Abstract
Type IB topoisomerases unwind positive and negative DNA supercoils and play a key role in removing supercoils that would otherwise accumulate at replication and transcription forks. An interesting question is whether topoisomerase activity is regulated by the topological state of the DNA, thereby providing a mechanism for targeting the enzyme to highly supercoiled DNA domains in genomes. The type IB enzyme from variola virus (vTopo) has proven to be useful in addressing mechanistic questions about topoisomerase function because it forms a reversible 3′-phosphotyrosyl adduct with the DNA backbone at a specific target sequence (5′-CCCTT-3′) from which DNA unwinding can proceed. We have synthesized supercoiled DNA minicircles (MCs) containing a single vTopo target site that provides highly defined substrates for exploring the effects of supercoil density on DNA binding, strand cleavage and ligation, and unwinding. We observed no topological dependence for binding of vTopo to these supercoiled MC DNAs, indicating that affinity-based targeting to supercoiled DNA regions by vTopo is unlikely. Similarly, the cleavage and religation rates of the MCs were not topologically dependent, but topoisomers with low superhelical densities were found to unwind more slowly than highly supercoiled topoisomers, suggesting that reduced torque at low superhelical densities leads to an increased number of cycles of cleavage and ligation before a successful unwinding event. The K271E charge reversal mutant has an impaired interaction with the rotating DNA segment that leads to an increase in the number of supercoils that were unwound per cleavage event. This result provides evidence that interactions of the enzyme with the rotating DNA segment can restrict the number of supercoils that are unwound. We infer that both superhelical density and transient contacts between vTopo and the rotating DNA determine the efficiency of supercoil unwinding. Such determinants are likely to be important in regulating the steady-state superhelical density of DNA domains in the cell.
The free energy stored in the form of negative DNA supercoils is essential for many genomic DNA transactions.1,2 Topological strain and the associated DNA strand separation promote the initiation of DNA replication3−5 and RNA transcription6−10 and facilitate homologous recombination.11 In addition, because the free energy trapped within an entire DNA superhelical domain must be partitioned between twist and writhe, local unwinding events caused by protein binding or DNA strand breaks can be rapidly propagated over large distances within the domain by the associated changes in writhe. Such local events that bring about changes in the three-dimensional topology of DNA can promote interactions between bound proteins that would otherwise be separated by large distances in the linear DNA sequence, and they could also be used as a signaling event for DNA damage.12,13 The essential processes described above occur in all eukaryotic cells and rely on the maintenance of a steady-state level of DNA supercoiling regulated in part by type I DNA topoisomerase enzymes.14,15 These catalysts serve to remove the excess supercoils generated during the cellular processes of DNA replication and transcription and allow the steady-state superhelical density to be optimally maintained.15
Given the global importance of negative supercoiling, it is of interest to understand if eukaryotic type IB topoisomerases sense features of DNA superhelical topology. Such a mechanism could target these enzymes to highly supercoiled DNA domains and also preclude these abundant catalysts from completely unwinding genomic DNA. Type IB topoisomerases bind nonspecifically to DNA and form a C clamp structure that allows them to interact with the phosphate backbone of both strands of the DNA duplex, but these enzymes cause little overall distortion of the linear duplex structure.2,16,17 Structural and mechanistic studies have not established whether type IB topoisomerases recognize superhelical nodes,18 bind more tightly to destabilized negatively supercoiled duplex structures,19 or detect other subtle features of supercoiled DNA in preference to relaxed DNA.
Although biased affinity for DNA of high superhelical density would be an attractive mechanism for targeting topoisomerases to where they are needed in the genome, it is not the only possible specificity mechanism. An attractive alternative would be for the enzyme to take advantage of the intrinsic structural or dynamic properties of supercoiled DNA after it has formed a covalent phosphotyrosyl linkage.15 Because the lifetime of the covalent intermediate and the swivel rate of the mobile portion of the DNA determine the number of supercoils that are removed each time the enzyme cleaves DNA,20 this kinetic intermediate is especially well-suited to regulate supercoil unwinding. In this regard, both ensemble and single-molecule measurements have established that type IB topoisomerases remove multiple supercoils each time a DNA strand is cleaved.20,21 Moreover, the number of supercoils that are removed during the lifetime of the covalent intermediate is influenced by interactions between the enzyme and the DNA and the intrinsic superhelical torque.21,22
The specificity and efficiency for unwinding highly supercoiled DNA could be obtained in several ways that are related to the unwinding mechanism. First, the lifetime of the covalent intermediate could increase with higher superhelical density, resulting in a larger time window for DNA rotation for supercoils to be unwound. Second, the efficiency of supercoil unwinding could be enhanced in highly supercoiled DNA substrates because of faster duplex rotation resulting from the high superhelical torque that is present. Finally, the enzyme–DNA interactions that restrict rotation could be weaker in the context of a highly mobile DNA segment.
The mechanistic questions mentioned above have been difficult to address using standard plasmid supercoiled substrates because most type IB topoisomerases cleave DNA nonspecifically and each cleavage site has a different lifetime for the covalent intermediate and an unknown rate of supercoil unwinding. Here we describe the synthesis and characterization of plasmid minicircles that contain only a single cleavage site for the sequence specific topoisomerase from variola virus (vTopo). In addition to allowing the study of sequence specificity in the context of supercoiled DNA, these substrates can be supercoiled to different densities, allowing mechanistic investigation of the effect of supercoiling and torque on enzyme activity. These engineered minicircles also have features that allow for the insertion of unnatural nucleotides containing base or backbone substitutions, which will foster further informative studies on topoisomerase action on supercoiled DNA.
Materials and Methods
Cloning, Expression, and Purification of vTopo
Wild-type vTopo was purified as previously described using phosphocellulose chromatography.23 The K271E mutant vTopo was made in a similar manner and purified as described previously.22
Minicircle Construction
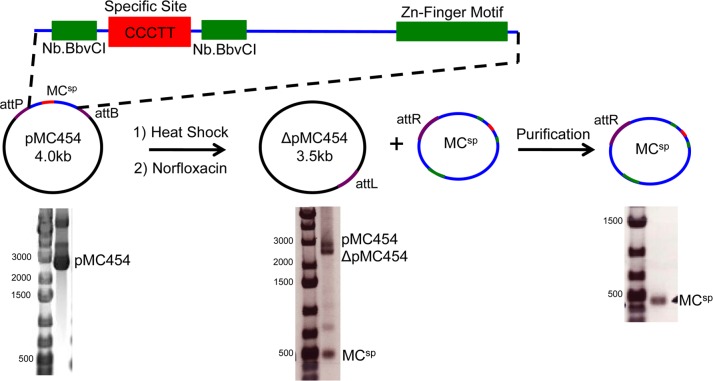
A 1000 bp sequence containing the 454 bp minicircle (MCsp) sequence and flanked by NcoI and XhoI restriction sites was generated by gene synthesis (Integrated DNA Technologies) in the pIDTSMART-Amp vector (Supporting Information). The 1000 bp sequence was cloned into the 3000 bp pattD vector using its NcoI and XhoI sites to produce pMC454 (Supporting Information).24 The size and sequence of pMC454 were confirmed via agarose gel electrophoresis and DNA sequencing (Figure 1).
Figure 1.
Minicircle (MC) preparation. MCs containing a single vTopo cleavage site (CCCTT) were prepared by inserting an engineered DNA sequence between the attP and attB λ integration sites in plasmid pattD to give pMC454 (the attP and attB sites are in the direct orientation). The engineered plasmid sequence was devoid of any other pentapyrimidine sequences that might serve as good cleavage sites but also includes engineered nicking sites (Nb. BbvC1) and a zinc finger binding motif for future mechanistic explorations (see the Discussion). For the propagation and recovery of the MC plasmids, pMC454 was transformed into bacterial strain LZ54, which is lysogenic for λ bacteriophage, and integrase expression was induced by heat shock. The addition of norfloxacin results in decatenation of the circular products of the disintegration reaction, and MCs are purified by preparative agarose gel electrophoresis. The products of the integration reaction and purified MCsp (454 bp) are shown. Gel electrophoresis was performed using a 1% agarose gel with visualization by EtBr staining.
MCsp Expansion in Bacteria, Disintegration, and Decatenation
pMC454 was transformed into LZ54 cells via heat shock, conditioning, and growth at 30 °C as previously described.24 A 10 mL overnight culture grown in LB/Amp at 30 °C was used to inoculate 350 mL of modified TB (12 g of tryptone, 48 g of yeast extract, 30 mL of glycerol, 0.1 mL of antifoam 204, 2.32 g of KH2PO4, and 12.54 g of K2HPO4 per liter). Inoculated medium was incubated at 30 °C while being shaken at 225 rpm to an OD600 of 2.0. The culture was then heat shocked to induce λ-integrase activity via the addition of 350 mL of modified TB preheated to 60 °C, and cultures were shaken at 42 °C for 30 min before norfloxacin was added to a final concentration of 30 μg/mL. The temperature was returned to 30 °C, and cells were shaken for an additional 1 h.24
Cells were harvested via centrifugation at 6000g for 15 min, resuspended in Buffer P1 (Qiagen), and frozen at −20 °C. Plasmid DNA was purified using a HiSpeed Maxiprep Kit (Qiagen) according to the manufacturer’s instructions and eluted into 1 mL of buffer EB [10 mM Tris-HCl (pH 8.5)]. To confirm that the λ-integrase disintegration reaction and decatenation were complete, 1 μL of eluent was run on a 1% agarose gel (Figure 1).
MCsp Purification
Approximately 125 μg of eluted DNA was run on a 0.7 or 1% agarose gel to purify the MCs from all other DNA. The band containing the desired MC product was excised following staining with EtBr and destaining with distilled water. MCs were gel extracted in one of two ways. In the first method, a Qiaquick Gel Extraction Kit (Qiagen) was used. In the second, the agarose gel slice was incubated for at least 24 h in 3× (w/v) gel extraction buffer (300 mM NaOAc and 1 mM EDTA) while being shaken at 37 °C to allow the DNA to diffuse out of the gel. Isolated MCsp was concentrated with a Millipore concentrator (molecular weight cutoff of 10 kDa), buffer exchanged using a polymerase chain reaction (PCR) purification kit (Qiagen), and eluted with distilled water. Concentrations were determined using a Nanodrop spectrophotometer with calculated extinction coefficients at 260 nm absorbance measurements. The purity was assessed using agarose gel electrophoreses (1% agarose) (Figure 1).
