestans from the Atacama (III) region (middle point among locations of Alto del Carmen and Tierra Amarilla, −27°27′S, −70°15′W), Valparaiso (V) region (mainly from Petorca location −32° 16′S, −70° 58′W) and Metropolitan (M) region (Calera de Tango location, −33°37′S, −70°47′W) regions of Chile (Table 1).
Table 1. Genetic characteristics of Trypanosoma cruzi samples isolated from Triatoma infestans insects captured by the Chilean Vector Control Program of Health Ministry determined by three microsatellite markers.
| Triatomine | Minimal number of clonesa | Lineage of predominant T. cruzi cloneb | ||
| Code | Region | Location | ||
| III | ||||
| 1 | " | Alto del Carmen (AC) | 4 | TcIII |
| 9 | " | AC | 2 | TcI |
| 10 | " | AC | 2 | ND |
| 44 | " | AC | 2 | ND |
| 55 | " | Tierra Amarilla (TA) | 2 | TcI |
| 86 | " | TA | 3 | TcIII |
| 106 | " | TA | 3 | ND |
| 107 | " | TA | 4 | TcIII |
| 108 | " | TA | 5 | TcIII |
| 109 | " | TA | 6 | ND |
| 110 | TA | 6 | TcIII | |
| 111 | " | TA | 4 | TcIII |
| 112 | " | TA | 5 | TcIII |
| 8–120 | " | TA | 7 | TcI |
| 51–163 | " | Mallorca | 4 | TcI |
| V | ||||
| 23 | " | Alto del Puerto | 2 | ND |
| 101 | " | Chincolco | 3 | TcIII |
| 3 | " | San Antonio | 2 | TcI |
| 51 | " | Pedernal (PED) | 2 | TcI |
| 89 | " | PED | 2 | TcIII |
| 90 | " | PED | 4 | TcIII |
| 11 | " | Petorca (PET) | 2 | ND |
| 9 | " | PET | 2 | TcI |
| 24 | " | PET | 2 | ND |
| 25 | " | PET | 2 | TcIII |
| 27 | " | PET | 4 | ND |
| 43 | " | PET | 1 | TcIII |
| 56 | " | PET | 2 | TcIII |
| 59 | " | PET | 1 | TcI |
| 92 | " | PET | 3 | Hybrid |
| 102 | " | PET | 3 | ND |
| 103 | " | PET | 8 | TcI |
| 11–123 | V | PET | 1 | ND |
| 18–130 | V | PET | 7 | TcI |
| 31–143 | V | PET | 3 | ND |
| 58–170 | V | PET | 3 | ND |
| 60–172 | V | PET | 2 | TcIII |
| 61–173 | V | PET | 2 | ND |
| 63–175 | V | PET | 5 | TcIII |
| 64–176 | V | PET | 4 | TcI |
| 65–177 | V | PET | 2 | TcIII |
| 69–181 | V | PET | 4 | Hybrid |
| 72–184 | V | PET | 5 | TcIII |
| 75–187 | V | PET | 2 | TcIII |
| 76–188 | V | PET | 1 | TcI |
| 78–190 | V | PET | 1 | TcIII |
| 14 | V | Sobrante (SO) | 2 | TcI |
| 15 | V | SO | 2 | TcI |
| 16 | V | SO | 1 | TcI |
| 17 | V | SO | 2 | TcI |
| 64 | V | SO | 3 | Hybrid |
| 88–200 | V | San Felipe | 1 | TcI |
| M | ||||
| 68 | M | Calera de Tango (CT) | 1 | TcI |
| 69 | M | 2 | TcI | |
| 2–114 | M | CT | 3 | TcIII |
| 33–145 | M | CT | 9 | TcI |
| 34–146 | M | CT | 4 | TcIII |
| 35–147 | M | CT | 3 | Hybrid |
| 37–149 | M | CT | 3 | TcI |
| 39–151 | M | CT | 4 | TcIII |
| 45–157 | M | CT | 5 | TcIII |
| 49–161 | M | CT | 2 | TcIII |
| 50–162 | M | CT | 5 | TcI |
| 2 | M | Sanatorio | 5 | Hybrid |
aBased on the genotype of predominant T. cruzi clones and total number of alleles, as described (the section on ‘Materials and methods’).
bLineage assignation was performed using three different methods, as described (the section on ‘Materials and methods’). The nomenclature is according to Zingales et al. (2009): TcI (DTU I), TcII (DTU IIb), TcIII (DTU IIc), TcIV (DTU IIa), TcV (DTU IId) and TcVI (DTU IIe). Hybrid corresponds to TcV and TcVI (de Freitas et al., 2006).
T. cruzi DNA Samples from T. infestans
Samples were obtained by the dissection of the complete digestive apparatus of each triatomine. These samples were boiled, centrifuged and the supernatants were used to obtain DNA for the PCR test, as described (Venegas et al., 2010).
PCR Assay
PCR was used to detect the presence of DNA of T. cruzi and was performed in duplicate with the samples of triatomines, according to a described protocol (Dorn et al., 1999). The kinetoplast primers used for the reaction of PCR were S35 and S36 and the nuclear primers were TcZ1 and TcZ2. The products of the PCR were electrophoresed in 2% agarose gel. Gels were stained with ethidium bromide (5 μg/ml) and photographed.
Analysis of Microsatellites
Three microsatellite loci were analysed (SCLE10, SCLE11 and MCLE01) with a modification of the technique described by Oliveira et al. (1998), using a second amplification with an aliquot of 1 μl from the first PCR and the same amplification conditions. The alleles were detected using primers stained with fluorescence (Oliveira et al., 1998). The amplification products were sent to the Roy J. Carver Biotechnology Center, University of Illinois, USA, for analysis by capillary electrophoresis and fluorescence detection with an automatic sequencer and an appropriate software program. The number of base pairs (bps) of each allele was determined using cloned alleles from each marker as controls. In this analysis, the minimal detectable peak height was set to 80 arbitrary fluorescence units (FUs).
Genotyping of the Predominant T. cruzi Clone in Each DNA Sample
The determination of the predominant T. cruzi clone genotype in samples containing multiple alleles per locus was conducted taking into account the distance and the height in FU of the highest two peaks, the major and second peaks, directly in the electropherogram, as described (Venegas et al., 2010). Briefly, when the height ratio between the second and the major peak was less than 0.33, it was concluded that the predominant clone genotype in this sample has only a single allele, and therefore, is homozygous. By contrast, if the height ratio between the second and the major peak was greater than or equal to 0.33, it was analysed further, locus by locus. For instance, Fig. 1a, shows the electropherogram of the sample 24 analysed by the SCLE10 microsatellite marker. Four alleles of 207, 217, 251 and 255 bp were detected. The major and the second peaks correspond to the alleles 207 and 251 with a height of 400 and 210 FU, respectively. The ratio of the heights of the second peak divided by the major one is 0.53, but the distance between them is 44 bp. This distance is beyond the 32 bp that separates two alleles of a heterozygous clone, according to the data previously found by de Freitas et al. (2006) and Venegas et al. (2009a). For this reason, when this happens, it is necessary to look for a third allele that is within the maximum range expected. In this sample, the allele 217 meets this requirement, whose heights ratio among the 217 and 207 alleles is 0.33, just the minimum ratio accepted for a heterozygous clone. The minimum ratio was decided based on those found in heterozygous clones previously described (de Freitas et al., 2006; Venegas et al., 2009a). So, the genotype of the predominant clone of this sample is 207/217, and the minimum number of clones detected in it is 2. In the triatomine sample 25, the major and second alleles are the 207 and 217, respectively, which are within the allowed distance for heterozygous T. cruzi clone (Fig. 1b). As well as the 24 sample, the minimal number of clones detected is 2.
Fig. 1.
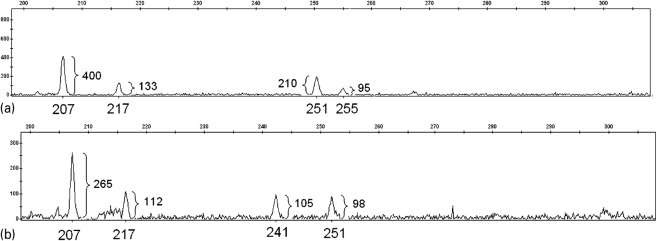
Electropherogram of two midgut Triatoma infestans samples analysed by the SCLE10 microsatellite marker. The panels a and b correspond to the samples code 24 and 25 from the V region. The number below each peaks are the corresponding allele in base-pair length. The numbers beside each peak represent the height of each one in fluorescence units (FU). The genotype of predominant clones of both samples was estimated as 207/217 with two minimal clones (see the section on ‘Materials and method’).
