Abstract
Introduction:
The genetic make-up of malaria parasite is potent for understanding the parasite virulence, designing antimalarial vaccine and evaluating the impact of malaria control measures. There is a paucity of information on genetic structure of Plasmodium falciparum in Jharkhand, India where malaria is rampant and this study aimed to establish molecular characterization of P. falciparum field isolates from Jharkhand measured with two highly polymorphic genetic markers, i.e. the merozoite surface proteins (MSPs) 1 and 2.
Methods:
The genetic diversity of P. falciparum population from low transmission area, Ranchi, Bokaro and Hazaribagh and highly malarious area, Latehar and Palamau districts of Jharkhand were evaluated by polymerase chain reaction-sequencing analyzing msp-1 and msp-2 genes to explore the genetic structure of parasite from this understudied region.
Results:
A total of 134 P. falciparum isolates were analyzed by polymorphic regions of msp-1 and msp-2 and classified according to prevalence of allelic families. The majority of patients from all the five sites had mean monoclonal infections of 67.1 and 60.4% of P. falciparum for msp-1 and msp-2, respectively, whereas, mean multiple genotypes of 32.8 and 39.5% for msp-1 and msp-2, respectively. Interestingly, we observed higher multiclonal infection in low transmission area as compared to highly malarious area in the case of msp-1 genotypes, whereas in msp-2 higher multiclonal infection was observed in highly malarious area compared to low transmission area. The overall multiplicities of infection of msp-1 and msp-2 were 1.38 and 1.39, respectively.
Conclusion:
This is the first report on molecular characterization of P. falciparum field isolates from Jharkhand. The genetic diversity and allelic distribution found in this study is somewhat similar to other reports from India and Southeast Asian countries. However, P. falciparum infection can be highly complex and diverse in these disease-endemic regions of Jharkhand, suggesting continual genetic mixing that could have significant implications for the use of antimalarial drugs and vaccines.
INTRODUCTION
Malaria is a disease of global importance that results in 300–600 million cases annually and an estimated 2.2 billion people at risk of infection (Snow et al., 2005). Of the four species known to infect humans, Plasmodium falciparum is the most virulent and contributes to the majority of deaths associated with the disease (Gupta et al., 1994). Despite enormous efforts for malaria control and prevention, multiple factors, including insecticide resistance in the mosquito vectors, the lack of effective vaccines, and the emergence and rapid spread of drug-resistant strains, are contributing to the global worsening of the malaria situation (Mwingira et al., 2011). Understanding the genetic structure of malaria parasite is essential to predict how fast phenotypes of interest, such as novel antigenic variants or drug resistance, originate and spread in populations (Zhong et al., 2007). Further elucidating the mechanisms generating variation in malaria surface antigens is essential for designing immunization strategies to circumvent the emergence of novel polymorphisms (Hartl et al., 2002). The extensive genetic diversity in natural parasite populations is a major obstacle for the development of an effective vaccine against the human malaria parasite, since antigenic diversity limits the efficacy of acquired protective immunity to malaria (Healer et al., 2004; Takala et al., 2006). Genetic diversity is one of the prominent features of P. falciparum infections. Natural infections often contain mixtures of several genotypes and the human and mosquito hosts are exposed to heterogeneous parasite populations (Zakeri et al., 2005). P. falciparum population diversity is commonly assessed by polymerase chain reaction (PCR)-based typing of the highly polymorphic parasite merozoite surface proteins (MSPs) 1 and 2. MSP-1 of P. falciparum is a major surface protein, with an approximate molecular size of 190 kDa that plays an important role in erythrocyte invasion by the merozoite (Holder and Blackman, 1994). The protein is a principal target of human immune responses (Ferreira et al., 2003; Woehlbier et al., 2006), and is a promising candidate for a blood stage subunit vaccine (Ferreira et al., 2003; Woehlbier et al., 2006). The MSP-1 gene has seven variable blocks that are separated either by conserved or semi-conserved regions. Block 2, a region near the N-terminal of the MSP-1 gene, is the most polymorphic part of the antigen and appears to be under the strongest diversifying selection within natural populations (Holder and Blackman, 1994). Up to now, four different allelic types of block 2 have been identified: MAD20, K1, RO33 and MR (Takala et al., 2002; Happi et al., 2004). MR is a recombinant type allele, made up of sequence from the 5′ end of Mad20 and the 3′ end of RO33, and thus termed as ‘MR’. The most common MR allele detected was 140 bp in length, containing 72 bp of Mad20 sequence followed by 69 bp of RO33 sequence (Takala et al., 2002). MSP-2 of P. falciparum is another leading candidate antigen for subunit malaria vaccine (Lawrence et al., 2000). It comprises highly polymorphic central repeats flanked by unique variable domains and conserved N- and C-terminal domains (Ferreira and Hartl, 2007). The MSP-2 alleles generally fall into two allelic types, FC27 and 3D7, which differ considerably in the dimorphic structure of the variable central region, block 3. Due to their polymorphic features, the MSP-1 and MSP-2 genes have been employed as polymorphic markers in studies of malaria transmission dynamics in natural isolates of P. falciparum. Population genetic analyses of P. falciparum in Southeast Asia have been reported from Thailand, Malaysia, Myanmar, Philippines, Iran and Pakistan (Anderson et al., 2000; Anthony et al., 2005; Zakeri et al., 2005; Iwagami et al., 2009; Kang et al., 2010; Khatoon et al., 2010). In India, several epidemiological surveys and some molecular biological studies on drug-resistant malaria and antigenic molecules have been reported (Joshi et al., 2007; Mamillapalli et al., 2007). The genetic diversity among the P. falciparum population is an important indicator of the malaria transmission intensity in an area (Paul et al., 1998). The genotyping of P. falciparum at the surface antigens MSP-1 and MSP-2 having single copy genes with extensive polymorphism, both with regards to sequence and size, makes them attractive candidate for studies where identification of genetically distinct P. falciparum parasitic subpopulation is of interest (Fernert et al., 2001). A high endemic area is generally characterized by extensive parasite diversity and often carries multiple genotypes (Haddad et al., 1999; Cattamanchi et al., 2003).