Construction of MCns
A MC where the specific
vTopo site was removed (MCns) was constructed from pMC454
using the following mutagenesis primers (altered nucleotides are shown
in bold) to form pMC454I in a QuikChange mutagenesis kit according
to the manufacturer’s instructions: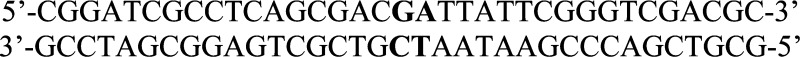 Mutations
were confirmed by DNA sequencing. MCns was obtained by
bacterial expansion of pMC454I and disintegration
as described above for MCsp. The identity of MCns was confirmed using vTopo supercoil relaxation assays in addition
to agarose gel electrophoresis to determine purity.
Mutations
were confirmed by DNA sequencing. MCns was obtained by
bacterial expansion of pMC454I and disintegration
as described above for MCsp. The identity of MCns was confirmed using vTopo supercoil relaxation assays in addition
to agarose gel electrophoresis to determine purity.
Construction of MCsp2
Plasmid pMC1752 was designed to generate a larger 1752 bp MC with a single vTopo recognition site (MCsp2). pMC1752 was constructed using a 661 bp sequence of DNA constructed via gene synthesis (IDT) (Supporting Information). This sequence contains no specific vTopo recognition sites and no other pentapyrimidine sequences. The 661 bp sequence was cloned out of the IDT vector and into phosphatase-treated pMC454 using PstI sites to create pMC1103. The identity of pMC1103 was confirmed by agarose gel electrophoresis. pMC1103 contains a NheI site between the two PstI sites, in the 661 bp sequence segment that was inserted, and this site conflicts with subsequent cloning strategies. Therefore, it was removed by mutagenesis before the second round of cloning to form pMC1752 from pMC1103 (Supporting Information). In the second round of cloning, the 661 bp sequence was cut out of the IDT vector using NheI sites (rather than the PstI sites used above) and ligated into NheI-digested pMC1103 that had been pretreated with phosphatase to create pMC1752. The identity of pMC1752 was confirmed via 1% agarose gel electrophoresis. Following confirmation, pMC1752 was expanded and MCsp2 was obtained as stated above for MCsp. The identity of MCsp2 was confirmed using vTopo activity assays in addition to agarose gel electrophoresis to determine its purity (data not shown).
Ethidium Bromide Supercoiling of Minicircles
Intercalating agents such as ethidium bromide (EtBr) can be used to introduce negative supercoils into circular DNA.25 This is accomplished by creating a transient nick in the DNA backbone in the presence of EtBr, resealing the backbone, and then removing the EtBr to restore the native twist of the DNA.26 Purified MCsp and MCns were supercoiled by reacting each MC (10 nM) with 100 nM vTopo in the presence of 1.5 μg/mL EtBr for 1 h to make MCsp* and MCns*, respectively. Following incubation, EtBr was removed via two rounds of phenol/chloroform extraction and the MCs were purified using two PCR purification kits (Qiagen). After the second column, each MC was eluted with distilled water and the concentration was determined using a Nanodrop spectrophotometer. The supercoiling and purity of each MC were ascertained using electrophoresis through a 5% native polyacrylamide gel (19:1) in the presence of TBM buffer (90 mM Tris, 90 mM boric acid, and 10 mM MgCl2) at 4 W for 6 h or 1 W overnight (Figure 2, rightmost lane). Gels were stained with SYBR Green (Life Technologies) for 30 min and imaged using a Typhoon imaging system (GE Healthcare).
Figure 2.
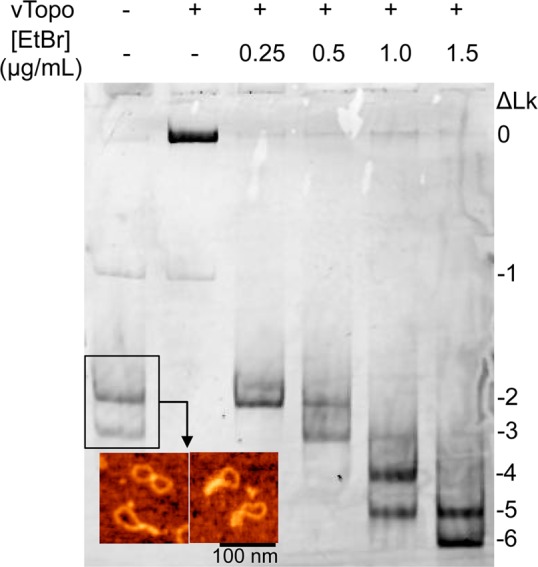
Minicircle supercoiling using ethidium bromide intercalation. Following purification from bacterial cells, MCsp has a superhelical density of −0.062. The inset shows an atomic force microscopy (AFM) image of MCsp. The images show the expected topological features of a supercoiled minicircle with a superhelical density of approximately −0.06. AFM was performed by deposition of MCs onto APS-functionalized mica in air as described previously.52 Lane 2 shows that reaction of MCsp with vTopo collapses all supercoils into a single relaxed band with lower mobility. Increasing numbers of negative supercoils were introduced using in vitro reaction mixtures containing vTopo and increasing concentrations EtBr as indicated. After extraction of vTopo and the EtBr intercalater, the DNA was isolated and subjected to polyacrylamide gel electrophoresis in the presence of MgCl2 (shown). The negative linking numbers are indicated.
Superhelical Density and Linking Number Determinations
To determine the linking number and superhelical density of MCsp, MCns, and MCsp*, electrophoresis was performed using a 6% native polyacrylamide gel (75:1) containing chloroquine concentrations between 1 and 20 μg/mL. The running buffer was 1× TBE (90 mM Tris base, 90 mM boric acid, and 2 mM EDTA) containing the same concentration of chloroquine. Electrophoresis was performed for 18 h at 3 V/cm (Figure S1 of the Supporting Information).27 A similar process was used to determine the number of supercoils present in MCsp2 except that 2% agarose gels containing chloroquine concentrations between 1 and 5 μg/mL were used. Electrophoresis was performed for 18 h at 4 V/cm in the cold room using TAE buffer (40 mM Tris base, 20 mM acetic acid, and 1 mM EDTA) containing the same concentration of chloroquine (Figure S2 of the Supporting Information). Gels were stained with SYBR Green and imaged.
Following imaging, the number of supercoils present in each MC was determined.28 First, the bands in each gel lane corresponding to the topoisomer distribution generated at a given chloroquine concentration were fit to Gaussian peak shapes using QuantityOne. Bands with identical intensities with different chloroquine concentrations were aligned to allow reliable counting of linking number differences. All intensities were then plotted against the number of negative supercoils they contained and fit to a Gaussian distribution to determine the mean number of negative supercoils and standard deviation (Table 1 and Figures S1 and S2 of the Supporting Information).28 Once the average number of supercoils was determined, the superhelical density (σ) was calculated using eq 1.
| 1 |
Table 1. Superhelical Densities, Binding Constants, and Single-Turnover Kinetic Constants for Relaxation of MCs by WT vTopo.
| no. of supercoilsa | superhelical densityb | klim′ (M–1 s–1) | KDc (nM) | |
|---|---|---|---|---|
| MCsp | –2.7 ± 1.1 | –0.062 | 1.7 × 106 | NDd |
| MCsp* | –5.4 ± 1.7 | –0.125 | NDd | NDd |
| i–1 | –1e | –0.023 | NDd | NDd |
| MCsp2 | –11.2 ± 1.4 | –0.066 | NDd | NDd |
| pool I1 | –7 ± 1e | –0.042 | NDd | NDd |
| pool I2 | –4 ± 1e | –0.024 | NDd | NDd |
| MCns | –2.7 ± 1.1 | –0.062 | 5.9 × 104 | 0.38 |
| MCns* | –5.4 ± 1.7 | –0.125 | NDd | 0.54 |
| MCnsL | 0 | 0 | NDd | 0.70 |
Determined by band counting using chloroquine gel electrophoresis (Figures S1 and S2 of the Supporting Information). Reported as the mean ± the standard deviation of a Gaussian fit.
Determined using eq 1.
Determined using eq 3. The units are nanomolar minicircle.
Not determined.
These values were calculated using the most prevalent topoisomer in the intermediate pool, which was then used to calculate the average superhelical density of the pool.
Competition Binding Experiments
A fluorescein (FAM) fluorescence-based continuous steady-state DNA vTopo cleavage assay was performed as described by Kwon et al.,29 except that the Dabsyl quencher was replaced with Iowa Black (IABK, IDT). Briefly, the 18U-FAM/18-IABK ribonucleotide substrate was annealed in buffer A [20 mM Tris (pH 9) and 200 mM NaCl] overnight using 110 μM 18U-IABK strand and 100 μM 18U-FAM strand. The ribonucleotide substrate at 1 μM and various concentrations of competitor DNAs were combined in 148 μL of buffer A. The sample was placed into a Quartz microcuvette (0.3 cm, Starna Cells) at 37 °C, and readings were taken every 15 s for 5–10 min at an excitation wavelength of 492 nm and an emission wavelength of 522 nm with an integration time of 2 s in a fluorometer (Fluoromax-3). Excitation slits were set at 0.5 nm and emission slits at 7 nm. Following the initial scan, 1.5 μL of 1 μM vTopo was added to the cuvette to give a final concentration of 10 nM and the increase in fluorescence intensity was monitored continuously after a 30 s equilibration period. Readings were taken every 15 s for at least 135 s in triplicate.