A similar analysis was conducted using the SCLE11 and MCLE01 markers with the exception that the maximum distances of separation between the major and the second peaks were 16 and 20 bp, respectively. This approach was applied with corresponding SCLE10, SCLE11 and MCLE01 locus in each T. cruzi sample (Table 1).
The minimum number of T. cruzi clones in each triatomine sample was estimated based on the genotype of the predominant clone plus the total number of alleles found in each sample divided by 2. Thus, if the predominant clone is homozygous and the sample has three additional alleles (a total of four alleles), it is concluded that the MNC is 3, because an odd number of alleles is considered as another clone. By contrast, in the same sample, if the predominant clone is heterozygous, it is concluded that the MNC is 2, as occur for samples 24 and 25 (Fig. 1).
Determination of Genetic Variation within Each Population
After the genotype of each predominant T. cruzi clone was determined in the different midgut samples of triatomines, the genetic parameters within each population were estimated (Table 2). Allele number (a) was determined using the MSA software (Dieringer and Scholötterer, 2003); the observed heterozygosis (Ho), expected heterozygosis (He) and Hardy–Weinberg equilibrium (HWE) were determined using the Guo and Thompson (1992) method and the program Arlequin (Excoffier et al., 2005).
Table 2. Microsatellite diversity of three Chilean T. cruzi population samples found in T. infestans midgut from the III, V and Metropolitan regionsa.
| Population sample | Parameter | Locus | ||
| SCLE10 | SCLE11 | MCLE01 | ||
| III | n | 12 | 12 | 12 |
| ab | 10 | 10 | 10 | |
| Hoc | 0.000 | 0.083 | 0.384 | |
| Hec | 0.978 | 0.952 | 0.941 | |
| HWEc | 0.000* | 0.000* | 0.000* | |
| V | N | 35 | 32 | 33 |
| a | 17 | 24 | 16 | |
| Ho | 0.241 | 0.414 | 0.393 | |
| He | 0.874 | 0.953 | 0.904 | |
| HWE | 0.000* | 0.000* | 0.000* | |
| M | N | 8 | 11 | 8 |
| a | 7 | 15 | 11 | |
| Ho | 0.250 | 0.818 | 0.666 | |
| He | 0.766 | 0.9480 | 0.954 | |
| HWE | 0.000* | 0.174 | 0.015* | |
| Total number of different alleles observed | 27 | 34 | 26 | |
a III, V and M: groups of midgut samples from 15, 37 and 12 Triatoma infestans captured in the III, V and Metropolitan Chilean regions. Samples with information on at least two loci were considered.
bUsing MSA program (Dieringer and Scholötterer, 2003).
cUsing Arlequin program (Excoffier et al., 2005).
n = individual number genotyped by locus; a = allele number; He = expected heterozygosis; Ho = observed heterozygosis; HWE = P value from the Hardy–Weinberg equilibrium test; asterisk = significant differences.
Analysis of Genetic Differentiation
This was conducted by three approaches: locus by locus Fisher method, multilocus analysis by the Fisher method across three loci and analysis of molecular variance (AMOVA). The genetic markers used in the three approaches were SCLE10, SCLE11 and MCLE01, described previously (Oliveira et al., 1998; de Freitas et al., 2006). The genotypic differentiation by the first and the second approach were analysed with the Genepop program using the Markov algorithm (Raymond and Rousset, 1995a, b). The AMOVA studies were performed with the Arlequin software using the genotype data determined from the predominant T. cruzi clone in each sample with unknown gametic phase, inferring the haplotypes from the distance matrix, using FST as genetic distance and a significant P value of genetic differences among population samples of 0.05 (Weir and Cockerham, 1984; Excoffier et al., 1992).
Lineage Determination of Predominant T. cruzi Clones
The nomenclature used in the present paper for the different lineages, according to the most recent agreement of several researchers, was: TcI (DTU I), TcII (DTU IIb), TcIII (DTU IIc), TcIV (DTU IIa), TcV (DTU IId) and TcVI (DTU IIe) (Brisse et al., 2000; Zingales et al., 2009). However, since that by several criteria, the lineages TcV and TcVI are hybrids, and by microsatellite markers, correspond to the same phylogenetic cluster, in the present paper, they were named as Hybrid (de Freitas et al., 2006).
In order to have a robust assignation of the different T. cruzi lineages, three different methods were used simultaneously with the Geneclass program: the method based on microsatellite lengths described by Goldstein et al. (1995), the method based on allelic frequencies described by Paetkau et al. (1995) and the Bayesian method described by Rannala and Mountain (1997). At least two coincidences in the same lineage with different methods were necessary to assign the final lineage of each predominant T. cruzi clone. Otherwise, the predominant clone was considered as not determined (ND, Table 1).
Correlation Analysis between Genetic and Geographical Distances
Correlation among genetic and geographical distances was estimated by the Mantel test using the Arlequin program and the linear genetic distance FST/(1−FST) (Mantel, 1967; Schneider et al., 2000).
Phylogenetic Reconstruction
In order to determine the genetic relationships between the Chilean T. cruzi sample populations and its foreign counterparts, a phylogram tree was constructed by the program PHYLIP 3.5c (Felsenstein, 1989) using the allele frequencies obtained with the SCLE10, SCLE11 and MCLE01 microsatellite loci. In order to determine the best phylogenetic reconstruction according to the published tree topology obtained with microsatellite markers (de Freita et al., 2006; Venegas et al., 2009a), three measures of genetic distances were tested. Two measurements based on the infinite allelic model, the chord distance Dc of Cavalli–Sforza and Edwards (1967) and the shared allele distance of Bowcock et al. (1994). In addition, genetic distance based on the stepwise mutation model of Goldstein et al. (1995) was also used. The ‘Neighbour-Joining’ algorithm (Saitou and Nei, 1987) was employed for tree construction and 1000 bootstrap iterations were used to test the confidence of the nodes (Felsenstein, 1989).
Published Data of Laboratory Cultured T. cruzi Clones and Strains Genotyped with Different Microsatellite Markers
This was taken from the published data of de Freitas et al. (2006) and Venegas et al. (2009a). The several T. cruzi clones and strains were grouped based on their geographic origin (Table 3) in which the majority corresponded to regions of Brazil, such as Amazon (AM) (−4°30′S, −64°41′W), Espiritu Santos (ES), Minas Gerais (−19°42′S, −44°1′W), Para (PA), Rio de Janeiro (RJ), Sao Paulo (SP), Goias (GO) (−14°53′S, −49°34′W) and Piauí (PI).
Table 3. Data previously published by de Freitas et al. (2006) and Venegas et al. (2009a) corresponding to T. cruzi clones and strains cultured in the laboratory and typed with five microsatellite markers. The groups analysed in the present manuscript are shown in the last column.