Jharkhand, an understudied and tribal dominant region with perennial malaria transmission zone where malaria is rampant and causing 20×103 annual malaria deaths, second to Orissa in India as per the latest observation published by Dhingra et al. (2010) in Lancet, which reflects the importance of the area and its necessity of undertaking extensive investigation in terms of malarial pathology, is concerned. To the best of our knowledge, our investigation is the first report attempting to investigate the multiplicity of infection and genetic diversity of P. falciparum populations to understand the relationship of these factors in varied level of malaria transmission in this region. Information on the nature and extent of population diversity within malaria parasites prevalent in the country is essential not only for understanding the mechanism underlying the pathology of malaria but also for establishing a proper control strategy. However, only limited data are available on the genetic diversity of P. falciparum populations of the country in general and particularly from Jharkhand region. Thus, this study was designed to analyze the genetic diversity of MSP-1 and MSP-2 in field isolates of P. falciparum collected in five districts of Jharkhand with different transmission intensities, namely Palamau, Latehar, Bokaro, Ranchi and Hazaribag. Investigating the geographic variation and frequencies of these polymorphisms using population genetic analyses infers the existence of natural selection. Such knowledge will help in the rational prioritization of vaccine candidates.
MATERIALS AND METHODS
Selection and Description of Study Area
The province of Jharkhand in eastern India is one such area where malaria is rampant. The complexity and magnitude of malaria in the central eastern part of India deserves special mention and attention as the central eastern state contributes 15–20% of total malaria cases in the country as per the Draft on National Policy on Tribals by Govt of India, 2005. Jharkhand had a yearly average slide positivity rate (SPR) for symptomatic individuals of 7.4% over the last three years with P. falciparum accounting for 44% of the cases as per the State Malaria Control Program Annual Report Ranchi, Jharkhand, Directorate of Health Services in 2008. The investigation was conducted in the Jharkhand state emphasizing tribal dominant area (total population according to 2001 census is 31 463 866), and the state of Jharkhand is selected to represent an endemic with stable transmission of malaria with a total of 230 686 malaria cases reported in 2009, of which 39.53% were due to P. falciparum (State Malaria Control Program Annual Report Ranchi, Jharkhand, Directorate of Health Services, 2009). Study is conducted in Jharkhand state emphasizing tribal dominant area as shown in Fig. 1, including Palamau, semi-urban mix population of tribal, rural and urban, accounting highest malaria prevalence and malaria-associated morbidity having SPR of 18% in 2008 with P. falciparum accounting for 21.31% of the cases as per the Malaria Control Program Annual Report Ranchi, Jharkhand, Directorate of Health Services, 2008, in Jharkhand, and Latehar, rural and ethnic tribal population, geographically adjacent to Palamau, accounting severe malaria anemia, cerebral malaria and deaths associated with malaria having SPR of 21.6% in 2008 with P. falciparum accounting for 14.56% of the cases; Bokaro and Ranchi district was selected to represent an urban district with low malaria transmission and the SPRs of Ranchi and Bokaro district were 7.8 and 3.6%, respectively in 2008 with P. falciparum accounting for 50.33 and 36.62% of the cases, respectively as per the Ministry of Health and Family Welfare, National Vector Borne Disease Control Program, Government of India 2008 (Hamer et al., 2009). Hazaribagh, a semi-urban district, had a yearly average SPR for symptomatic individuals of 7.3% over the last three years with P. falciparum accounting for 14% of the cases as per the State Malaria Control Program Annual Report Ranchi, Jharkhand, Directorate of Health Services, 2008. Moreover, the state lies in the tropical zone with an annual rainfall of 1234.5 mm with favorable geo-climatic and ecological conditions conducive for perennial malaria transmission. Thus, the selected sites were meant to provide a reasonable representation of typical conditions that would be found in Jharkhand.
Fig. 1.
Map of Jharkhand with study sites.
Blood Samples and Genomic DNA Extraction
A total of 134 P. falciparum infected blood samples used in this study were collected from patients attending the hospitals at Sadar Hospital Ranchi, Sadar Hospital Hazaribag, Sadar Hospital Palamau, Government PHC at Latehar and Bokaro General Hospital in Ranchi, Hazaribag, Palamau, Latehar and Bokaro districts, respectively in Jharkhand during 2007–2009. Blood spots collected from microscopically positive P. falciparum patients on Whatmann 3 mm filter paper strips were analyzed for the polymorphic forms of MSP-1 (block 2) and MSP-2 (block 3). DNA was extracted from blood spots by using QIAamp Blood Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instruction. This study was carried out in accordance with the protocol approved by the Ethical Review Board of the Vinoba Bhave University as reflected in the guidelines of the medical ethics committee, Ministry of Health, Government of India. Blood specimens were collected of all age groups during different transmission periods of the year from the positive cases of P. falciparum malaria, and confirmed on the basis of clinical symptoms and the parasite blood film was checked after staining with JSB stain (Singh, 1956).