Time courses were fit to a linear regression line in Prism (GraphPad). Corrections were made for the background fluorescence at 522 nm. The fluorescence change at each time (Ft – F0) was converted to the percent reaction by normalization to the maximal fluorescence change (Fmax – F0) corresponding to complete reaction for the substrate according to eq 2 (Figure S3 of the Supporting Information):
| 2 |
The Ki for each competitor (in base pairs) was calculated using eq 3 using the known Km of 7920 nM bp and an [S] of 18000 nM bp:29
| 3 |
The competitor DNAs used in these experiments were MCns, linearized MCns (MCnsL), MCns*, a nonspecific 206mer, and a nonspecific 35mer. The construction of MCns and MCns* is described above. MCnsL was obtained by digestion with PstI-HF, and the nonspecific 206mer was constructed by PCR using pMC454I as the template (Supporting Information). The nonspecific 35mer was obtained by annealing the mutagenesis primers that were used for construction of MCns (see above). All constructs were confirmed by gel electrophoresis. To account for differences in DNA length, we express binding in base pair molarity and not molar concentration of the DNA molecules.
Single-Turnover Kinetics
MCsp or MCns (10 nM each) was reacted with 20, 40, or 80 nM vTopo in a single 80 μL reaction mixture in supercoil release buffer [50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT, 20 μg/mL BSA, 0.01% Brij-35, and 5 mM MgCl2] by adding 40 μL of 2× vTopo to 40 μL of 20 nM MC with vigorous mixing. At desired time points (Figures 4 and 5), 10 μL was removed from the reaction mixture and added to 10 μL of 2× quench buffer (1% SDS and 20% glycerol in 1× Tris-glycine). Fifteen microliters of the quenched reaction mixture was loaded onto a 2% agarose gel run in 1× TAE for 1 h at 100 V. Gels were stained for 10 min with 0.5 μg/mL EtBr and destained with distilled water before being imaged. All experiments were performed in triplicate.
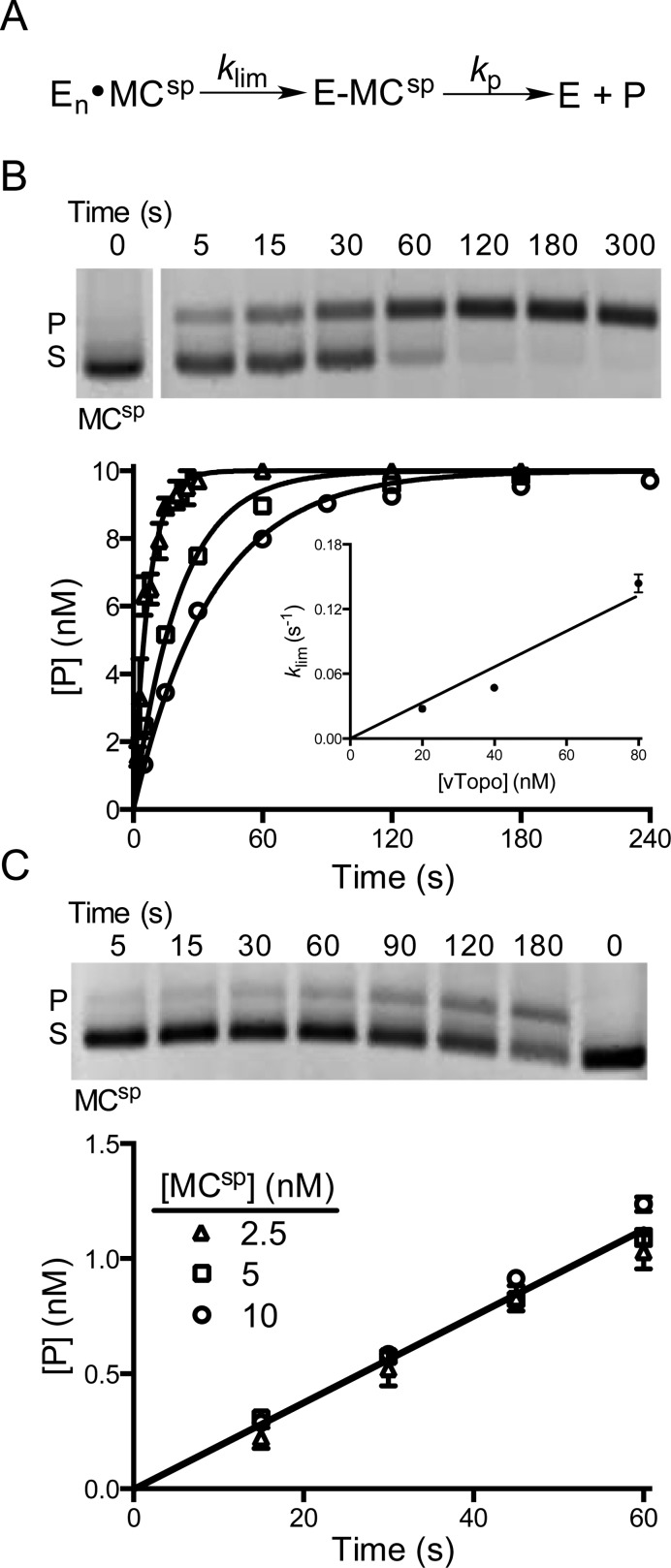
Figure 4.
Cleavage activity of vTopo with MCsp. (A) Cleavage and supercoil unwinding reactions involve a binding step, an intramolecular search mechanism, and reversible strand cleavage (klim) prior to the formation of product through supercoil unwinding (kp). (B) Single-turnover cleavage and supercoil unwinding. Supercoil unwinding of 10 nM MCsp in the presence of 20 (○), 40 (□), or 80 nM vTopo (△). The gel image shows the time course for relaxation of MCsp by 40 nM vTopo (S is the supercoiled substrate and P the relaxed product). The data were fit to a first-order rate equation for the appearance of product and confirmed using Dynafit 3. The values of klim were plotted against enzyme concentration and gave a linear response with respect to enzyme concentration over the accessible range (inset). Some error bars have been omitted for the sake of clarity but are similar to those shown. (C) Steady-state turnover under initial rate conditions. The relaxation of 5 nM MCsp in the presence of 1 nM vTopo is shown in the gel image. The rate was independent of DNA concentration in the range from 2.5 to 10 nM, indicating that the enzyme is saturated with DNA and the maximal steady-state velocity is being measured.
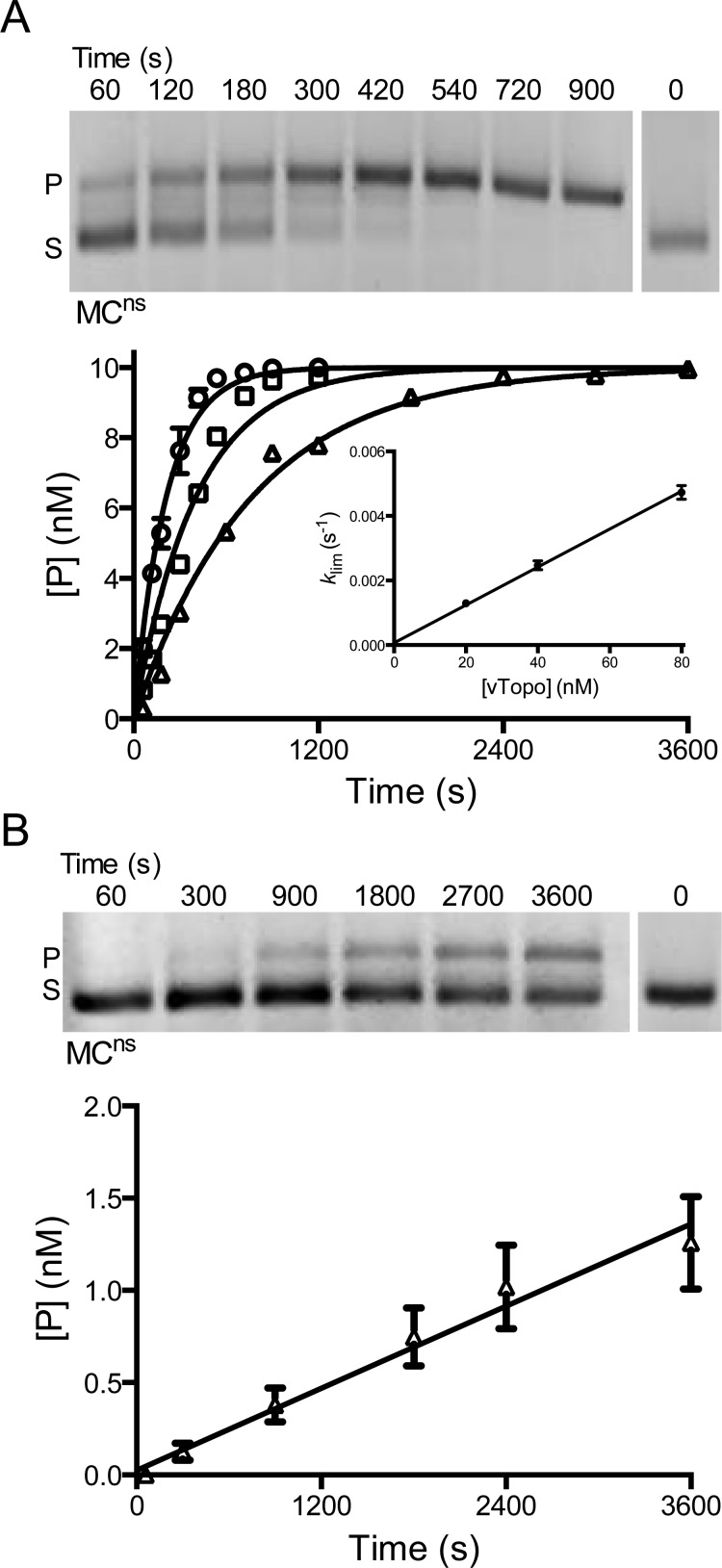
Figure 5.