| Lineagesa | Strains | SCLE10 | SCLE11 | MCLE01 | Originb | Reference | Code: population samples studied here |
| Rb1 | 255/259 | 139/139 | 128/136 | AM/Brazil | de Freitas | TcI-Br-A | |
| 1502 | 255/255 | 141/143 | 136/136 | AM/" | de Freitas | " | |
| TcI | SE | 287/297 | 143/151 | 136/140 | AM/" | de Freitas | " |
| (Z/Z) | Rb2 | 251/255 | 139/141 | 136/142 | AM/" | de Freitas | " |
| 1523 | 255/255 | 143/147 | 144/144 | AM/" | de Freitas | " | |
| Cutia cl1 | 255-255 | 143/143 | 132/132 | ES/Brazil | Venegas | TcI-Br-B | |
| 1004 | 253/255 | 139/139 | 130/130 | MG/" | de Freitas | " | |
| 1006 | 253/255 | 139/141 | 130/130 | MG/" | de Freitas | " | |
| 1001 | 255/255 | 143/143 | 130/130 | MG/" | de Freitas | " | |
| SilvioX10 cl1 | 235/275 | 153/153 | 130/130 | PA/Brazil | de Freitas | " | |
| D7 | 245/253 | 143/143 | 134/142 | RJ/Brazil | de Freitas | " | |
| Gamba cl1 | 253/255 | 143/143 | 130/130 | SP/Brazil | de Freitas | " | |
| 13379 cl7 | 239/251 | 139/143 | 132/132 | Bolivia | Venegas | TcI-Br-C | |
| Colombiana | 235/257 | 139/139 | 136/136 | Colombia | de Freitas | " | |
| Col18/05 | 253/257 | 139/139 | 136/136 | Colombia | de Freitas | " | |
| A83 | 253/253 | 141/141 | 134/138 | FG | de Freitas | " | |
| A87 | 253/253 | 141/141 | 142/150 | FG | de Freitas | " | |
| TcII | GOCH | 271/281 | 153/159 | 128/150 | GO/Brazil | de Freitas | TcII-Br-G |
| (Y/Y) | 580 | 279/279 | 149/149 | 130/134 | GO/" | de Freitas | " |
| 183744 | 281/281 | 149/149 | 130/134 | GO/" | de Freitas | " | |
| 578 | 281/281 | 149/155 | 130/148 | GO/" | de Freitas | " | |
| 581 | 277/277 | 153/161 | 130/148 | GO/" | de Freitas | " | |
| 577 | 273/281 | 151/155 | 152/154 | GO/" | de Freitas | " | |
| Esm cl3 | 289/289 | 151/159 | 126/138 | MG/Brazil | Venegas | TcII-Br-M | |
| Mas 1 cl1 | 271/289 | 151/151 | 130/130 | MG/" | Venegas | " | |
| Tu 18 cl11 | 289/289 | 149/163 | 124/132 | MG/" | Venegas | " | |
| GMS | 273/285 | 149/157 | 126/134 | MG/" | de Freitas | " | |
| 84 | 267/271 | 153/155 | 126/134 | MG/" | de Freitas | " | |
| JSM | 267/275 | 143/151 | 128/128 | MG/" | de Freitas | " | |
| Tu18 cl11 | 255/285 | 149/163 | 128/130 | MG/" | de Freitas | " | |
| 1043 | 275/281 | 157/157 | 128/134 | MG/" | de Freitas | " | |
| 169/1 | 273/275 | 149/149 | 128/138 | MG/" | de Freitas | " | |
| JAF | 269/275 | 143/149 | 130/134 | MG/" | de Freitas | " | |
| 1005 | 275/275 | 153/155 | 130/156 | MG/" | de Freitas | " | |
| 209 | 269/275 | 153/155 | 132/132 | MG/" | de Freitas | " | |
| Ig539 | 281/281 | 151/153 | 132/150 | MG/" | de Freitas | " | |
| JG | 273/275 | 145/149 | 134/136 | MG/" | de Freitas | " | |
| JHF | 273/273 | 149/149 | 134/136 | MG/" | de Freitas | " | |
| 207 | 245/251 | 151/155 | 134/142 | MG/" | de Freitas | " | |
| 239 | 271/275 | 151/157 | 134/150 | MG/" | de Freitas | " | |
| 84Ti | 271/271 | 143/147 | 136/136 | MG/" | de Freitas | " | |
| 200pm | 277/277 | 153/155 | 136/136 | MG/" | de Freitas | " | |
| Be62 | 273/275 | 155/155 | 136/138 | MG/" | de Freitas | " | |
| Gil | 271/271 | 155/157 | 136/144 | MG/" | de Freitas | " | |
| 803 | 271/281 | 145/145 | 136/152 | MG/" | de Freitas | " | |
| MPD | 267/271 | 143/153 | 138/138 | MG/" | de Freitas | " | |
| 1931 | 275/275 | 141/153 | 140/140 | MG/" | de Freitas | " | |
| 1014 | 269/275 | 149/149 | 140/142 | MG/" | de Freitas | " | |
| CPI95/94 | 277/277 | 149/155 | 124/132 | PI/Brazil | de Freitas | TcII-Br-X | |
| OPS27/94 | 287/297 | 149/159 | 132/136 | PI/" | de Freitas | " | |
| GLT593 | 271/285 | 151/151 | 136/136 | RJ/" | de Freitas | " | |
| GLT564 | 275/275 | 151/151 | 136/136 | RJ/" | de Freitas | " | |
| Y | 273/273 | 153/153 | 130/130 | SP/" | de Freitas | " | |
| TcIII | M5631 cl5 | 245/245 | 155/155 | 134/134 | PA/Brazil | Venegas | TcIII-Br |
| (X/X) | M6241 cl6 | 255/255 | 151/153 | 130/132 | PA/" | Venegas | " |
| 3663 | 245/245 | 155/155 | 128/132 | AM/" | de Freitas | " | |
| 222 | 215/251 | 151/155 | 132/132 | MG/" | de Freitas | " | |
| 115 | 245/251 | 153/153 | 132/132 | MG/" | de Freitas | " | |
| 226 | 245/251 | 153/155 | 132/132 | MG/" | de Freitas | " | |
| 3869 | 249/249 | 153/157 | 132/132 | AM/" “ | de Freitas | " | |
| 231 | 245/251 | 151/155 | 134/134 | MG/" | de Freitas | " | |
| Hybrid | 167 | 237/275 | 153/153 | 130/130 | MG/Brazil | de Freitas | H-Bra |
| TcV–TcVI | 1022 | 233/275 | 153/153 | 130/134 | MG/" | de Freitas | " |
| (X/Y) | 182 | 237/275 | 153/157 | 130/146 | MG/" | de Freitas | " |
| CLBrener | 237/275 | 153/159 | 132/148 | RS/Brazil | Venegas | " | |
| MN cl2 | 255/289 | 149/149 | 130/132 | IV/Chile | Venegas | H-Chi | |
| NR cl3 | 255/289 | 149/165 | 128/130 | Salvador/Chile | Venegas | " | |
| Tula cl2 | 289/289 | 149/151 | 130/130 | IV/Chile | de Freitas | " | |
| Tula 1954 | 237/273 | 153/159 | 132/132 | IV/Chile | Venegas | " |
aEach lineage is composed of T. cruzi clones and strains isolated and cultured in the laboratory using different microsatellite markers (de Freitas et al., 2006; Venegas et al., 2009a). The nomenclature used corresponds to the recent agreement: TcI (DTU I), TcII (DTU IIb), TcIII (DTU IIc), TcIV (DTU IIa), TcV (DTU IId) and TcVI (DTU IIe) (Brisse et al., 2000; Zingales et al., 2009). Hybrid corresponds to TcV and TcVI (de Freitas et al., 2006).
bBrazil regions: Amazon (AM) (−4°30′S, −64°41′W), Espiritu Santos (ES), Minas Gerais (−19°42′S, −44°1′W), Para (PA), Rio de Janeiro (RJ), Sao Paulo (SP), Goias (GO) (−14°53′S, −49°34′W) and Piauí (PI). France Guyana (FG).
RESULTS
Genetic Variation within Each T. cruzi Population
This study was performed in order to determine whether geographic origin has any impact on the genetic characteristics of T. cruzi populations. According to this goal, T. cruzi Chilean and foreign population samples were analysed. The Chilean population samples were from triatomine midguts collected in the III, V and Metropolitan regions (see the section on ‘Materials and methods’ and Table 1). The III region has a hyper-arid climate and is separated by several hundred kilometres of desert from the V and M regions. However, both the V and M regions have similar ecological conditions with a Mediterranean climate characterised by a moderate average temperature (20°C), occasional rain during the winter and close one from another. The sampling point of III region was located 560 and 707 km from the corresponding points in the V and M regions, respectively. In addition, the sampling points in these two last regions were located at 147 km.
The foreign T. cruzi population samples correspond to published data by de Freitas et al. (2006) and Venegas et al. (2009a). These published data are composed of T. cruzi clones and strains isolated and cultured in the laboratory, mainly from regions of Brazil. Most T. cruzi samples were from the Amazon (−4°30′S, −64°41′W), Goias (−14°53′S, −49°34′W) and Minas Gerais (−19°42′S, −44°1′W) (de Freitas et al., 2006) (Table 3).
The intestinal contents of 15, 37 and 12 triatomines from the III, V and M regions positive by conventional PCR detection for T. cruzi DNA (Dorn et al., 1999), were analysed with the SCLE10, SCLE11 and MCLE01 microsatellite markers (Oliveira et al., 1998; de Freitas et al., 2006; Venegas et al., 2009a), as shown in Table 1. The genotype of the predominant T. cruzi clone was determined as described (Venegas et al., 2010) and the genetic parameters within each population were estimated (Table 2). As shown, the total number of alleles observed in the Chilean population samples for SCLE10, SCLE11 and MCLE01 markers were 27, 34 and 26, respectively, which means that the total number of possible haplotypes was 23 868. In other words, in theory, the present methodology using these three microsatellite markers has the potential resolving power to differentiate 23 868 T. cruzi clones. In addition, as expected for clonal populations almost all loci were not found in Hardy–Weinberg equilibrium, as was evidenced by the significant differences among observed and expected heterozygosis. Interestingly, thought this is the first glance, these genetic characteristics were more similar among the V and M regions than among III and V, or III and M regions. For instance, a clear difference in heterozygosis between the III and V regions and between the III and M regions was observed.