PCR Amplification of the MSP-1 and MSP-2 Genes
Primers and PCR protocols were followed as previously described by Snounou et al. (1999) for family-specific allele analysis of msp-1 (block 2) and msp-2 (block 3). In brief, in the primary reaction, the oligonucleotide primers span the entire genetic segments, block 2 for msp-1 and block 3 for msp-2. In the nested reaction, separate primer pairs target the respective allelic types of msp-1 (K1, MAD20 and RO33) and msp-2 (FC27 and 3D7) as described in detail by Snounou et al. (1999). PCR amplification was performed on a thermal cycler in a final volume of 25 μl. PCR product was electrophoresed on 1.5% agarose gels using ×0.5 TBE buffer at 80–100 V and the DNA was visualized by ultraviolet transillumination after staining with ethidium bromide. The number and size of the resulting amplified products were analyzed using Genetool programme.
Allelic Distribution and Multiplicity of Infection
The prevalence of each allelic type was determined as the presence of PCR products for the type in the total number of amplified bands for the corresponding locus. The number of genotypes per infection, or the multiplicity of the infection (MOI) was estimated by dividing the total number of fragments detected in the individual system by the number of samples positive in the particular system (either msp-1 or msp-2). The MOI, or complexity of infection, was estimated by the average number of PCR fragments per infected individual, as described previously (Zwetyenga et al., 1999).
Sequencing Analysis of MSP-1 and MSP-2
A limited number of representative isolates from different families of msp-1 and msp-2 were sequenced. Sequencing reactions were performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit in an ABI 377 automatic DNA sequencer (Applied Biosystems, Foster City, CA, USA). Analysis of the primary structures of the deduced amino acid sequences was done with DNASTAR (DNASTAR, Madison, WI, USA). Nucleotide sequences reported in this paper are available in the GenBank database under accession numbers HQ821869-72 for MSP-1 and HQ836366-67 for MSP-2, respectively.
RESULTS
Allelic Polymorphism of MSP-1 and MSP-2
Molecular typing revealed the highly polymorphic nature of P. falciparum field isolates from Jharkhand with respect to msp-1 and msp-2. All the three reported allelic families of msp-1 (K1, MAD20 and RO33) and two of msp-2 (FC27 and 3D7) were observed among the isolates of all the five study sites, as shown in Table 1. All family-specific allelic types of msp-1 were observed in the studied isolates and the ranges of prevalence of K1, MAD20 and RO33 types were 16–24, 30.7–48 and 3.3–11.5%, respectively, whereas the highest prevalences of K1 in Palamau, MAD20 in Palamau and RO33 in Bokaro were 24, 48 and 11.5%, respectively. Ranging from 3.5 to 20% of the infections carried two allelic types (K1+MAD20, K1+RO33 and MAD20+RO33), whereas 3.8–7.1% of infections contained all the three allelic types of msp-1. For msp-2, allelic prevalences of 3D7 and FC27 types were in the ranges of 35.7–56 and 12–23.3%, respectively, whereas 24–48% of the infection harbored both allelic types. The highest prevalences of 3D7 in Ranchi and FC27 in Hazaribag were 56 and 23.3%, respectively as shown in Table 1.
Table 1. Allelic distribution of Pfmsp-1 and Pfmsp-2 among Indian isolates from Ranchi, Hazaribag, Bokaro, Palamau and Latehar districts of Jharkhand.
| Gene | Ranchi | Hazaribag | Bokaro | Palamau | Latehar | |
| n = 25 | n = 30 | n = 26 | n = 25 | n = 28 | ||
| Pfmsp-1 | Genotype | Frequency | ||||
| K1 | 16% (4/25) | 23.3% (7/30) | 19.2% (5/26) | 24% (6/25) | 17.8% (5/28) | |
| MAD20 | 36% (9/25) | 36.6% (11/30) | 30.7% (8/26) | 48% (12/25) | 46.4% (13/28) | |
| RO33 | 8% (2/25) | 3.3% (1/30) | 11.5% (3/26) | 4% (1/25) | 10.7% (3/28) | |
| K1+MAD20 | 20% (5/25) | 13.3% (4/30) | 11.5% (3/26) | 8% (2/25) | 7.1% (2/28) | |
| K1+RO33 | 12% (3/25) | 6.6% (2/30) | 15.3% (4/26) | 4 (1/25) | 7.1% (2/28) | |
| MAD20+RO33 | 4% (1/25) | 10% (3/30) | 7.6% (2/26) | 8% (2/25) | 3.5% (1/28) | |
| K1+MAD20+RO33 | 4% (1/25) | 6.6% (2/30) | 3.8% (1/26) | 4% (1/25) | 7.1% (2/28) | |
| Multiclonal isolates | 40% | 36.6% | 38.4% | 24% | 25% | |
| Pfmsp-2 | ||||||
| 3D7 | 56% (14/25) | 36.6% (11/30) | 42.3% (11/26) | 40% (10/25) | 35.7% (10/28) | |
| FC27 | 20% (5/25) | 23.3% (7/30) | 15.3% (4/26) | 12% (3/25) | 21.4% (6/28) | |
| FC27+3D7 | 24% (6/25) | 40% (12/30) | 42.3% (11/26) | 48% (12/25) | 42.8% (12/28) | |
| Multiclonal isolates | 24% | 40% | 42.3% | 48% | 42.8% | |
A total of 186 distinct msp-1 block 2 and 187 msp-2 block 3 fragments, defined by the size and allelic type were detected in the 134 patients. For msp-1, the length variants of the PCR product were 175–600 base pair (bp) for MAD20 and 160–400 bp for K1. Only three different sizes, 170, 200 and 220 bp, were observed for RO33 alleles. For msp-2, the length variants of the PCR product were 240–600 bp for the FC27 and 450–600 bp for the 3D7 allelic families in all the five endemic regions including low transmission and highly malarious zones. However, observed proportions, numbers and size range of alleles among the isolates of different study sites are given in Table 2.