Activity of vTopo with MCns, a MC that does not contain a consensus cleavage site. (A) Single-turnover cleavage and supercoil unwinding. The gel image shows the time course for relaxation of 10 nM MCns by 80 nM vTopo (S is the supercoiled substrate and P the relaxed product). MCns was also reacted with 20 (△), 40 (□), or 80 nM vTopo (○). The data were fit to a first-order rate equation. The values of klim were plotted vs enzyme concentration and were linear with respect to enzyme concentration over the accessible range (inset). Some error bars have been omitted for the sake of clarity but are similar to those shown. (B) Steady-state turnover under initial rate conditions. The relaxation of 2.5 nM MCsp in the presence of 1 nM vTopo is shown in the gel image.
Gel bands were quantified using QuantityOne (Bio-Rad), and areas were determined by nonlinear least-squares fitting to Gaussian peak shapes. Because of unequal EtBr staining between the relaxed and supercoiled bands on agarose gels, the raw fluorescence of the relaxed product bands was multiplied by a normalization factor of 1.4 obtained from a standard curve (Figure S4 of the Supporting Information). Following normalization, the concentration of product at each time ([P]t) was calculated according to eq 4, where Fp represents the normalized fluorescence of the product and Fs the fluorescence of the substrate.
| 4 |
The time courses for product formation were fit to a first-order rate equation (GraphPad Prism). The assumption of pseudo-first-order kinetics was confirmed using Dynafit 3 (Biokin).
Steady-State Kinetics
Steady-state kinetic experiments were performed like those described above for single-turnover conditions, including the quantification of gel images and normalization for the product species fluorescence. In these experiments, 2.5, 5, or 10 nM MCsp or MCns was reacted with 1 nM vTopo in a reaction volume of 80 μL. The slope of the initial linear rates was used to determine vmax.
Supercoil Unwinding Mechanism of MCsp, MCsp*, and MCsp2
All reactions for investigating the kinetics of supercoil unwinding were performed in supercoil release buffer by adding 2× vTopo (WT or K271E) to 2× MC to achieve final concentrations of 5 nM DNA and 5 nM enzyme. Reactions were conducted for the desired times, and 10 μL samples were removed and quenched by adding 10 μL of quench buffer. For MCsp and MCsp*, 30 μL of distilled water was added to each quenched time point followed by two extractions with a phenol/chloroform mixture. Samples were dried under vacuum and resuspended in 10 μL of distilled water and 2 μL of 6× loading dye before 6 μL was loaded onto a 6% native PAGE gel (75:1 acrylamide:bisacrylamide) in 1× TBE. Gels were run for 18 h in the cold room at 4 V/cm. For MCsp2, no further manipulation was necessary. An 8 μL reaction sample was loaded onto a 3% agarose gel and then run in 1× TAE buffer in the cold room for 24 h at 3 V/cm. All gels were stained for at least 1 h with SYBR Green and imaged using a Typhoon imaging system.
Bands were quantified using QuantityOne with the Gaussian peak shape fitting routine. The measured Gaussian peak areas were used to determine the molar amounts of each band at each time point in the supercoil unwinding reaction. Standard curves were used to confirm that there were no staining differences between different topological forms of DNA (Figures S5 and S6 of the Supporting Information). To account for possible inconsistencies in staining across the gel and loading of the lanes, all bands were quantified as a fraction of the total DNA loaded in a lane. The molar concentration of each species was calculated by multiplying the fractional contribution of an individual band by the total concentration of substrate. If necessary, the amount of nicked DNA initially present in the substrate was subtracted from overlapped product bands. All reactions were performed in duplicate.
Kinetic Simulations of Supercoil Unwinding
The data were fit by numerical integration methods using Dynafit 3 and employed the simplest models that preserved the information content of the data sets. The procedures and controls used for model selection and the Dynafit input script files can be found in the Supporting Information.
Results
Minicircle Design, Purification, Supercoiling, and Linking Number Determination
An efficient technology for minicircle synthesis has recently been reported that uses in vivo integrase activity on plasmids having integrase recombination sites in the direct orientation.24,30 Taking advantage of this technology, we designed a custom minicircle sequence that contained a single vTopo recognition site (5′-CCCTT-3′) and no other similar pentapyrimidine sequences (Figure 1). The sequence was incorporated into a plasmid containing the bacteriophage λ-integrase sites attB and attD in the direct orientation to allow disintegration of the MC upon transformation of strain LZ54.24 Upon successful disintegration and decatenation, two circular DNA molecules consisting of the MC and a larger circle containing the accessory elements necessary for growth and replication were obtained (Figure 1). The minicircle was successfully purified, yielding between 5 and 10 μg of MC per 350 mL of bacterial culture depending on the size of the MC.
The 454 bp minicircle containing a single recognition site (MCsp) was visualized using atomic force microscopy and consisted of topological forms with a mean of 2.7 negative supercoils [standard deviation (SD) = 1.1], corresponding to an average superhelical density of −0.057, which matches the expected superhelical density of closed circular DNA isolated from Escherichia coli cells.31 The superhelical density of MCsp was increased beyond this basal level using reversible strand nicking by vTopo in the presence of EtBr to make MCsp* (Figure 2). Highly supercoiled MCsp* was optimally generated using 1.5 μg/mL EtBr and contained an average of 5.4 negative supercoils (SD = 1.7), corresponding to an average superhelical density of −0.125 (twice that of MCsp). Although all MCs migrated as single bands during agarose gel electrophoresis, they consisted of a Gaussian distribution of topoisomers when electrophoresis was performed using 5% polyacrylamide gels in the presence of 10 mM MgCl2, or using a 6% native gel in the presence of chloroquine (Figure 2 and Figure S1 of the Supporting Information).20,22,24 The 454 bp minicircle that lacked the specific site (MCns), as well as its corresponding highly supercoiled form MCns*, followed the same topological distributions as MCsp and MCsp*.
We also prepared a larger 1752 bp minicircle with a specific site (MCsp2). Using agarose gel electrophoresis in the presence of chloroquine, we determined that MCsp2 isolated from cells had an average superhelical density of approximately −0.067 and a Gaussian topoisomer distribution with a mean of 11.2 negative supercoils (SD = 1.4) (Figure S2 of the Supporting Information).
DNA Topology Does Not Impact vTopo Nonspecific DNA Binding Affinity
In the absence of unreactive supercoiled substrates, it has not been previously possible to study the contributions of DNA topology to the nonspecific binding affinity of vTopo.32 Using a continuous fluorescence steady-state kinetic assay, we measured the inhibition of DNA cleavage by vTopo using various competitor DNAs that lacked the specific 5′-CCCTT-3′ site. In this assay,29 the action of vTopo causes a linear time-dependent increase in fluorescence intensity as DNA strand cleavage releases a FAM-labeled 7mer strand (Figure 3A,B). As the concentration of supercoiled MCns competitor DNA was increased from 0 to 10 nM, the cleavage of the reporter substrate was completely inhibited (Figure 3C). In addition to MCns, this assay was used to test the inhibitory potentials of MCnsL, MCns* (ρ = −0.125), and shorter lengths of linear nonspecific DNAs (Figure 3D). Control experiments showed that MCns was not relaxed by vTopo over the course of these competition binding measurements (Figure S7 of the Supporting Information).
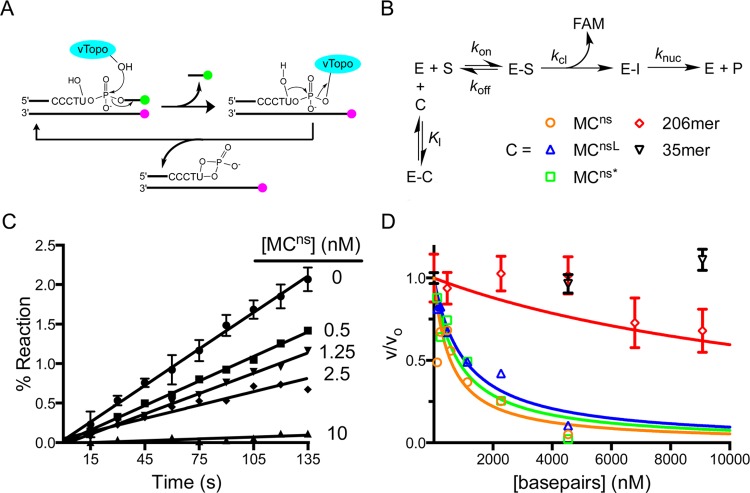
Figure 3.
Competition assay for measuring the binding affinity of vTopo for various topological isoforms of MCns and nonspecific linear DNAs of varying lengths. (A) Schematic of the continuous multiple-turnover fluorescence assay for measuring vTopo-catalyzed strand cleavage. The formation of a covalent complex between vTopo and the substrate releases a highly fluorescent 3′-FAM-labeled 7mer that is otherwise quenched in the duplex substrate by the IABK quench group on the opposite strand’s 5′-end. The presence of the uracil ribonucleotide in the cleavage sequence (CCCTU) results in the release of vTopo by nucleophilic attack by the 2′-OH group.29 Thus, multiple steady-state turnovers are possible, and initial linear rates can be accurately measured. (B) Competition binding mechanism. Because the Km for the fluorescent substrate is known, the Ki for each competitive inhibitor (C) can be determined from the concentration dependence of the decrease in fractional velocity (vi/v0) using eq 3. (C) Steady-state initial rates as a function of MCns concentration. Error bars have been removed for the sake of clarity but are similar in magnitude to those shown. The concentrations indicated are based on plasmid molecular weight. (D) Fractional velocity as a function of nucleic acid inhibitor concentration. To normalize for the different lengths of DNA used, the concentration is noted in molar base pairs. Most error bars have been omitted for the sake of clarity but are similar to those shown.