Interpopulation Analysis
In order to determine whether the three T. cruzi Chilean population samples from triatomine midgut were genetically differentiated, three analytical methods were used: a locus by locus and multilocus analysis using the Fisher’s exact test ((Raymond and Rousset, 1995a, b), and the AMOVA (Weir and Cockerham, 1984; Excoffier et al., 1992). With the first approach and the SCLE10 locus, a significant genetic differentiation was detected both between III and V and between III and M T. cruzi population samples (Table 4).
Table 4. Analysis of genotypic differentiation using the SCLE10, SCLE11 and MCLE01 loci for each Trypanosoma cruzi group pair.
| Locus | Groups compareda | P valueb | SE | |
| SCLE10c | III | V | 0.00025* | 0.00010 |
| III | M | 0.02763* | 0.00113 | |
| V | M | 0.65085 | 0.00432 | |
| SCLE11 | III | V | 0.29116 | 0.00542 |
| III | M | 0.32907 | 0.00327 | |
| V | M | 0.44513 | 0.00487 | |
| MCLE01 | III | V | 0.12652 | 0.00313 |
| III | M | 0.07590 | 0.00132 | |
| V | M | 0.06904 | 0.00211 | |
| Across three loci (Fisher method) | ||||
| III | V | 0.000745* | ||
| III | M | 0.023990* | ||
| V | M | 0.251287 | ||
aIII, V and MR: groups of midgut samples from 15, 37 and 12 Triatoma infestans captured in the third, fifth and Metropolitan regions, respectively. Samples with information on at least two loci were considered.
bMarkov chain parameters: dememorisation = 10 000; Batches = 100; iterations per batch = 5000.
cLocus by locus using the exact G test (see the section on ‘Materials and methods’).
*P<0.05.
With the second approach, the Fisher’s exact method across those three loci, a significant genetic differentiation was detected between the III and V as well as between III and M (Table 4). The third approach confirmed the result obtained with the second one, although the contribution to the variance among the populations samples was low (4.12%), the FST of 0.0412 was statistically significant (P = 0.018) (Table 5).
Table 5. Multilocus AMOVA analysis of predominant Trypanosoma cruzi genotypes found in the midgut of Triatoma infestans from the III, V and Metropolitan regions.
| Source of variation | df | Sum of squares | Variance componentsb | Percentage of variation |
| Among populationsa | 2 | 4.281 | 0.03560Va | 4.12 |
| Within populations | 125 | 103.586 | 0.82869Vb | 95.88 |
| Total | 127 | 107.867 | 0.86429 | |
| Fixation index | FST | 0.04119 |
aPopulations: III, V and M: groups of midgut samples from 15, 37 and 12 Triatoma infestans captured in the III, V and Metropolitan regions. Samples with information on at least two loci were considered. Molecular distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
bSignificance tests (1023 permutations):
Va and FST: P(rand. value>obs. value) = 0.01760
P(rand. Value = obs. value) = 0.00000
P(rand. value> = obs. value) = 0.01760±0.00338.
Based on the previous results which showed that the T. cruzi population sample from the III region was genetically differentiated from its V and M counterparts, it was interesting to determine whether other biological characteristics could also be different among these T. cruzi population samples. Thus, we analysed the lineage and the minimal number of T. cruzi clones (MNC). In order to have a robust assignation of the lineages, three dissimilar methods were used (see the section on ‘Materials and methods’). The results showed that no significant differences were observed among the predominant T. cruzi lineages of the III, V and M regions (Table 6). Interestingly, the most frequent lineage in regions was TcIII, followed by TcI, while the TcII and Hybrid (TcV–TcVI) lineages were less frequent (Fig. 2). In addition, no significant statistical differences were detected among the minimal number of T. cruzi clones comparing the III, V and M regions (Fig. 3).
Table 6. Lineages of predominant Trypanosoma cruzi clones detected in the midgut of T. infestans triatomines from three Chilean regions (III, V and M).
| Sample groupsa | No. (%) of T. cruzi clones found of each lineageb | ||||
| TcI | TcII | TcIII | H | NDd | |
| III (nc = 15) | 4 (26.7) | 0.0 | 7 (46.7) | 0.0 | 4 (26.7) |
| V (n = 37) | 13 (35.1) | 0.0 | 12 (32.4) | 3 (8.1) | 9 (24.3) |
| M (n = 12) | 5 (41.7) | 0.0 | 5 (41.7) | 2 (16.7) | 0.0 |
| Total (n = 64) | 22 (34.4) | 0.0 | 24 (37.5) | 5 (7.8) | 13 (20.3) |
| Pair | P valuee | ||||
| III/V | 0.3478; 0.5 | 1 | 0.9326; 0.334 | 0.548 | 0.0312; 0.861 |
| III/M | 0.448 | 1 | 0.675; 0.795 | 0.188 | 0.106 |
| V/M | 0.683; 0.16 | 1 | 0.584 | 0.584 | 0.09 |
aIII, V and M: groups of midgut samples from 15, 37 and 12 Triatoma infestans captured in the III, V and Metropolitan regions, respectively.
bDetermined using the Geneclass program using simultaneously the methods of: Goldstein et al. (1995), Paetkau et al. (1995) and Rannala and Mountain (1997) (see the section on ‘Materials and methods’). The lineage nomenclature used corresponds to the recent agreement (Zingales et al., 2009).
c Triatomine number.
dND = not determined.
eSignificant (*P<0.05). Determined by Fischer’s exact method (one value) or Chi-squared (two values).
Fig. 2.
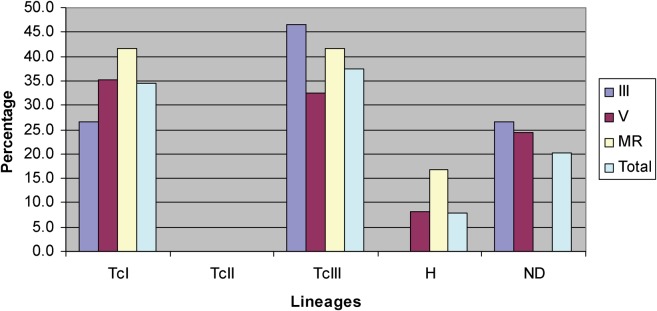
Lineage frequency of predominant T. cruzi clones found in T. infestans midgut collected in the III, V and M regions. The lineage were determined using the microsatellite markers SCLE10, SCLE11 and MCLE01 using the Geneclass program by the methods of Goldstein et al. (1995), Paetkau et al. (1995) and Rannala and Mountain (1997) (see the section on ‘Materials and methods’). The lineage nomenclature used corresponds to the recent agreement (Zingales et al., 2009). Sample sizes were 15, 37 and 12 from III, V and M regions, respectively.
Fig. 3.
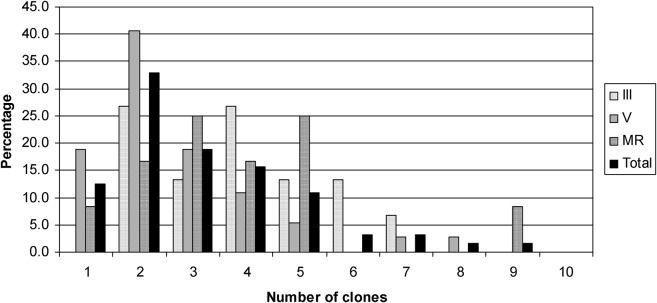
Frequency of minimum number of Trypanosoma cruzi clones detected in T. infestans midguts by three microsatellite markers in population samples from the III, V and M regions. The markers used were SCLE10, SCLE11 and MCLE01, previously described (Oliveira et al., 1998). The population samples included 15, 37 and 12 triatomine insects from the III, V and M regions, respectively (see the section on ‘Materials and methods’).