Table 2. Observed proportions of various families of msp-1 and msp-2, allele numbers and size range among P. falciparum isolates of Ranchi, Hazaribag, Bokaro, Palamau and Latehar districts of Jharkhand.
| Ranchi | Hazaribag | Bokaro | Palamau | Latehar | ||
| Gene | Genotype | |||||
| Pfmsp-1 | ||||||
| K1 | ||||||
| Observed nos. | 13 | 15 | 13 | 10 | 12 | |
| Observed allele nos. | 4 | 5 | 3 | 7 | 6 | |
| Size range (bp) | 160–400 | 160–350 | 170–300 | 180–400 | 160–400 | |
| MAD20 | ||||||
| Observed nos. | 16 | 20 | 14 | 17 | 18 | |
| Observed allele nos. | 5 | 9 | 6 | 8 | 7 | |
| Size range (bp) | 175–500 | 180–600 | 175–400 | 175–550 | 175–600 | |
| RO33 | ||||||
| Observed nos. | 7 | 8 | 10 | 5 | 8 | |
| Observed allele nos. | 2 | 1 | 2 | 3 | 2 | |
| Size range (bp) | 170, 220 | 220 | 170, 220 | 170, 220, 200 | 170, 220 | |
| Proportion of unique alleles | 11/36 = 30.5% | 15/43 = 34.8% | 11/37 = 29.7% | 18/32 = 56.2% | 15/38 = 39.4% | |
| Multiplicity of infection | 1.44 | 1.43 | 1.42 | 1.28 | 1.35 | |
| Pfmsp-2 | ||||||
| FC27 | ||||||
| Observed nos. | 11 | 19 | 15 | 15 | 18 | |
| Observed allele nos. | 7 | 4 | 5 | 8 | 9 | |
| Size range (bp) | 240–550 | 250–600 | 240–500 | 250–550 | 250–600 | |
| 3D7 | ||||||
| Observed nos. | 20 | 23 | 22 | 22 | 22 | |
| Observed allele nos. | 9 | 3 | 5 | 8 | 11 | |
| Size range (bp) | 450–600 | 460–650 | 450–550 | 450–650 | 460–600 | |
| Proportion of unique alleles | 16/31 = 51.6% | 7/42 = 16.6% | 10/37–27% | 16/37 = 43.2% | 20/40 = 50% | |
| Multiplicity of infection | 1.24 | 1.4 | 1.42 | 1.48 | 1.42 | |
Complexity of the Infection
The overall MOIs of msp-1 and msp-2 were 1.38 and 1.39, respectively. Interestingly, in a low malaria transmission area Ranchi, the highest MOI of 1.44 was observed as compared to other study sites in the case of msp-1, whereas in a high malaria transmission area Palamau, the highest MOI of 1.48 was observed as compared to other study sites in the case of msp-2. However, the mean MOI for msp-1 in low transmission area is slightly higher as compared to that in high transmission area and the difference in MOI is found to be statistically significant as shown in Fig. 2a, whereas in the case of msp-2, the MOI for higher transmission area is marginally higher but the difference is not significant as shown in Fig. 2b.
Fig. 2.
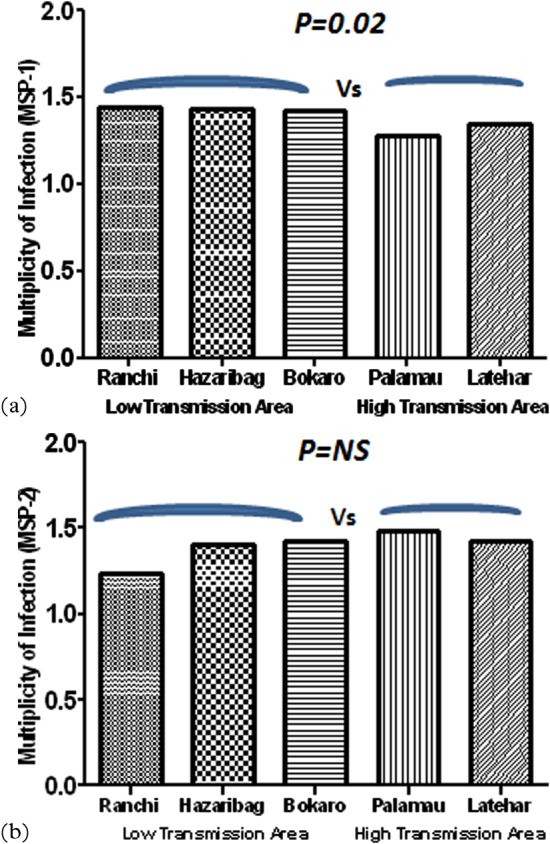
Multiplicity of Plasmodium falciparum infection as assessed with the (a) msp-1 and (b) msp-2 markers in different geographical locations with varied transmission area from malaria endemic districts of Jharkhand.
Sequence Analysis
To understand the diversity of Jharkhand isolates with respect to isolates of other regions, sequence data available in public domains were downloaded for alignment of allelic families of msp-1 and msp-2 and details are given in figure. Comparison of the sequences showed that all these isolates belong to one of these three alleles. Analysis of msp-1 and msp-2 sequence data revealed more than 80% identity of study isolates with isolates of other countries with a few exceptions. In K1 family, 88–100% similarity was observed with K1 allelic sequences reported for isolates of Thailand, Vietnam and Brazil. Similar identity was observed in MAD20 family with isolates of Thailand, Vietnam, Brazil and Iran. RO33 allelic sequences of Jharkhand isolates were very identical and had shown above 70% similarity with sequences reported from isolates of Thailand, Iran and Western Africa and sequence comparisons are shown in Fig. 3a–c. Allelic families of msp-2, 3D7 and FC27 showed above 70% identity with isolates of Vietnam, Brazil, Thailand and Iran. Precisely, with Iran, Vietnam and Brazil isolates, identity ranged between 60 and 70% for Jharkhand FC27 allele and more than 80% with Thailand, Vietnam and Brazil isolate for 3D7 sequences of the present study (Fig. 4a and b).