For supercoiled MCns (ρ = −0.057), we detected tight binding with a Ki of 167 ± 47 nM bp. Linearized MCnsL and highly supercoiled MCns* were found to compete similarly, with Ki values of 310 ± 50 and 240 ± 50 nM bp, respectively. Thus, there is no evidence that DNA topology significantly impacts nonspecific DNA binding by vTopo. In contrast, the length of the competitor DNA was very important. While an identical base pair concentration of a nonspecific 35mer competed poorly, an equivalent concentration of a nonspecific 206mer had an intermediate competitive capacity (Ki = 4.5 ± 1 μM bp) (Figure 3D). Because vTopo forms a C-clamp around duplex DNA,33 these findings are most likely attributed to facilitated diffusion of the enzyme on longer DNAs and the additional possibility of enhanced capture of the clamp on circular DNA, which does not possess free ends for escape. Neither of these interaction modes is possible with short oligomers.
Site Specific Cleavage Kinetics of MCsp
The presence of multiple vTopo recognition sites on plasmid DNA substrates has hindered a quantitative understanding of DNA cleavage because each site has a different rate constant for cleavage and ligation.20 Using MCsp, we now have the opportunity to measure vTopo cleavage of a single, defined specific site in supercoiled substrates (Figure 4A). In these measurements, we found that the overall rate of DNA relaxation was limited by cleavage or a nonchemical step that preceded cleavage. This step is represented by the single irreversible rate constant (klim) that leads to the covalent E–MCsp complex in Figure 4A (the assumption of irreversibility is justified by the rapid rate of supercoil unwinding that follows cleavage, as described below).
For MCsp with few superhelical turns, we expected that the substrate would be directly converted to product because ligation cannot effectively compete with supercoil unwinding when only a few supercoils are present (i.e., no partially relaxed topoisomer intermediates were expected). This expectation was confirmed in single-turnover measurements using excess enzyme, where substrate was directly relaxed to product in a single-exponential manner (Figure 4B). We observed a linear concentration dependence of klim for MCsp, corresponding to a second-order rate constant (klim′) of 1.7 × 106 M–1 s–1 (Figure 4B). The largest accessible klim was 0.15 s–1 at 80 nM vTopo. This value may be compared with the previously measured concentration-independent klim of 0.3 s–1 for pUC19, a plasmid that contains 17 vTopo specific cleavage sites.20
The concentration-dependent rate of cleavage of MCsp was intriguing given the opposite result obtained with the multiple-site plasmid. Because the binding measurements indicate exceedingly tight DNA binding (Figure 3), the concentration dependence for MCsp could not be attributed to a simple increase in the concentration of the ES complex as the enzyme concentration was increased; under the conditions of these experiments, each MC DNA was saturated with one or more enzyme molecules [En·MCsp (Figure 4A)]. Accordingly, the concentration dependence for MCsp must arise from rate-limiting movement of the vTopo C clamp along the DNA until its recognition sequence is encountered.16 As more vTopo molecules are added, the probability of finding the cleavage site increases linearly because more enzyme molecules (n) are scanning each substrate molecule for the single recognition site [En·MCsp (Figure 4A)]. For pUC19, the problem of finding a site is alleviated because a recognition site is present at an average density of approximately one in every 150 bp of DNA.20
We also measured initial rates for reaction of MCsp under steady-state conditions (Figure 4C). Using three different substrate concentrations in the range of 2.5–10 nM, we found that the rate constant was concentration-independent (0.02 s–1), providing an upper limit estimate for the Km (MCsp) of ≤0.5 nM (Figure 4C). The low Km value is consistent with the low KD for nonspecific DNA binding and supports the contention, described above, that the concentration dependence of the single-turnover reaction of MCsp arises from multiple enzyme molecules performing an intramolecular transfer step.
Cleavage Kinetics of the Nonspecific Minicircle (MCns)
It is known from DNA cleavage studies using small duplex oligomers that the specific 5′-CCCTT-3′ pentapyrimidine sequence is required for vTopo activity.32 Using MCns, we sought to determine whether a specific sequence was equally important for cleavage of a supercoiled substrate. Although the time-dependent cleavage was much slower for MCns than for MCsp, it was easily detectable because rapid supercoil unwinding efficiently traps rare cleavage events (Figure 5A). Single-turnover rates were concentration-dependent as observed for the specific substrate, but the apparent second-order rate constant (klim′ = 5.9 × 104 M–1 s–1) was reduced by 30-fold (Figure 5A, inset). As a point of comparison, we note that this apparent sequence specificity of vTopo is much lower than that of restriction enzymes (105–106).34
Under limiting enzyme steady-state conditions, we were able to measure an observed rate constant of 3.7 × 10–4 s–1 using 1 nM vTopo and 2.5 nM MCns, which is ∼50-fold less than that of MCsp under the same conditions (Figure 5B). Further steady-state measurements were not possible because higher concentrations of MCns proved to be inhibitory with respect to the reaction. This inhibition is likely due to significant substrate inhibition. Thus, under limiting enzyme conditions, the apparent specificity of vTopo increases as the S:vTopo concentration ratio increases.
Supercoil Unwinding of MCsp, MCsp*, and MCsp2
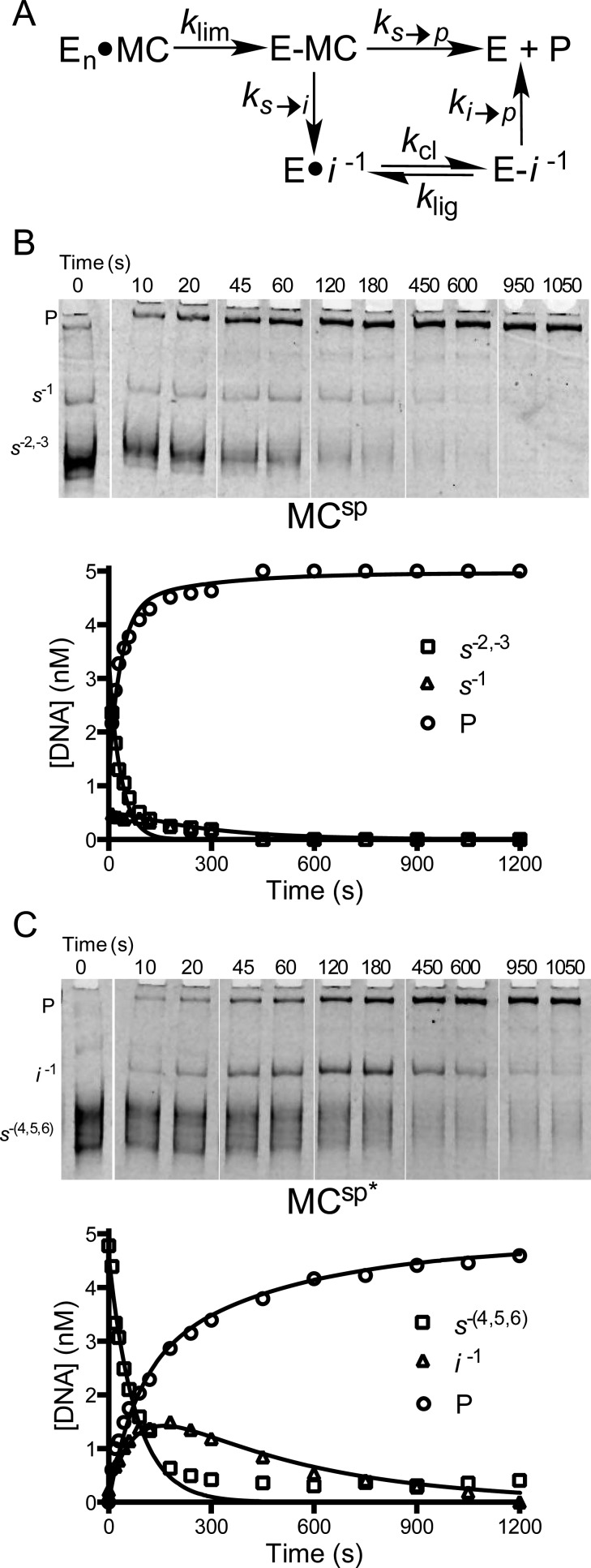
To explore whether supercoil unwinding proceeds through the generation of topoisomer intermediates, we extended the relaxation experiments to MCs that contained increasing numbers of supercoils. The expectation was that highly supercoiled substrates would provide a greater opportunity for detection of intermediates because the finite probability for strand ligation increases with the number of unwinding events needed to reach the relaxed product (Figure 6A).
Figure 6.
Mechanism of supercoil unwinding using MCsp and MCsp*. (A) The use of small minicircles with the potential to form a limited number of topoisomer intermediates facilitates resolution and allows use of a simple kinetic model for the partitioning of the enzyme–minicircle covalent complex (E–MC) between complete unwinding to form product (P) and formation of intermediates (I). Intermediates result from kinetic competition between strand ligation (ks→i) and supercoil unwinding (ks→p). The competition between these processes determines the number of supercoils that are released for each rate-limiting cleavage event. For the sake of brevity, only selected time points are shown. (B) Supercoil unwinding kinetics of MCsp that contains 2 ± 1 negative supercoils. The reaction mixture contained 5 nM MCsp and 5 nM vTopo. (C) Supercoil unwinding kinetics of highly supercoiled MCsp* that contains an average of 6 ± 2 negative supercoils. The reaction mixture contained 5 nM MCsp* and 5 nM vTopo. For both reactions, the time dependencies of the substrate (S), product (P), and topoisomer intermediate (I) concentrations were fit by numerical integration with least-squares optimization to the data using Dynafit 3 and employing the model in panel A.
Unlike previous supercoil unwinding studies using the multiple-site substrate pUC19 with excess enzyme,20,22 our studies were performed with an E:DNA concentration ratio of ∼1 and concentrations of enzyme and DNA that exceeded the estimated KD value by at least 10-fold. Therefore, the DNA is saturated with enzyme, and the rate-limiting step (klim) reflects an intramolecular event leading to site specific cleavage. Accordingly, the rate constants for rapid steps that follow cleavage (unwinding and ligation) are not revealed in these kinetic measurements. However, the measurements robustly report on partitioning of the initial substrate pool (s) between complete unwinding to form product (p) or partial unwinding with ligation to form topoisomer intermediates (i). Partitioning is quantified by the ratio ks→p/ks→i (Figure 6A), which is the key descriptor reflecting the average number of supercoils that are unwound before a ligation event traps a topoisomer intermediate. Because cleavage is rate-limiting, topoisomer intermediates that accumulate must react in subsequent cleavage–ligation events that ultimately yield relaxed product (ki→p). For topoisomer intermediates with lower superhelical torques, the assumption of irreversible cleavage followed by rapid unwinding was not appropriate. Accordingly, the model in Figure 6A allows for ligation of cleaved intermediates (klig), which can compete with the unwinding step (ki→p). Thus, multiple rounds of cleavage (kcl) and ligation are allowed before a successful unwinding event to give product. Further explanation of the simplifications and constraints used in the kinetic analyses is found in the Supporting Information.