With the aim to conduct a deeper study to try to identify the possible source of the genetic differentiation among these three Chilean T. cruzi population samples, further analyses were performed with the AMOVA method and the determination of the FST index among all corresponding subpopulation pairs. Thus, the T. cruzi subpopulation samples within the TcI and TcIII lineages were analysed. The result showed that there was no significant differentiation among the TcI subpopulation samples from the III, V and M regions. The percentage of variation among samples was 7.35%, but this was not statistically significant (P = 0.235) (data not shown). This result was confirmed determining the corresponding FST among all subpopulation pairs, since all P values were superior to 0.05 (data not shown).
A significant differentiation was observed among TcIII subpopulations, as is observed by the statistically significant variation and FST among T. cruzi subpopulation samples, 10.41 and 0.104, respectively (P = 0.012, Table 7). It is worth mentioning that only the FST values among the III and V and the III and M samples were statistically significant, among samples V and M FST was not significant (Table 8).
Table 7. Multilocus AMOVA analysis of TcIII predominant T. cruzi genotypes found in the midgut of T. infestans from the III, V and Metropolitan regions.
| Source of variation | df | Sum of squares | Variance componentsb | Percentage of variation |
| Among populationsa | 2 | 4.311 | 0.09162Va | 10.41 |
| Within populations | 45 | 35.501 | 0.78892Vb | 89.59 |
| Total | 47 | 39.812 | 0.88054 | |
| Fixation index | FST | 0.10406 |
aPopulations III, V and M: subgroups of TcIII predominant T. cruzi clones found in the midgut of T. infestans captured in the III, V and Metropolitan regions; each subgroup have 7, 12 and 5 individuals, respectively. Samples with information on at least two loci were considered. Molecular distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
Significance tests (1023 permutations):
Va and FST: P(rand. value>obs. value) = 0.01173
P(rand. value = obs. value) = 0.00000
P(rand. value> = obs. value) = 0.01173±0.00389.
Table 8. Genetic distances FST and corresponding P values among TcIII Trypanosoma cruzi population samples from the III, V and M Chilean regions.
| FST | |||
| Regionsa | III | V | MR |
| III | 0.00000 | ||
| V | 0.14036 | 0.00000 | |
| MR | 0.14829 | 0.02028 | 0.00000 |
| FST P valuesb | |||
| III | … | ||
| V | 0.00879* | … | |
| M | 0.01074* | 0.58008 | … |
aPopulation samples: III, V and M: predominant T. cruzi clones found in the midgut of 7, 12 and 5 Triatoma infestans captured in the third, fifth and Metropolitan Chilean regions, respectively. Molecular distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
bSignificant values with 1023 permutations (*P<0.05).
Genetic Relationship of Chilean T. cruzi Populations with Their Counterparts from Some Other Latin American Regions
In order to determine whether the Chilean T. cruzi population samples were genetically differentiated from their counterparts in other countries, AMOVA analyses and their corresponding FST distances among pairs of subpopulations were performed. To obtain this goal, the first step was to determine whether the T. cruzi population samples available from the literature could be genetically differentiated within each lineage. Therefore, T. cruzi clones and strains from the published data of de Freitas et al. (2006) and Venegas et al. (2009a) were separated by geographical origin. Three groups within the TcI (TcI-A, TcI-B and TcI-C), three groups within TcII (TcII-Br-G, TcII-Br-M and TcII-Br-X), one group of TcIII (TcIII-Br) and two groups of TcV–TcVI (H-Bra and H-Chi) were produced (Table 3). The results showed that within TcI as well as the TcV–TcVI lineages, T. cruzi subpopulation samples are genetically differentiated, as is evidenced by their statistically significant FST P values (P<0.05) (Tables 9 and 10, respectively). In contrast, no significant differentiation was detected among the TcII T. cruzi subpopulation samples (data not shown).
Table 9. Multilocus AMOVA analysis of TcI T. cruzi subpopulations from Brazil.
| Source of variation | df | Sum of squares | Variance componentsb | Percentage of variation |
| Among populationsa | 2 | 6.847 | 0.21309Va | 16.98 |
| Within populations | 31 | 32.300 | 1.04194Vb | 83.02 |
| Total | 33 | 39.147 | 1.25503 | |
| Fixation index | FST | 0.16979 |
aPopulations: TcI Brazilian subpopulations TcI-Br-A, TcI-Br-B and TcI-Br-C (Table 3). Distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
Significance tests (1023 permutations):
Va and FST: P(rand. value>obs. value) = 0.00196
P(rand. value = obs. value) = 0.00000
P(rand. value> = obs. value) = 0.00196±0.00136.
Table 10. Multilocus AMOVA analysis of TcV–TcVI T. cruzi subpopulations from Brazil and Chile.
| Source of variation | df | Sum of squares | Variance componentsb | Percentage of variation |
| Among populationsa | 1 | 2.938 | 0.23772Va | 18.67 |
| Within populations | 14 | 14.500 | 1.03571Vb | 81.33 |
| Total | 15 | 17.438 | 1.27344 | |
| Fixation index | FST | 0.18668 |
aPopulations: TcV–TcVI T. cruzi subpopulations H-Bra and H-Chi (Table 3). Distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
Significance tests (1023 permutations):
Va and FST: P(rand. value>obs. value) = 0.00000
P(rand. value = obs. value) = 0.02444
P(rand. value> = obs. value) = 0.02444±0.00495.
AMOVA analysis and the corresponding FST determinations within the TcI lineage showed that the Chilean subpopulation from the III region is not differentiated from any Brazilian counterpart, since in all pairs of comparisons the P values were superior to 0.05 (data not shown). On the contrary, both the V and M Chilean T. cruzi subpopulation samples are differentiated from the Brazilian TcI-Br-B sample (P<0.05).
The AMOVA and FST determinations of the Chilean and Brazilian TcIII subpopulation samples showed that in all pairs of comparisons, the Chilean samples are significantly differentiated from their Brazilian counterparts, since the P values of FST for each subpopulation pair is much less than 0.05 (Table 11).
Table 11. Genetic distances FST and corresponding P values among TcIII Trypanosoma cruzi population samples from three Chilean regions and one Brazilian region.
| FST | ||||
| Regionsa | III | V | M | Brazil |
| III | 0.00000 | |||
| V | 0.14036 | 0.00000 | ||
| M | 0.14829 | 0.02028 | 0.00000 | |
| Brazil | 0.17918 | 0.17148 | 0.16412 | 0.00000 |
| FST P valuesb | ||||
| III | … | |||
| V | 0.00684* | … | ||
| M | 0.01758* | 0.60254 | … | |
| Brazil | 0.00195* | 0.00000* | 0.00195* | … |
aPopulation samples: III, V and M: predominant T. cruzi clones found in 7, 12 and 5 Triatoma infestans captured in the III, V and Metropolitan Chilean regions. A fourth population sample of eight Brazilian T. cruzi clones mainly from the Minas Gerais region (MG) was included. Molecular distance: number of different alleles (FST) (see the section on ‘Materials and methods’).
bSignificant values with 1023 permutations (P<0.05).
Phylogenetic Analysis of Chilean and Foreign T. cruzi Subpopulation Samples
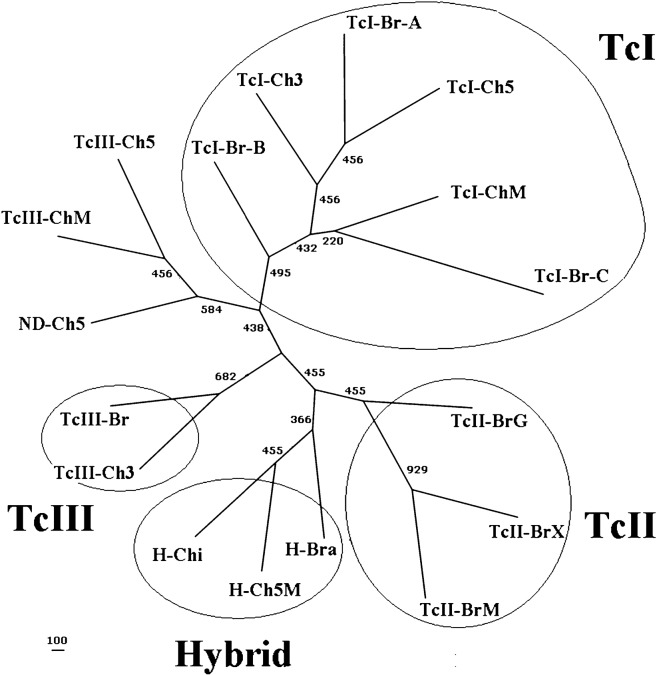
To estimate the phylogenetic relationships among the Chilean and foreign T. cruzi subpopulation samples, phylogenetic trees were constructed based on four different genetic distances; the Cavalli–Sforza and Edwards (1967) chord distance, the Nei et al. (1983) distance, the Bowcock et al. (1994) distance based on the proportion of shared alleles and the Goldstein et al. (1995) distance based on the stepwise mutation model. According to the known T. cruzi phylogeny, the best result was obtained with the Bowcock method (Fig. 4), which is characterized by the main monophyletic clusters TcI and TcII located at the extremes of the tree. In addition, as indicated in the literature (Westenberger et al., 2005; de Freitas et al., 2006; Venegas et al., 2009a), the hybrid lineage (TcV–TcVI) is located between the nodes of TcIII and TcII. In agreement with the results of AMOVA and FST determinations, the three TcI Chilean T. cruzi subpopulation samples are located in the TcI monophyletic node, while the TcIII Chilean subpopulation samples are located in two separate clusters: on the one hand, the TcIIICh3 sample forms an internal node with the single sample from Brazil TcIII-Br, whereas the TcIIICh5 and TcIIIChM Chilean samples form another cluster with the unknown ND-V sample. Interestingly, the hybrid sample including T. cruzi clones from the V and M regions (H-Ch5M) is grouped together with the Chilean laboratory cultured T. cruzi clones belonging to TcV–TcVI lineage (H-Chi).