Fig. 3.
Alignment for the amino acid sequences corresponding to (a) MAD20, (b) K1 and (c) RO33 families of msp-1. Sequences shown are collected during the study and as compared to the sequences from the GenBank database. Gaps are represented by minus sign (−), missing amino acids are represented by question mark sign (?) and alphabets represent a change in amino acid at the position.
Fig. 4.

Alignment for the amino acid sequences corresponding to (a) 3D7 and (b) FC27 families of msp-2. Sequences shown are collected during the study and as compared to the sequences from the GenBank database. Gaps are represented by minus sign (−), missing amino acids are represented by question mark sign (?) and alphabets represent a change in amino acid at the position.
DISCUSSION
The genetic structure of P. falciparum populations plays a highly important role in the natural acquisition of immunity in malarial infections (Healer et al., 2004). Knowledge of the nature and extent of genetic diversity within P. falciparum is essential in understanding the mechanism underlying the pathology of malaria, the acquisition of immunity, the spread of the drug resistance and the condition of transmission. The genetic complexity of the parasite has been well established by demonstrating the occurrence of geographic variation within the species, and of multiple infections with more than one genetically distinct parasite (Babiker et al., 1995). Therefore, an insight in the genetic structure of these populations is necessary to develop strategies to control the disease, including the design of effective vaccines against P. falciparum.
Our results reveal a diverse nature of P. falciparum isolates of malarious districts of Jharkhand in respect of length as well as sequence motifs with prevalence of all the allelic families of msp-1 and msp-2 which are in agreement with the earlier reports from different regions on Indian Isolates (Raj et al., 2004; Ranjit et al., 2005; Joshi et al., 2007). Despite of substantial regional variations, recent increases in P. falciparum incidence and malaria-associated death, Jharkhand represents a low malaria transmission setting with mesoendimicity (Hamer et al., 2009; Dhingra et al., 2010). Our findings of 32.8 and 39.5% of patients with more than one genotype in msp-1 and msp-2, respectively showed lower degree of multi-strain infection in comparison to isolates from Assam, Orissa and West-Bengal as reported by Joshi et al. (2007). It seems that the decrease in immigration of laborer from neighboring states that occurred in recent years is a reason for this finding. The presence of more than one parasitic gene type in a single human host may lead to cross-fertilizations, meiotic recombination and generation of new strains during the developmental stage in the mosquito vector (Snounou et al., 1999; Raj et al., 2004). Furthermore, it seems that non-reciprocal recombination events, such as replication slippage and gene conversion, during the mitotic (asexual) replication of the parasite also play a plausible role in creating allele variation (Rich and Ayala, 2000; Ferreira et al., 2003), allelic diversity of P. falciparum MSP-1 and MSP-2 is mainly generated by meiotic recombination events involving genetically distinct parasite clones that infect the same mosquito vector (Kerr et al., 1994; Babiker and Walliker, 1997). Therefore, the proportion of mixed infections and the number of clones per individual are one of the pre-requisites to generate new genotypes and to increase the diversity of the parasitic population (Da Silveira et al., 1999). These may be the probable reasons that Jharkhand isolates showed rich polymorphism in each gene. MAD20 is observed to be predominant allele in the P. falciparum population in Jharkhand, which is consistent with the situations in Thailand, Iran, Pakistan and Colombia (Snounou et al., 1999; Gόmez et al., 2002; Zakeri et al., 2005; Ghanchi et al., 2010). Interestingly, the prevalence of K1+MAD20 mixed genotype in India (Ranjit et al., 2005), and in the present study is substantially higher than what was observed in Pakistan by Khatoon et al. (2010). On the other hand, MSP-2 in both 3D7 and FC27 allele types were identified among the isolates from Jharkhand. However, the frequency and proportion of 3D7 allele is higher as compared to FC27 in Jharkhand isolates. Similar frequency patterns are observed in Thailand, Iran, Pakistan and Cameroon (Snounou et al., 1999; Basco et al., 2004; Zakeri et al., 2005; Ghanchi et al., 2010), but not in Brazil, where FC27 type is more prevalent (Sallenave–Sales et al., 2007). In view of differences in transmission intensities in various areas, the observed differences in parasite types could be attributed to the factors such as sampling biases, host immune selective pressure on particular types and/or spatiotemporal changes in the availability of different mosquito species that can transmit specific parasite types in a particular area over different times or seasons, with areas that share the same mosquito types tending to have similar parasite types (Zakeri et al., 2010; Branch et al., 2011).
A majority of infections were monoclonal, and should be emphasized that majority of the patients presented with symptomatic malaria infection at a tertiary level of care, i.e. from various hospitals, consequently some of these patients may receive antimalarial treatment prior to enrollment in the study, as treatment is likely to reduce the number of genotypes in an infected individual, and the P. falciparum identified in these hospitals may not be representative for the Jharkhand parasite population. Moreover, the reports of genetic diversity from low endemic areas generally study symptomatic P. falciparum infection. Since symptomatic carriers are rarely observed in setting where semi-immunity cannot be acquired (Beck et al., 1997). This is of importance since data indicate that symptomatic infection generally appears to harbor a lower MOI as compared to asymptomatic patients residing in high transmission areas (Farnert et al., 1999). Lastly, the inability to detect all parasite subpopulations present in an individual, with one single blood sample, either due to inborn limitations of the PCR technique to detect minority clones or parasitic population dynamics (Martensson et al., 2007), may also be considered as a factor potentially underestimating the MOI retrieved in our study. Thus, the result of this study may reflect a significant underestimation of the genetic diversity of the P. falciparum population from Jharkhand.