The expectation that intermediates would be detected with more highly supercoiled MCs was borne out when the relaxation of MCsp* (mean of 5.4 negative supercoils) was compared with that of MCsp (mean of 2.7 negative supercoils) using high-resolution polyacrylamide gel electrophoresis to resolve topoisomers. Consistent with the results using low-resolution agarose gel electrophoresis described above (Figure 4), the unwinding of MCsp did not generate any observable topoisomer intermediates (Figure 6B). However, we did resolve a minor substrate topoisomer (s–1) with one negative supercoil that relaxed much more slowly than the more highly supercoiled substrate topoisomers (s–2,–3) (Figure 6B and Table 1). We attributed the slower relaxation rate of s–1 to its reduced superhelical torque, which would reduce the driving force for progressing forward to the relaxed product (ks-1→p).21,35 Our attribution of this effect to ks-1→p (and not kcl or klig) is supported by previous observations that cleavage (kcl) and ligation (klig) are similar in linear and supercoiled substrates.20 Thus, topoisomer s–1 disappears more slowly because it spends more time undergoing repetitive cycles of cleavage and ligation before a productive unwinding event occurs. Similarly, MCsp* showed a slowly relaxing intermediate (i–1) that contained a single negative supercoil. An intermediate defined as a distinct topoisomer not present in the initial substrate pool that accumulates and then proceeds to product over time (Figure 6C). The simulations of these two data sets led to the rate constants listed in Table 2 and discussed further below. Qualitatively, these data demonstrate that ligation is poorly competitive with supercoil unwinding when 5–6 supercoils are present in the starting substrate topoisomer pool, but not when 2–3 supercoils are present. Another salient point is that ligation can efficiently compete with unwinding under low-superhelical density conditions.
Table 2. Kinetic Parameters for Supercoil Unwinding of MCs by WT and K271E vTopoa.
| klim (s–1) | ks→p/ks→i | ki→p/kligc | |
|---|---|---|---|
| MCsp | 0.029 ± 0.0012 | not determined | 0.0074 ± 0.0016 |
| MCsp* | 0.014 ± 0.0006 | 1.2 ± 0.12 | 0.008 ± 0.00088 |
| MCsp2 | 0.019 ± 0.0004 | ||
| (0.0023 ± 0.000074)b | |||
| pool I1 | 2.1 ± 0.22 | 0.0084 ± 0.0013 | |
| (7.7 ± 1.5)b | (0.017 ± 0.0094)b | ||
| pool I2 | 3.8 ± 0.26 | 0.0032 ± 0.00026 | |
| (15.1 ± 5.1)b | (0.013 ± 0.017)b |
Determined using numerical simulations (Supporting Information).
Fitted parameters for K271E are shown in parentheses.
klig was set to 4 and 0.17 s–1 for WT and K271E vTopo, respectively (Supporting Information).
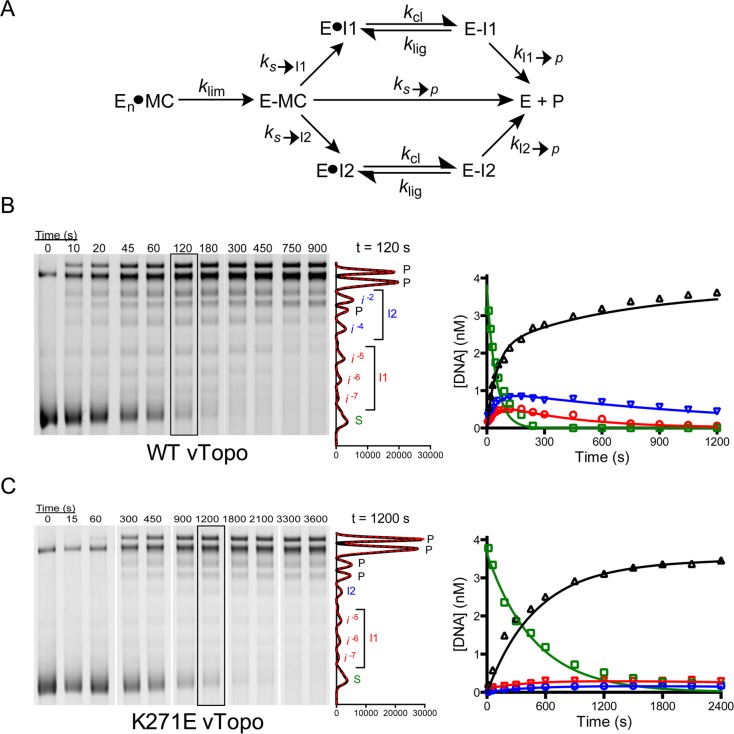
We extended the investigation to a larger minicircle (MCsp2) that contained 1752 bp and an average of 11.2 negative supercoils (Figure 7). With MCsp2, substrate topoisomers s–14 to s–9 were overlapped in a normal 3% agarose gel, but all other topoisomer species generated during relaxation were resolved. The situation of overlapped substrate bands required confirmation that relaxation of the most highly supercoiled substrate topoisomers did not generate relaxed topoisomers that overlapped with other substrate topisomers (e.g., s–14 did not relax to form s–9 to s–13). This point was established using a chloroquine gel that resolved substrate bands, which were observed to disappear at a uniform rate into resolved intermediate topoisomers and not other substrate topoisomers (Figure S8 of the Supporting Information). Thus, the entire substrate pool comprised of s–14 to s–9 can be economically treated as a single species in the kinetic analysis (Figure 7A).
Figure 7.
Processivity of supercoil unwinding by wild-type vTopo and the K271E mutant using highly supercoiled MCsp2. For the sake of clarity, only selected time points are shown in the gel figures. (A) Kinetic model for the disappearance of substrate into product and two intermediate pools (I1 and I2), followed by reversible cleavage and unwinding of intermediate pools to product. (B) Time course for unwinding using 5 nM MCsp2 and 5 nM wild-type vTopo. Substrate (S), product (P), and intermediate bands (i–2 to i–7) were resolved by gel electrophoresis, visualized by SYBR Green staining, imaged, and quantified using the Gaussian curve fitting routine in QuantityOne. A representative plot of the Gaussian fits for the time point at 120 s is shown at the right. The time courses for the appearance and disappearance of substrate (green), products (black), and intermediate pools (blue and red) were fit by numerical integration and least-squares minimization to a stepwise supercoil unwinding model shown in panel A and the Supporting Information. (C) Time course for unwinding using 5 nM MCsp2 and 5 nM K271E mutant. All steps in data acquisition and analysis are identical to those described in panel A.
While supercoil unwinding of MCsp* proceeded through a single detectable intermediate (Figure 6C), we now observed five intermediates that accumulated and then disappeared during relaxation of MCsp2 [i–2 to i–7 (Figure 7B)]. As shown in the kinetic model in Figure 7A, these intermediates were divided into two pools consisting of i–5 to i–7 (pool I1) and i–2 to i–4 (pool I2). This simplification was justified because all of the intermediates within each pool appeared and disappeared with equal rates, and pool I1 proceeded directly to product (kI1→p) without generating detectable amounts of pool I2 intermediates. The maximal level of pool I1 was lower than that of pool I2, which is attributed to the different ratios (ks→p/ks→I1 = 3.8 ± 0.26, and ks→p/ks→I2 = 2.1 ± 0.22) for the formation of the pools, as well as the different partitioning ratios for the formation of product (kI1→p/klig = 0.0084 ± 0.0013, and kI2→p/klig = 0.0032 ± 0.00026). The lower partitioning ratios for pool I2, which has a lower average superhelical density than pool I1, is consistent with the more efficient capture and ligation of the rotating DNA segment as superhelical torque is decreased. In other words, for pool I2, more rounds of cleavage and ligation occur before a successful unwinding event generates product.
In summary, pool I2 accumulates to a greater extent than pool I1 for two reasons. First, it takes more time to unwind the initial substrate supercoils to generate pool I2 intermediates (i.e., ΔLk is greater), which in turn increases the opportunity for a successful ligation event. Also, the reduced unwinding torque for intermediates with superhelical densities contained in pool I2 allows for more efficient strand ligation. The optimal fitted curves for wild-type vTopo are shown in Figure 7B, and the optimized rate constants are listed in Table 2.
Supercoil Unwinding by K271E vTopo
Previous studies indicated that cationic residues on vTopo interact with the negatively charged DNA backbone of the rotating DNA segment and impact the number of supercoils that are released per cleavage event.21,22,36 A previously characterized vTopo mutant (K271E) was known to increase the number of supercoils that were removed per cleavage event and was a desirable mutant to test using MCsp2.22 Indeed, relaxation experiments with MCsp2 showed slower cleavage and fewer intermediates with K271E than with wild-type vTopo (compare panels B and C of Figure 7). The numerical simulations indicated that the probability of completely unwinding the substrate pool by K271E was increased compared to that with wild-type vTopo (for K271E, ks→p/ks→I1 = 7.7 ± 1.5 and ks→p/ks→I2 = 15.1 ± 5.1), but unwinding of the two intermediate pools was similar (for K271E, kI1→p/klig = 0.017 ± 0.0094 and kI2→p/klig = 0.013 ± 0.017). The increased substrate partitioning ratio for K271E compared to that of WT vTopo suggests potential roles for this interaction. This mutation could result in less efficient capture of the rotating DNA segment, an increased lifetime of the covalent phosphotyrosyl intermediate (lower klig), and/or suboptimal positioning for strand ligation when high superhelical torque is present.