Fig. 4.
Unrooted phylogenetic tree of the T. cruzi population samples from the midgut of triatomines from Chilean and Brazilian regions. The phylogenetic reconstruction was performed using the allele sharing distance method (Bowcock et al., 1994) and the SCLE10, SCLE11 and MCLE01 microsatellite markers (Oliveira et al., 1998). The T. cruzi population samples from triatomine midgut were directly genotyped and correspond to groups of insects collected in the III (TcI-Ch3 and TcIII-Ch3), V (TcI-Ch5, TcIII-Ch5 and ND-Ch5) and Metropolitan (TcI-ChM and TcIII-ChM) Chilean regions. The lineage nomenclature corresponds to the recent agreement (Zingales et al., 2009); they were assigned based on three simultaneous methods and correspond to the following T. cruzi population samples: TcI lineage (TcI-Ch3, TcI-Ch5 and TcI-ChM) and TcIII lineage (TcIII-Ch3, TcIII-Ch5 and TcIII-ChM). H-Ch5M corresponds to a T. cruzi population sample of the TcV–TcVI lineage from triatomine midguts collected in the V and Metropolitan Chilean regions. The laboratory cultured T. cruzi population samples correspond to published data of de Freita et al. (2006) and Venegas et al. (2009a) and are the following: TcI from Brazil Amazon (TcI-Br-A), TcI mainly from Brazil-Minas Gerais (TcI-Br-B), TcI from Bolivia, Colombia and France Guyana (TcI-C), TcII from Brazil-Goias (TcII-Br-G), TcII from Brazil-Minas Gerais (TcII-Br-M), TcII from several Brazilian regions (TcII-Br-X), TcIII mainly from Minas Gerais (TcIII-Br), TcV–TcVI from Minas Gerais (H-Bra) and TcV–TcVI from Chile (H-Chi) (see Table 3 and the section on ‘Materials and methods’ for more details).
Correlation among Chilean and Brazilian TcIII Subpopulations
In order to determine whether the geographic distances have played an important role in the differentiation of these T. cruzi subpopulation samples, we performed the Mantel test (Mantel, 1967). A correlation coefficient of 0.71 was obtained with a significant P value of 0.038.
Table 12 shows the matrices of lineal genetic distance and geographical distance of the Chilean and Brazilian TcIII T. cruzi subpulations.
Table 12. Matrix of Slatkin linearized FST genetic distance [FST/(1−FST)] and geographic distances of TcIII Trypanosoma cruzi population samples from three Chilean regions and one Brazilian region.
| FST/(1−FST)a | ||||
| Regionsb | III | V | M | Brazil |
| III | 0.00000 | |||
| V | 0.16328 | 0.00000 | ||
| M | 0.17411 | 0.02070 | 0.00000 | |
| Brazil | 0.21829 | 0.20697 | 0.19635 | 0.00000 |
| Geographic distance (km)b | ||||
| Regions | ||||
| III | 0 | |||
| V | 560 | 0 | ||
| M | 707 | 147 | 0 | |
| Brazil | 4056 | 4616 | 4763 | 0 |
aReference: Slatkin et al. (1995).
bPopulation samples: III, V and M: predominant T. cruzi clones found in 7, 12 and 5 Triatoma infestans captured in the III, V and Metropolitan Chilean regions, respectively. A fourth population sample of eight Brazilian T. cruzi clones mainly from the Minas Gerais region (MG) was included. The geographic distances were determined as the smallest distance between two regions with the Google heart online map (see the section on ‘Materials and methods’).
DISCUSSION
The evidence presented in this study strongly suggests that the Chilean T. cruzi populations from the midgut of domestic T. infestans are geographically differentiated. The following evidences support this conclusion.
First, the significant differences detected with the SCLE10 marker among the T. cruzi populations from the III and V regions as well as between the III and Metropolitan regions. It is very interesting that using only a single locus, it is possible to detect significant genotypic differentiation, although methods based on a single locus have less power of resolution than those based on multiple loci. One possible explication that SCLE10 could detect this differentiation may be the high polymorphism we detected, as is illustrated by its 27 total alleles. Considering that T. cruzi is meanly a diploid organism (Tibayrenc et al., 1986; Oliveira et al., 1998; Brisse et al., 2000), this means that using this single SCLE10 marker, in theory, it is possible to distinguish n(n−1)/2 = 27(27−1)/2 = 351 different combinations or genotypes, which may explain why it was possible to detect subtle genetic differences between these two Chilean T. cruzi population samples.
Second, the significant genetic differences detected by the multilocus Fisher’s exact method, using the microsatellite markers SCLE10, SCLE11 and MCLE01 between the III and V regions, as well as between the III and M regions of T. cruzi population samples. This result confirms the above evidence using a more powerful multilocus method. Considering that the total alleles observed with each microsatellite marker (27, 34 and 26 with SCLE10, SCLE11 and MCLE01, respectively), the total number of different haplotypes would be 27×34×26 = 23 868, in absence of linkage disequilibrium, which could explain the resolving power to detected genetic differences among these T. cruzi populations, apparently much higher than the resolving power of isoenzymes, traditionally used in the study of T. cruzi populations (Tibayrenc et al., 1986; Tibayrenc et al., 1990; Brisse et al., 2000).
Third, the AMOVA of haplotype distribution among the T. cruzi population samples from the III and V regions, and from III and M regions. This result reinforces that the genetic differentiation between the III and the V and M regions most probably is real and not due to a fortuitous event or an experimental artefact, because the AMOVA and the Fisher’s exact methods are based on dissimilar principles: the first on haplotype variance and the second on genotype frequences (Raymond and Rousset, 1995a, b).
Fourth, the determination of the genetic distance FST among the T. cruzi population samples from III, V and M regions. The fact that the FST values comparing III and V, and III and M T. cruzi population samples were statistically significant reinforced the first observations which showed that the T. cruzi population from the III region is genetically differentiated from its V and M counterparts. It is worth mentioning that important different geographic characteristics exist between the two triatomine population niches formed by the III region and V–M. They are separated by about 700 km and have dissimilar geographic, climatic and demographic conditions (Apt et al., 1987). These different ecological niches also are associated with different peridomestic mammals as possible reservoirs (Rozas et al., 2007). We think that these ecological and geographical factors of T. cruzi have favoured the conditions for population structuring, originating two dissimilar genetic T. cruzi subpopulations. On the other hand, the V and M regions are separated by only 150 km and they have similar climate and a high rate of human migration; for these reasons, it is very probable that these two regions may really constitute a single ecotope and in consequence, have a single T. cruzi ‘deme’ or subpopulation.
In order to determine whether the genetic differentiation detected between the T. cruzi population samples III and V or between III and M is manifested in another genetic characteristic, we studied the distribution of clones in these samples. Unexpectedly, no significant difference in distribution was observed. However, it is worth recalling the high percentage of multiclonal T. cruzi samples observed in all populations, illustrated by the presence of more than one T. cruzi clone in each triatomine midgut sample. This evidence is concordant with the previous results in showing that in the triatomine intestine the T. cruzi parasites have a more favourable environment for growth and development than in the highly selective environment of the mammalian host (Oliveira et al., 1998; Coronado et al., 2006; Venegas et al., 2010).