Our observation of higher multiplicity of infection in low transmission area does corroborate with recent finding from Iran where Zakeri et al., (2010) also reported a relatively high MOI in msp-1 from a presumably low endemic area. However, there is evidence of high genetic diversity of MSP-1 and MSP-2 in areas of low malaria transmission (Sakihama et al., 2007). Furthermore, our observations are also in accordance with the mean MOI values for MSP-1 across the five sub-Saharan African countries as reported by Mwingira et al. (2011). However, genetic differentiation exists between parasite populations in Asian and African isolates (Volkman et al., 2007; Neafsey et al., 2008). Our observation signifies that in highly endemic areas, MOI is not directly correlated with exposure to P. falciparum (Engelbrecht et al., 2000). Furthermore, the extent of allelic diversity is determined not only by the transmission intensity but also by the number of alleles prevalent in the local parasite population and the extent of MOIs. Intragenic meiotic recombination in the mosquito is a major mechanism of generation of allelic variation of P. falciparum msp-1. The frequency of recombination in P. falciparum generally depends on the intensity of malaria transmission, which varies greatly in different endemic areas. This is consistent with the concept that genetic diversity decreases as levels of transmission decrease (Mu et al., 2005).
Finally, these studies have implications for the design of blood stage malaria vaccine based on MSP-1 because an effective vaccine should induce immune response specific for these dominant related to the MSP-1 allelic sequences. As the findings are novel and well aware of the small number of subjects, it deserves to be investigated in a large study group. Future studies will be designed with large number of field isolates from these regions of Jharkhand and analyses of the genetic markers related to antimalarial drug resistance will also be included to gain a more thorough insight of P. falciparum molecular epidemiology from Jharkhand, India.
In conclusion, these data provide link between the infection and diversity through complex interplay of transmission cascade in population exposed to falciparum malaria and would be of great help in gaining an insight into the molecular and epidemiological aspects of malaria infection and possible control measures. Since msp-1 and msp-2 genes are under strong natural selection, interpretation of population structure using data derived from these loci is not easy, since it is not clear whether the patterns observed reflect population history or natural selection. MSP-1 and MSP-2 are non-neutral markers and may differ in their ability to discriminate between populations, even when they are equivalent in their ability to discriminate between clones and strains within the same population, especially when natural selection is a major source of variation in allele frequencies. Lastly, these results also suggested the highly complex population structure of the parasite in Jharkhand. The P. falciparum msp-1 and msp-2 markers used for antigenic variation studies in endemic areas appear to be highly polymorphic with varying transmission intensities. These observations also reinforce the value of these genotyping markers in classifying recurrent post-treatment P. falciparum episodes as recrudescence or new infections and it will facilitate policy makers to plan and to improve antimalarial treatment guidelines for this region.
Acknowledgments
We would like to thank Dr Kumar Abhishek, PJLN Medical College, Raipur, India, for helpful comments on parts of the manuscript and reviewing. We also wish to acknowledge Professor R. N. Bhagat, Vice Chancellor of Vinoba Bhave University for support and kind assistance for the work.
REFERENCES
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP.(2000)Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Molecular Biology and Evolution 171467–1482. [DOI] [PubMed] [Google Scholar]
- Anthony TG, Conway DJ, Cox-Singh J, Matusop A, Ratnam S, Shamsul S, Singh B.(2005)Fragmented population structure of plasmodium falciparum in a region of declining endemicity. Journal of Infectious Diseases 1911558–1564. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Charlwood JD, Smith T, Walliker D.(1995)Gene flow and cross-mating in Plasmodium falciparum in households in a Tanzanian village. Parasitology 111433–442. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Walliker D.(1997)Current views on the population structure of Plasmodium falciparum: implications for control. Parasitology Today 13262–267. [DOI] [PubMed] [Google Scholar]
- Basco LK, Tahar R, Escalante A.(2004)Molecular epidemiology of malaria in Cameroon. XVIII. Polymorphisms of the Plasmodium falciparum merozoite surface antigen-2 gene in isolates from symptomatic patients. The American Journal of Tropical Medicine and Hygiene 70238–244. [PubMed] [Google Scholar]
- Beck HP, Felger I, Huber W, Steiger S, Smith T, Weiss N, Alonso P, Tanner M.(1997)Analysis of multiple Plasmodium falciparum infections in Tanzanian children during the phase III trial of the malaria vaccine SPf66. Journal of Infectious Diseases 175921–926. [DOI] [PubMed] [Google Scholar]
- Branch OH, Sutton PL, Barnes C, Castro JC, Hussin J, Awadalla P, Hijar G.(2011)Plasmodium falciparum genetic diversity maintained and amplified over 5 years of a low transmission endemic in the Peruvian Amazon. Molecular Biology and Evolution 281973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattamanchi A, Kyabayinze D, Hubbard A, Rosenthal PJ, Dorsey G.(2003)Distinguishing recrudescence from reinfection in a longitudinal antimalarial drug efficacy study: comparison of results based on genotyping of msp-1, msp-2, and glurp. The American Journal of Tropical Medicine and Hygiene 68133–139. [PubMed] [Google Scholar]