Discussion
DNA Topology and vTopo Binding
The superhelical density of DNA could influence the topoisomerase reaction at one or more steps along a reaction coordinate that involves DNA binding, site recognition, strand cleavage and religation, and supercoil unwinding. In this regard, human type IB topoisomerase (hTopo) has been suggested to sense DNA topology and localize to regions of increased superhelical density or node regions where double helices cross themselves.18,37 Using two nonspecific 454 bp substrates with varying levels of superhelical density, as well as the corresponding linear sequence, we found no topological dependence of nonspecific DNA binding to vTopo (Figure 3 and Table 1). Although previous studies using very high vTopo:DNA ratios detected cooperative protein interactions that resulted in DNA synapse formation,19,38 such interactions were minimized in our competitive binding studies because of the very low concentration of free enzyme and the use of a [DNA]free:[protein]free ratio much greater than one. Therefore, the dissociation constants reflect single enzyme binding events.
Binding to superhelical DNA nodes would be facilitated by the presence of a secondary DNA binding site on vTopo. For hTopo, such a site was localized to a linker region and solvent-exposed basic residues in its core subdomain III.37 Although the linker element is absent from the vTopo enzyme structure, its C-terminal catalytic domain contains the same conserved basic residue motif.16 In one study, these residues were mutated on vTopo, resulting in disruption of plectonomic supercoiling where two duplexes bind to multiple vTopos and intertwine in a right-handed helix.39 We do not expect that this weak DNA binding site is relevant under the conditions of our experiments for the reasons stated above. Instead, the weak site likely becomes important during DNA synapsis, taking advantage of an avidity effect when multiple vTopo molecules act cooperatively.19,39
Topological Effects on DNA Strand Cleavage in Single-Site MCs
Although previous studies with supercoiled pUC19 provided no evidence that DNA strand cleavage was enhanced by DNA supercoiling as compared to the rates observed with small DNA duplexes,20 we wanted to explore this explicitly using single-site MCs. Using MCsp with superhelical densities of −0.062 and −0.125 (Table 1), we observed a 3-fold decrease in the cleavage rate for the more supercoiled substrate. Although it would be tempting to conclude that an increased extent of DNA supercoiling reduced the cleavage rate, this interpretation is confounded by the additional observation that the single-turnover cleavage–relaxation reactions were dependent on enzyme concentration. This concentration dependence was unexpected because direct DNA binding affinity measurements established that all vTopo molecules were saturated with MCs (Figure 3). Similarly, the steady-state rates were saturable with low nanomolar concentrations of DNA (Figure 4C). Thus, the concentration-dependent single-turnover rates must be attributed to a concentration-dependent intramolecular rate-limiting step that precedes DNA cleavage. We surmise that in the case of excess enzyme over DNA, increasing the number of bound enzyme molecules increases the probability that a vTopo C clamp will find the cleavage site. Because of this slow intramolecular process, the DNA strand cleavage step is not directly observed with these MCs.
Rate-limiting intramolecular transfer should result in slower apparent cleavage rates for larger single-site substrates such as MCsp2 because of the larger amount of nonspecific decoy DNA. Although MCsp2 does show a cleavage rate that is 50% of that of MCsp, this is less than their size ratio in base pairs (MCsp2:MCsp ∼ 4). The lack of a direct proportionality between rate and size could result from DNA supercoiling, which serves to compact MCsp2 and reduce the volume that must be searched by intramolecular “hopping” steps. An improved understanding of these aspects of the vTopo site recognition mechanism is beyond the scope of these studies and methods.
Sequence-Dependent DNA Cleavage Using Supercoiled MCs
Historically, pox virus type IB topoisomerases have been useful in elucidating mechanistic aspects of topoisomerase reactions because of their specificity for cleavage at 5′-C/TCCTT-3′ pentapyrimidine sequences.40,41 The structural basis for their specificity has in part been suggested from the observed structural transition that occurs between the noncovalent and covalent enzyme–DNA complexes.16 In this transition, α-helix 5 docks in the major groove upstream of the cleavage site, forming direct interactions with the DNA bases of the pentapyrimidine sequence. Accordingly, DNA cleavage studies using short DNA duplexes containing sequences that diverge from the consensus sequence show markedly reduced rates in the range 10–5000-fold for single-base changes within the sequence.40,42 To construct MCns from the specific substrate MCsp, two base changes were made (CCCTT → CGATT). These changes would be expected to decrease the rate of cleavage by ∼105-fold if the known effects of the corresponding single-base changes were energetically additive.42 MCns also contains four other tetrapyrimidine sequences (also present in MCsp) that should be cleaved at least 104-fold slower on the basis of the sequence dependence of site cleavage with small duplexes. With these considerations, we anticipated that MCns would be at least 104-fold less reactive than MCsp.
We were surprised to find that single-turnover cleavage of MCns occurred with a second-order rate that was only 1/30th of that of MCsp (Figure 5A). In part, the reduced specificity with supercoiled MCs can be attributed to different rate-limiting steps for cleavage of MCsp and MCns. For MCsp, intramolecular transfer of the vTopo C clamp to the cleavage site is fully rate-limiting, while for MCns, the slow step is most certainly cleavage. However, other factors could also contribute to the apparent reduction in sequence specificity for supercoiled DNA. Most notably, the dense concentration of nonspecific cleavage sites allows multiple vTopo molecules to act nonspecifically on the same substrate. It is also possible that a subtle structural effect of DNA supercoiling could enhance nonspecific cleavage relative to sequence specific cleavage.
Superhelical Density (torque) and the Mechanism of Supercoil Unwinding
Single-molecule DNA extension measurements have become the premiere method of investigating supercoil unwinding by type 1B topoisomerases.43,44 Despite the unique insights provided by these approaches, there are merits to exploring such questions with ensemble methods using defined substrates such as those employed here. First, the large (∼25 kb) DNA substrates used in the single-molecule methods contain ∼30 sequences that are permutations of the vTopo cleavage recognition sequence, each of which has a unique cleavage and ligation rate that can affect the observed unwinding originating from that site.21 A similar situation applies to ensemble measurements with random plasmid sequences, but unlike single-molecule methods, the ensemble average is not sensitive to rare relaxation events that deviate substantially from the ensemble mean. Second, because the single-molecule measurements are made under applied force, it is more difficult to study negatively supercoiled DNA because the DNA tends to denature. Thus, the most comprehensive single-molecule measurements have been performed using positively supercoiled DNA with extrapolations to zero force to mimic the natural condition of supercoiled DNA in solution.21 In contrast, the ensemble measurements presented here, and those previously made with pUC19, use negatively supercoiled minicircles, which could behave differently. Given these considerations, we think it is unfounded to expect quantitative agreement between these experimental approaches. Nevertheless, both ensemble and single-molecule methods have converged on a processive mechanism for supercoil removal that involves removal of ∼5–19 supercoils during the lifetime of the covalent complex.20,21,45
The most informative new parameters derived from our current supercoil relaxation experiments with MCsp2 (1752 bp, 11.2 negative supercoils) are partitioning ratios ks→p/ks→I1,2 and kI1,2→p/klig (Table 2). Taken together, the ks→p/ks→I1 of 3.8 and the ks→p/ks→I2 of 2.1 indicate that once cleavage occurs there is a net 0.5 probability that a substrate topoisomer of MCsp2 will unwind all 11 negative supercoils without forming any intermediate. It is noteworthy that our ability to observe intermediates is limited to two pools with low average superhelical densities. This suggests that the increased superhelical torque present in more highly supercoiled topoisomer intermediates prevents their efficient trapping by strand ligation. Although the DNA rotation rate is driven by superhelical torque, which is expected to result in a decrease in the unwinding rate over the course of relaxation, this effect would not be directly observable in ensemble measurements because rotation is always more rapid than the rate of covalent complex formation and ligation. In contrast, single-molecule experiments can directly detect unwinding by measuring the time-dependent increase in the level of DNA extension that results from supercoil relaxation.21 Nevertheless, ensemble measurements can provide an estimate of the average duplex unwinding rate (in rotations per second) using the ligation rate constant as a clock. The kinetic basis for this estimate is that intermediate pools cannot be formed any faster than klig = 4 s–1 (i.e., klig ≥ ks→I1,2). Using this upper limit for ks→I1, and the larger product partitioning ratio ks→p/ks→I1 of 3.8, an upper limit for ks→p of ≤16 s–1 is obtained. Because 11 negative supercoils were removed in this time, the average unwinding rate is kuw ≤ (11 rotations)(16 s–1) ≤ 180 rotations s–1.
In contrast to the high unwinding rate for highly supercoiled substrate topoisomers, the low values of kI→p/klig in the range of 0.003–0.0085 for pool I1 and I2 topoisomers indicate that cleavage and religation occur ∼100–350 times for every unwinding event when the torque is small. This kinetic regime amounts to rapid equilibrium cleavage and ligation followed by slow unwinding. To the best of our knowledge, this is the first report of slow topoisomer unwinding in a regime of low superhelical torque and indicates that DNA domains with low superhelical density may persist longer in the presence of topoisomerases.
Controlled Rotation Mechanism
vTopo is known to make interactions with the rotating DNA segment downstream of the cleavage site, and such interactions have been previously implicated in supercoil unwinding.16,22 Crystal structures show that residue K271 sits within helix 10 of vTopo and could contribute to the positioning of the DNA segment downstream of the cleavage site to facilitate religation.36 We anticipated that reversing the charge on this amino acid might decrease the probability for religation, resulting in an increased ks→p/ks→i ratio. Indeed, we found that the K271E mutation increased this ratio 2- and 7-fold for pools I1 and I2, respectively, compared to that of the wild-type enzyme (Figure 7C). Even though duplex rotation is rapid with wild-type vTopo, the results with the K271E enzyme indicate that the enzyme does form interactions with the DNA during relaxation that serve to reduce the number of supercoils that are unwound per cleavage event.