At first glance, analysing simultaneously the distribution of the different T. cruzi lineages among the III, V and M regions, no statistically significant dissimilarities were detected. However, it is important to mention that the lineage distributions compared here correspond to lineages of predominant T. cruzi clones; therefore, we cannot discard that the distribution of minor clones may have greater differences among these T. cruzi populations samples or that there may exist subpopulations within each lineages with asymmetric genetic distribution. To determine whether this latter possibility could be occurring in the populations studied here, we proceeded to perform AMOVA studies and determinations of FST between subgroups or subpopulations consisting of a single lineage. The results obtained showed that statistically significant dissimilar haplotype distributions and FST values were found comparing TcIII subpopulation samples either between the III and V regions or between the III and M regions. However, no significant differences were found comparing the same subpopulation pairs only containing TcI individuals. This result not only confirms again the fact that the III region T. cruzi population is genetically different from its V and M region counterparts, but also strongly suggests that the III population of T. cruzi is a discrete entity, physically separated from the other two southern regions, and that this genetic differentiation is only due to the asymmetrical distribution of TcIII T. cruzi clones. To our knowledge, this is the first time that in Chile, it has been shown by population genetics methods that T. cruzi populations really are structured, and not only between lineages; we also demonstrated differential distributions within the same lineage. It is true that in other previous studies, analysing kDNA with restriction enzymes or/and with minicycle probes, some dissimilar distributions of lineages among different localities were observed. Nevertheless, in none of these studies, statistical analyses were conducted to support their observations, and they were mainly based on the analysis of laboratory cultured parasites (Sanchez et al., 1993; Solari et al., 2001; Torres et al., 2004). In this regard, there is a very interesting recent report from Brazil which used several microsatellite markers to show that between different regions, there is subtle but statistical significant genetic differentiation among TcIII T. cruzi populations and that this phylogeographic differentiation could have some impact in clinical manifestations of Chagas disease (Llewelyn et al., 2009a, b).
The very low frequency of the hybrid lineage TcV–TcVI detected in all the population samples studied here (7.8%) was similar to that found in a previous study of our group analysing T. cruzi samples from chronic chagasic patients (Venegas et al., 2010). However, this low percentage of the hybrid lineage is markedly different than published reports of T. cruzi stocks from triatomines, where 21–51% of stocks were characterized as TcIId, corresponding to the old Z2bol nomenclature and the current hybrid TcV lineage (Sanchez et al., 1993; Venegas et al., 1997; Torres et al., 2004; Zingales et al., 2009). Some of the causes which could explain this dissimilarity may be linked to the fact that here we are only typing the predominant T. cruzi clone found in each triatomine sample; therefore, minor T. cruzi clones such as the hybrid ones may not be detected. In addition, in the present paper, it was used as a direct method to type T. cruzi clones without previous isolating and culturing, as it was done in the cited literature (Sanchez et al., 1993; Venegas et al., 1997; Torres et al., 2004). Another explanation to this dissimilarity could be related with the differences among the nature of molecular makers (Junqueira et al., 2005; Coronado et al., 2006; Tillerias et al., 2006; Rozas et al., 2007; Venegas et al., 2009b).
The geographic distances separating these host insects may be a relevant factor involved in genetic differentiation of Chilean T. cruzi populations from triatomine midguts. The evidence which supports this suggestion is the correlation study using the Mantel test (Mantel, 1967; Schneider et al., 2000). Significant statistical correlation (correlation coefficient r = 0.71 and P = 0.038) was found between the Slatkin linearized FST genetic distance [FST/(1–FST)] and the geographic distances of the III, V, M and Brazil TcIII subpopulation samples. This result is concordant with recent studies analysing T. cruzi TcIII isolates from the north and centre of Latin America with a large panel of 49 microsatellite markers and sequences of the glucose-6-phophate isomerase locus (Llewellyn et al., 2009a). In this study, a significant statistical correlation was found between genetic and geographical distances among T. cruzi isolates from four localities of Latin America made up of population samples from the north (Brazil, Venezuela and Colombia), north of Bolivia, south of Bolivia and Paraguay.
With the aim to further obtain more information about the genetic relationship among the studied Chilean and foreign population samples, we conducted phylogenetic studies using different genetic distances. The best result, based on the expected topology and the high polymorphism of the three microsatellite markers used, was found using the infinite allelic model as genetic distance which corresponded to the shared genetic distance (Dc) and Neighbour-Joining method (Cavalli–Sforza and Edwards, 1967; Saitou and Nei, 1987). The tree topology was very similar to the described topology using five microsatellite markers, characterized by the most divergent TcI and TcII lineages at the each end of tree and the hybrid lineage node surrounded by the nodes of TCII and TCIII (de Freitas et al., 2006; Venegas et al., 2009a). The tree also reflects, shown by population genetics studies, the more homogenous nature of the TcI lineages from the III, V and M regions, because all these subpopulation samples were grouped in the same node. However, as expected, the TcIII from the III region was grouped in a distinct node in relation to the node in which the subpopulations in V and M regions were clustered. This phylogenetic construction strongly supports and validates the above population genetic results, giving more evidence that these results are probably not due to a biased sample or experimental error.
CONCLUSIONS
Taken together, the evidence shown in the present paper strongly supports the idea that the T. cruzi populations really are geographically structured in Chile and in other localities of Latin America. Second, in Chile, this geographical differentiation would be due to a dissimilar distribution of TcIII T. cruzi clones, at least in some regions. In addition, it was found that the genetic distance of TcIII populations is correlated with geographical distances among Chilean and foreign populations. The fact that the T. cruzi populations are geographical structured not only among lineages but even within one lineage could have relevant impact on the epidemiological and pathogenic characteristic of Chagas disease. These interesting aspects should be further investigated.
Acknowledgments
This study was supported by FONDECYT 1070837. We are grateful to Leonardo Venegas Hermosilla for the excellent technical support in the preparation of the figures.
REFERENCES
- Apt W, Aguilera X, Arribada A, Gomez L, Miles M, Widmer G.(1987)Epidemiology of Chagas’ disease in northern Chile: isozyme profiles in Trypanosoma cruzi from domestic and sylvatic transmission cycles and their association with cardiopathy. American Journal of Tropical Medicine and Hygiene 37302–307. [DOI] [PubMed] [Google Scholar]
- Bowcock AM, Ruíz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL.(1994)High resolution human evolutionary trees with polymorphic microsatellites. Nature 368455–457. [DOI] [PubMed] [Google Scholar]
- Brisse S, Barnabé C, Tibayrenc M.(2000)Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. International Journal for Parasitology 3035–44. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF.(1967)Phylogenetic analysis: models and estimation procedures. Evolution 32550–570. [DOI] [PubMed] [Google Scholar]
- Coronado X, Zulantay I, Albrecht H, Rosas M, Apt W, Ortiz S, Rodriguez J, Sanchez G, Solari A.(2006)Variation in Trypanosoma cruzi clonal composition detected in blood patients and xenodiagnosis triatomines: implications in the molecular epidemiology of Chile. American Journal of Tropical Medicine and Hygiene 741008–1012. [PubMed] [Google Scholar]
- Deane MP, Mangia RHR, Pereira NM, Momen H, Gonçalves AM, Morel CM.(1984)Trypanosoma cruzi: strain selection by different schedules of mouse passage of an initially mixed infection. Memories Institute Oswaldo Cruz 79495–497. [DOI] [PubMed] [Google Scholar]
- de Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SMR, Chiari E, Junqueira ACV, Fernandes O, Macedo AM, Machado C. R, Pena SDJ.(2006)Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathogen 2226–235. [Google Scholar]
- Dieringer D, Schlötterer C.(2003)Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes 3167–169. [Google Scholar]
- Dorn PL, Engelke D, Rods A, Rosales R, Melgar S, Braney B, Flores J, Monroy C.(1999)Utility of the polymerase chain reaction in detection of Trypanosoma cruzi in Guatemalan Chagas’ disease vectors. American Journal of Tropical Medicine and Hygiene 60740–745. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM.(1992)Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA data. Genetics 131479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S.(2005)Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolution on Bioinformatic Online 147–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.(1989)PHYLIP — Phylogeny Inference Package (Version 3.2). Cladistics 5164–166. [Google Scholar]