- Da Silveira LA, Dorta ML, Kimura EA, Katzin AM, Kawamoto F, Tanabe K, Ferreira MU.(1999)Allelic diversity and antibody recognition of Plasmodium falciparum merozoite surface protein 1 during hypoendemic malaria transmission in the Brazilian amazon region. Infection and Immunity 675906–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra N, Jha P, Sharma VP, Cohen AA, Jotkar RM, Rodriguez PS, Bassani DG, Suraweera W, Laxminarayan R, Peto R. & Million Death Study Collaborators(2010)Adult and child malaria mortality in India: a nationally representative mortality survey. Lancet 3761768–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht F, Tögel E, Beck HP, Enwezor F, Oettli A, Felger I.(2000)Analysis of Plasmodium falciparum infections in a village community in Northern Nigeria: determination of msp2 genotypes and parasite-specific IgG responses. Acta Tropica 7463–71. [DOI] [PubMed] [Google Scholar]
- Farnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, Bruce MC, Conway DJ, Day KP, Henning L, Mercereau-Puijalon O, Ranford-Cartwright LC, Rubio JM, Snounou G, Walliker D, Zwetyenga J, Do Rosario VE.(2001)Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Transactions of the Royal Society of Tropical Medicine and Hygiene 95225–232. [DOI] [PubMed] [Google Scholar]
- Farnert A, Rooth I, Svensson A, Snounou G, Bjorkman A.(1999)Complexity of Plasmodium falciparum infections is consistent over time and protects against clinical disease in Tanzanian children. Journal of Infectious Diseases 179989–995. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Hartl DL.(2007)Plasmodium falciparum: worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2). Experimental Parasitology 11532–40. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM.(2003)Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene 30465–75. [DOI] [PubMed] [Google Scholar]
- Ghanchi NK, Mårtensson A, Ursing J, Jafri S, Bereczky S, Hussain R, Beg MA.(2010)Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malaria Journal 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez D, Chaparro J, Rubiano C, Rojas MO, Wasserman M.(2002)Genetic diversity of Plasmodium falciparum field samples from an isolated Colombian village. The American Journal of Tropical Medicine and Hygiene 67611–616. [DOI] [PubMed] [Google Scholar]
- Gupta S, Hill AV, Kwiatkowski D, Greenwood AM, Greenwood BM, Day KP.(1994)Parasite virulence and disease patterns in Plasmodium falciparum malaria. Proceedings of the National Academy of Sciences of the United States of America 913715–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D, Snounou G, Mattei D, Enamorad GI, Figueroa J, Stahl S, Berzins K.(1999)Limited genetic diversity of Plasmodium falciparum in field isolates from Honduras. The American Journal of Tropical Medicine and Hygiene 6030–34. [DOI] [PubMed] [Google Scholar]
- Hamer DH, Singh MP, Wylie BJ, Yeboah-Antwi K, Tuchman J, Desai M, Udhayakumar V, Gupta P, Brooks MI, Shukla MM, Awasthy K, Sabin L, MacLeod WB, Dash AP, Singh N.(2009)Burden of malaria in pregnancy in Jharkhand State, India. Malaria Journal 8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi CT, Gbotosho GO, Sowunmi A, Falade CO, Akinboye DO, Gerena L, Kyle DE, Milhous W, Wirth DF, Oduola AM.(2004)Molecular analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. The American Journal of Tropical Medicine and Hygiene 7020–26. [PubMed] [Google Scholar]
- Hartl DL, Volkman SK, Nielsen KM, Barry AE, Day KP, Wirth DF, Winzeler EA.(2002)The paradoxical population genetics of Plasmodium falciparum. Trends in Parasitology 18266–272. [DOI] [PubMed] [Google Scholar]
- Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A.(2004)Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Molecular Microbiology 52159–168. [DOI] [PubMed] [Google Scholar]
- Holder AA, Blackman MJ.(1994)What is the function of MSP-I on the malaria merozoite? Parasitology Today 10182–184. [DOI] [PubMed] [Google Scholar]
- Iwagami M, Rivera PT, Villacorte EA, Escueta AD, Hatabu T, Kawazu S, Hayakawa T, Tanabe K, Kano S.(2009)Genetic diversity and population structure of Plasmodium falciparum in the Philippines. Malaria Journal 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi H, Valecha N, Verma A, Kaul A, Mallick PK, Shalini S, Prajapati SK, Sharma SK, Dev V, Biswas S, Nanda N, Malhotra MS, Subbarao SK, Dash AP.(2007)Genetic structure of Plasmodium falciparum field isolates in eastern and north-eastern India. Malaria Journal 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, Kim TS, Na BK.(2010)Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malaria Journal 9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr PJ, Ranford-Cartwright LC, Walliker D.(1994)Proof of intragenic recombination in Plasmodium falciparum. Molecular and Biochemical Parasitology 66241–248. [DOI] [PubMed] [Google Scholar]
- Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G.(2010)Genetic structure of Plasmodium vivax and Plasmodium falciparum in the Bannu district of Pakistan. Malaria Journal 9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence G, Cheng QQ, Reed C, Taylor D, Stowers A, Cloonan N, Rzepczyk C, Smillie A, Anderson K, Pombo D, Allworth A, Eisen D, Anders R, Saul A.(2000)Effect of vaccination with 3 recombinant asexual-stage malaria antigens on initial growth rates of Plasmodium falciparum in non-immune volunteers. Vaccine 181925–1931. [DOI] [PubMed] [Google Scholar]