- Fernandes O, Souto RP, Castro JA, Pereira JB, Fernandes NC, Junqueira ACV, Naiff RD, Barret TV, Degrave W, Zingales B, Campbell DA, Coura JR.(1998)Brazilian isolates of Trypanosoma cruzi from humans and triatomines classified into two lineages using mini-exon and ribosomal RNA sequences. American Journal of Tropical Medicine and Hygiene 58807–811. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, Taylor MC, Mena SS, Veazey P, Miles GAJ, Acosta N, de Arias AR, Miles MA.(2003)Mechanism of genetic exchange in American trypanosomes. Nature 421936–939. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz-Linares A, Cavalli-Sforza LL, Feldman MW.(1995)Genetic absolute dating based on microsatellites and the origin of modern humans. Proceeding of National Academic of Sciences 926723–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou S, Thompson E.(1992)Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48361–372. [PubMed] [Google Scholar]
- Junqueira AC, Degrave W, Brandao A.(2005)Minicircle organization and diversity in Trypanosoma cruzi populations. Trends in Parasitology 21270–272. [DOI] [PubMed] [Google Scholar]
- Llewelyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Segobia M, Vargas J, Torrico F, Diosque P, Valente V, Valente S, Gaunt MW.(2009a)Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathogens 51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewelyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, Vargas J, Torrico F, Miles MA, Gaunt MW.(2009b)Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Neglected Tropical Disease 31–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca M, García A, Bahamonde MI, Fritz A, Tassara R.(2001)Serological certification of the interruption of the vectorial transmission of Chagas disease in Chile. Revista Medica de Chile 129264–269. [PubMed] [Google Scholar]
- Macedo AM, Pena SDJ.(1998)Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas disease. Parasitology Today 14119–124. [DOI] [PubMed] [Google Scholar]
- Machado CA, Ayala FJ.(2001)Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proceeding of National Academy of Sciences 987396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N.(1967)The detection of disease clustering and a generalized regression approach. Cancer Research 27209–220. [PubMed] [Google Scholar]
- Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira ACV, Souza AI, Rosa JA, Campaner M, Lewis MD, Llewellyn MS, Miles MA, Teixeira MMG.(2009)Comparative phylogeography of Trypanosoma cruzi TCIIc: new host, association with terrestrial ecotopes, and spacial clustering. Infection Genetic and Evolution 91265–1274. [DOI] [PubMed] [Google Scholar]
- Miles MA, Toye PJ, Oswald SC, Godfrey DG.(1977)The identification by isoenzyme patterns of two distinct strain groups of Trypanosoma cruzi, circulating independently in a rural area of Brazil. Transaction of Royal Society for Tropical Medicine and Hygiene 71217–225. [DOI] [PubMed] [Google Scholar]
- Miles MA, Llewellyn MS, Lewis MD, Yeo M, Baleela R, Fitzpatrick S, Gaunt MW, Mauricio IL.(2009)The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology 1361509–1528. [DOI] [PubMed] [Google Scholar]
- Momen H.(1999)Taxonomy of Trypanosoma cruzi: a commentary on characterization and nomenclature. Memorias Instituto Oswaldo Cruz 94181–184. [DOI] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno T.(1983)Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution 19153–170. [DOI] [PubMed] [Google Scholar]
- Oliveira RP, Broude NE, Macedo AM, Cantor CR, Smith CL, Pena SDJ.(1998)Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proceeding of National Academy of Sciences 953776–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D, Calver W, Stirling I, Strobeck C.(1995)Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology 4347–354. [DOI] [PubMed] [Google Scholar]
- Rannala B, Mountain JL.(1997)Detecting immigration by using multilocus genotypes. Proceeding of National Academy of Sciences 949197–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas M, Botto-Mahan C, Coronado X, Ortiz S, Cattan PE, Solari A.(2007)Coexistence of Trypanosoma cruzi genotypes in wild and periodomestic mammals in Chile. American Journal of Tropical Medicine and Hygiene 77647–653. [PubMed] [Google Scholar]
- Raymond M, Rousset F.(1995a)An exact test for population differentiation. Evolution 491283–1286. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F.(1995b)GENEPOP Version 1.2: population genetics software for exact tests and ecumenicism. Journal of Heredity 86248–249. [Google Scholar]
- Saitou N, Nei M.(1987)The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4406–425. [DOI] [PubMed] [Google Scholar]
- Sanchez G, Wallace A, Muñoz S, Venegas J, Ortiz S, Solari A.(1993)Characterization of Trypanosoma cruzi populations by several molecular markers support a clonal mode of reproduction. Biological Research 26167–176. [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L.(2000)Arlequin ver. 2.000: a software for population genetics data analysis Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland [Google Scholar]
- Slatkin M.(1995)A measure of population subdivision based on microsatellite allele frequencies. Genetics 139457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari A, Campillay S, Ortiz S, Wallace A.(2001)Identification of Trypanosoma cruzi genotypes circulating in Chilean chagasic patients. Experimental Parasitology 97226–233. [DOI] [PubMed] [Google Scholar]
- Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B.(1996)DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Molecular Biochemistry and Parasitology 83141–152. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Ward P, Moya A, Ayala FJ.(1986)Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proceeding of National Academy of Sciences 83115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Kjellberg F, Ayala FJ. (1990)A clonal theory of parasitic protozoa: the population structure of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma, and its medical and taxonomical consequences. Proceeding of National Academic of Science 872414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M.(2003)Genetic subdivisions within Trypanosoma cruzi (Discrete Typing Units) and their relevance for molecular epidemiology and experimental evolution. Kinetoplastid Biology and Disease 32–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria J, Lafay B, Virreira M, Barnabé C, Tibayrenc M, Svoboda M.(2006)Trypanosoma cruzi: sequence analysis of the variable region of kinetoplast minicircles. Experimental Parasitology 114279–288. [DOI] [PubMed] [Google Scholar]
- Torres JP, Ortiz S, Munoz S, Solari A.(2004)Trypanosoma cruzi isolates from Chile are heterogeneous and composed of mixed populations when characterized by schizodeme and Southern analyses. Parasitology 128161–168. [DOI] [PubMed] [Google Scholar]
- Venegas J, Coñoepan W, Pichuantes S, Miranda S, Jercic MI, Gajardo M, Sánchez G.(2009a)Phylogenetic analysis of microsatellite markers further supports the two hybridization events hypothesis as the origin of the Trypanosoma cruzi lineages. Parasitology Research 105191–199. [DOI] [PubMed] [Google Scholar]
- Venegas J, Coñoepan W, Pichuantes S, Miranda S, Apt W, Arribada A, Zulantay I, Coronado X, Rodriguez J, Reyes E, Solari A, Sánchez G.(2009b)Differential distribution of Trypanosama cruzi clones in human chronic cardiopathic and non- cardiopathic individuals. Acta Tropica 109187–193. [DOI] [PubMed] [Google Scholar]
- Venegas J, Miranda S, Coñoepan W, Pichuantes S, Jercic MI, González C, Gajardo M, Apt W, Arribada A, Sánchez G.(2010)Microsatellite marker analysis shows differentiation among Trypanosoma cruzi populations of peripheral blood and dejections of Triatoma infestans fed on the same chronic chagasic patients. Parasitology Research 107855–863. [DOI] [PubMed] [Google Scholar]
- Venegas J, Ortiz S, Muñoz S, Solari A.(1997)Molecular karyotype and schizodeme analyses of Trypanosoma cruzi stocks from Chilean triatomines. Parasitology 11541–46. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.(1984)Estimating F-statistics for the analysis of population structure. Evolution 381358–1370. [DOI] [PubMed] [Google Scholar]
- Westenberger SJ, Barnabé C, Campbell DA, Sturm NR.(2005)Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). (2007). Report on the Chagas Disease, eds Guhl F, Lazdins-Helds J K.Special Program for the Research and Training of Tropical Diseases (TDR/SWG/09)Geneva: WHO [Google Scholar]
- Yeo M, Acosta N, Llewellyn M, Sanchez H, Adamson S, Miles GAJ, Lopez E, Gonzalez N, Patterson JS, Gaunt MW.(2005)Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. International Journal for Parasitology 35225–233. [DOI] [PubMed] [Google Scholar]
- Zingales B, Souto RP, Mangia RH, Lisboa CV, Campbell DA, Coura JR, Jansen A, Fernandes O.(1998)Molecular epidemiology of American trypanosomiasis in Brazil based on dimorphisms of rRNA and mini-exon gene sequences. International Journal for Parasitology 28105–112. [DOI] [PubMed] [Google Scholar]
- Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG.(2009)A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Memorias do Instituto Oswaldo Cruz 1041051–1054. [DOI] [PubMed] [Google Scholar]