- Mamillapalli A, Sunil S, Diwan SS, Sharma SK, Tyagi PK, Adak T, Joshi H, Malhotra P.(2007)Polymorphism and epitope sharing between the alleles of merozoite surface protein-1 of Plasmodium falciparum among Indian isolates. Malaria Journal 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martensson A, Ngasala B, Ursing J, Veiga IsabelM, Wiklund L, Membi C, Montgomery SM, Premji Z, Farnert A, Bjorkman A.(2007)Influence of consecutive-day blood sampling on polymerase chain reaction- adjusted parasitological cure rates in an antimalarial drug trial conducted in Tanzania. Journal of Infectious Diseases 195597–601. [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ.(2005)Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biolology 3e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck HP, Snounou G, Felger I, Olliaro P, Mugittu K.(2011)Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malaria Journal 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA, Jr, Lukens A., Rosen D., Daniels R., Houde N., Cortese J. F., Tyndall E., Gates C., Stange-Thomann N., Sarr O., Ndiaye D., Ndir O., Mboup S., Ferreira M. U., Sdo MoraesL., Dash A. P., Chitnis C. E., Wiegand R. C., Hartl D. L., Birren B. W., Lander E. S., Sabeti P.C., Wirth DF.(2008)Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biology 9R171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul RE, Hackford I, Brockman A, Muller-Graf C, Price R, Luxemburger C, White NJ, Nosten F, Day KP.(1998)Transmission intensity and Plasmodium falciparum diversity on the northwestern border of Thailand. The American Journal of Tropical Medicine and Hygiene 58195–203. [DOI] [PubMed] [Google Scholar]
- Raj DK, Das BR, Dash AP, Supakar PC.(2004)Genetic diversity in the MSP-1 gene of Plasmodium falciparum in different malaria endemic localities. The American Journal of Tropical Medicine and Hygiene 71285–289. [PubMed] [Google Scholar]
- Ranjit MR, Das A, Das BP, Das BN, Dash BP, Chhotray GP.(2005)Distribution of Plasmodium falciparum genotypes in clinically mild and severe malaria cases in Orissa, India. Transactions of the Royal Society of Tropical Medicine and Hygiene 9389–395. [DOI] [PubMed] [Google Scholar]
- Rich SM, Ayala FJ.(2000)Population structure and recent evolution of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America 976994–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakihama N, Nakamura M, Palanca AA, Jr, Argubano R. A., Realon E.P., Larracas A.L., Espina R.L., Tanabe K.(2007)Allelic diversity in the merozoite surface protein 1 gene of Plasmodium falciparum on Palawan Island, the Philippines. Parasitology International 56185–194. [DOI] [PubMed] [Google Scholar]
- Sallenave-Sales S, Faria CP, Zalis MG, Daniel-Ribeiro CT, Ferreira-da-Cruz Mde F.(2007)Merozoite surface protein 2 allelic variation influences the specific antibody response during acute malaria in individuals from a Brazilian endemic area. Memórias do Instituto Oswaldo Cruz 102421–424. [DOI] [PubMed] [Google Scholar]
- Singh J.(1956)J. S. B. stain; a review. Indian Journal of Malariology 10117–129. [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S.(1999)Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Transactions of the Royal Society of Tropical Medicine and Hygiene 93369–374. [DOI] [PubMed] [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI.(2005)The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala S, Branch O, Escalante AA, Kariuki S, Wootton J, Lal AA.(2002)Evidence for intragenic recombination in Plasmodium falciparum: identification of a novel allele family in block 2 of merozoite surface protein-1: Asembo Bay Area Cohort Project XIV. Molecular and Biochemical Parasitology 125163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala SL, Escalante AA, Branch OH, Kariuki S, Biswas S, Chaiyaroj SC, Lal AA.(2006)Genetic diversity in the Block 2 region of the merozoite surface protein 1 (MSP-1) of Plasmodium falciparum: additional complexity and selection and convergence in fragment size polymorphism. Infection, Genetics and Evolution 6417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr, Daily J. P., Sarr O., Ndiaye D., Ndir O., Mboup S., Duraisingh M. T., Lukens A., Derr A., Stange-Thomann N., Waggoner S., Onofrio R., Ziaugra L., Mauceli E., Gnerre S., Jaffe D. B., Zainoun J., Wiegand R.C., Birren B.W., Hartl D.L., Galagan J.E., Lander E.S., Wirth DF.(2007)A genome-wide map of diversity in Plasmodium falciparum. Nature Genetics 39113–119. [DOI] [PubMed] [Google Scholar]
- Woehlbier U, Epp C, Kauth CW, Lutz R, Long CA, Coulibaly B, Kouyaté B, Arevalo-Herrera M, Herrera S, Bujard H.(2006)Analysis of antibodies directed against merozoite surface protein 1 of the human malaria parasite Plasmodium falciparum. Infection and Immunity 741313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri S, Bereczky S, Naimi P, Pedro Gil J, Djadid ND, Färnert A, Snounou G, Björkman A.(2005)Multiple genotypes of the merozoite surface proteins 1 and 2 in Plasmodium falciparum infections in a hypoendemic area in Iran. Tropical Medicine & International Health 101060–1064. [DOI] [PubMed] [Google Scholar]
- Zakeri S, Kakar Q, Ghasemi F, Raeisi A, Butt W, Safi N, Afsharpad M, Memon MS, Gholizadeh S, Salehi M, Atta H, Zamani G, Djadid ND. (2010)Detection of mixed Plasmodium falciparum & P. vivax infections by nested-PCR in Pakistan, Iran & Afghanistan. Indian Journal of Medical Research 13231–35. [PubMed] [Google Scholar]
- Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, Temu EA, Yan G.(2007)Plasmodium falciparum genetic diversity in western Kenya highlands. The American Journal of Tropical Medicine and Hygiene 771043–1050. [PubMed] [Google Scholar]
- Zwetyenga J, Rogier C, Spiegel A, Fontenille D, Trape JF, Mercereau-Puijalon O.(1999)A cohort study of Plasmodium falciparum diversity during the dry season in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene 93375–380. [DOI] [PubMed] [Google Scholar]