Mechanistic Utility of DNA Minicircles
The frequency of use of DNA minicircles has increased in recent years as more convenient synthesis and purification schemes have become available.24,46 The majority of these uses have been in the area of gene therapy or for observing the local and long-range dynamics of short circularized DNA.47−49 In contrast, there has been limited use of minicircles as substrates for enzymes that interact with specific DNA sites.50 In this work, we showed how minicircle technology in combination with the ability to generate specific DNA sequences by gene synthesis allowed the creation of engineered supercoiled DNA substrates for the investigation of specific mechanistic questions. These engineered MCs have additional features that will allow exploration of other interesting aspects of topoisomerase action on supercoiled substrates. These features include (i) the introduction of specific zinc finger protein binding sites distal to the vTopo site for exploring the effects of the frictional drag of bound proteins on supercoil unwinding and (ii) the opportunity to introduce chemically modified oligonucleotide sequences between engineered Nb.BbvC1 nicking sites that flank the vTopo cleavage site.51 These modified sequences could include phosphorothioate or methylphosphonate backbone substitutions, or unnatural base substitutions at any site or multiple sites. Such substrates will expand the range of structure–function studies that are possible with topoisomerase enzymes.
Acknowledgments
We thank Helen Jun for purifying the K271E vTopo mutant, the Nanoscale Imaging Center at the University of Nebraska Medical Center for generating the AFM images, Dr. L. Zechiedrich at Baylor University (Waco, TX) for the pattD plasmid and LZ54 cells, and Dr. Zechiedrich’s student, D. J. Catanese, for helpful discussions regarding MC integration and purification.
Glossary
Abbreviations
- vTopo
variola topoisomerase
- EtBr
ethidium bromide
- nt
nucleotides
- Lk
linking number
- MC
minicircle.
Supporting Information Available
Supplemental sequences, eight supporting figures, and further explanation of kinetic models, including the corresponding script files. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
The work was supported by National Institutes of Health (NIH) F31 Diversity Predoctoral Fellowship GM100576 (B.G.A.), NIH T32 Training Grant 2T32GM080189 (B.G.A.), and NIH Grant RO1 GM068626 (J.T.S.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Vologodskii A. V.; Cozzarelli N. R. (1994) Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 23, 609–643. [DOI] [PubMed] [Google Scholar]
- Schoeffler A. J.; Berger J. M. (2008) DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101. [DOI] [PubMed] [Google Scholar]
- Postow L.; Crisona N. J.; Peter B. J.; Hardy C. D.; Cozzarelli N. R. (2001) Topological challenges to DNA replication: Conformations at the fork. Proc. Natl. Acad. Sci. U.S.A. 98, 8219–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P.; Krimer D. B. (2013) The benefit of DNA supercoiling during replication. Biochem. Soc. Trans. 41, 646–651. [DOI] [PubMed] [Google Scholar]
- Schvartzman J. B.; Stasiak A. (2004) A topological view of the replicon. EMBO Rep. 5, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A.; Biewen B.; Paulsen M. T.; Berg N.; Carmo de Andrade Lima L.; Prasad J.; Bedi K.; Magnuson B.; Wilson T. E.; Ljungman M. (2013) Genome-wide transcriptional effects of the anti-cancer agent camptothecin. PLoS One 8, e78190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I. F.; Yandava C. N.; Mabb A. M.; Hsiao J. S.; Huang H.-S.; Pearson B. L.; Calabrese J. M.; Starmer J.; Parker J. S.; Magnuson T.; Chamberlain S. J.; Philpot B. D.; Zylka M. J. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Bai L.; Wang M. D. (2013) Transcription Under Torsion. Science 340, 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternglanz R.; DiNardo S.; Voelkel K. A.; Nishimura Y.; Hirota Y.; Becherer K.; Zumstein L.; Wang J. C. (1981) Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc. Natl. Acad. Sci. U.S.A. 78, 2747–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Shyy S.; Wang J.; Liu L. (1988) Transcription generates positively and negatively supercoiled domains in the template. Cell 53, 433–440. [DOI] [PubMed] [Google Scholar]
- Wang J. C. (2002) Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440. [DOI] [PubMed] [Google Scholar]
- Norregaard K.; Andersson M.; Sneppen K.; Nielsen P. E.; Brown S.; Oddershede L. B. (2013) DNA supercoiling enhances cooperativity and efficiency of an epigenetic switch. Proc. Natl. Acad. Sci. U.S.A. 110, 17386–17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg J. M.; Randall G. L.; Pettitt B. M.; Sumners D. W. L.; Harris S. A.; Zechiedrich L. (2012) Bullied no more: When and how DNA shoves proteins around. Q. Rev. Biophys. 45, 257–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos S. M.; Tretter E. M.; Schmidt B. H.; Berger J. M. (2011) All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. (2001) DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Perry K.; Hwang Y.; Bushman F. D.; Van Duyne G. D. (2006) Structural basis for specificity in the poxvirus topoisomerase. Mol. Cell 23, 343–354. [DOI] [PubMed] [Google Scholar]
- Redinbo M. R.; Stewart L.; Kuhn P.; Champoux J. J.; Hol W. G. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E. L.; Osheroff N. (1990) Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 9, 4555–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S.; Bear D. G.; Sekiguchi J. (1997) Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J. 16, 6584–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers J. T.; Harris T. K.; Mildvan A. S. (1997) Vaccinia DNA Topoisomerase I: Evidence Supporting a Free Rotation Mechanism for DNA Supercoil Relaxation. Biochemistry 36, 5212–5222. [DOI] [PubMed] [Google Scholar]
- Koster D. A.; Croquette V.; Dekker C.; Shuman S.; Dekker N. H. (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674. [DOI] [PubMed] [Google Scholar]
- Jun H.; Stivers J. T. (2012) Diverse Energetic Effects of Charge Reversal Mutations of Poxvirus Topoisomerase IB. Biochemistry 51, 2940–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.; Stivers J. T. (2002) Fluorescence spectroscopy studies of vaccinia type IB DNA topoisomerase. Closing of the enzyme clamp is faster than DNA cleavage. J. Biol. Chem. 277, 345–352. [DOI] [PubMed] [Google Scholar]
- Fogg J. M.; Kolmakova N.; Rees I.; Magonov S.; Hansma H.; Perona J. J.; Zechiedrich E. L. (2006) Exploring writhe in supercoiled minicircle DNA. J. Phys.: Condens. Matter 18, S145–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M.; Vinograd J. (1976) The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell 8, 215–226. [DOI] [PubMed] [Google Scholar]
- Crawford L. V.; Waring M. J. (1967) Supercoiling of polyoma virus DNA measured by its interaction with ethidium bromide. J. Mol. Biol. 25, 23–30. [DOI] [PubMed] [Google Scholar]
- Gowetski D. B.; Kodis E. J.; Kahn J. D. (2013) Rationally designed coiled-coil DNA looping peptides control DNA topology. Nucleic Acids Res. 41, 8253–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. (1975) Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc. Natl. Acad. Sci. U.S.A. 72, 4876–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.; Nagarajan R.; Stivers J. T. (2004) Ribonuclease activity of vaccinia DNA topoisomerase IB: Kinetic and high-throughput inhibition studies using a robust continuous fluorescence assay. Biochemistry 43, 14994–15004. [DOI] [PubMed] [Google Scholar]
- Zhao N.; Fogg J. M.; Zechiedrich L.; Zu Y. (2011) Transfection of shRNA-encoding Minivector DNA of a few hundred base pairs to regulate gene expression in lymphoma cells. Gene Ther. 18, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A. V.; Lukashin A. V.; Anshelevich V. V.; Frank-Kamenetskii M. D. (1979) Fluctuations in superhelical DNA. Nucleic Acids Res. 6, 967–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S.; Prescott J. (1990) Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 265, 17826–17836. [PubMed] [Google Scholar]
- Carey J. F.; Schultz S. J.; Sisson L.; Fazzio T. G.; Champoux J. J. (2003) DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc. Natl. Acad. Sci. U.S.A. 100, 5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser D. R.; Kurpiewski M. R.; Jen-Jacobson L. (1990) The energetic basis of specificity in the EcoRI endonuclease–DNA interaction. Science 250, 776–786. [DOI] [PubMed] [Google Scholar]
- Lillian T. D.; Taranova M.; Wereszczynski J.; Andricioaei I.; Perkins N. C. (2011) A Multiscale Dynamic Model of DNA Supercoil Relaxation by Topoisomerase IB. Biophys. J. 100, 2016–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K.; Hwang Y.; Bushman F. D.; Van Duyne G. D. (2010) Insights from the structure of a smallpox virus topoisomerase-DNA transition state mimic. Structure 18, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Carey J. F.; Champoux J. J. (2009) Mutational analysis of the preferential binding of human topoisomerase I to supercoiled DNA. FEBS J. 276, 5906–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F.; Holtzer L.; Koster D. A.; Shuman S.; Dekker C.; Dekker N. H. (2005) Atomic force microscopy shows that vaccinia topoisomerase IB generates filaments on DNA in a cooperative fashion. Nucleic Acids Res. 33, 5945–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.; Yakovleva L.; Shuman S.; Mondragón A. (2010) Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure 18, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. (1998) Vaccinia virus DNA topoisomerase: A model eukaryotic type IB enzyme. Biochim. Biophys. Acta 1400, 321–337. [DOI] [PubMed] [Google Scholar]
- Baker N. M.; Rajan R.; Mondragón A. (2009) Structural studies of type I topoisomerases. Nucleic Acids Res. 37, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkah N.; Hwang Y.; Perry K.; Van Duyne G. D.; Hendrickson R.; Lefkowitz E. J.; Hannenhalli S.; Bushman F. D. (2007) Variola virus topoisomerase: DNA cleavage specificity and distribution of sites in Poxvirus genomes. Virology 365, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster D. A.; Crut A.; Shuman S.; Bjornsti M.-A.; Dekker N. H. (2010) Cellular strategies for regulating DNA supercoiling: A single-molecule perspective. Cell 142, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol Y.; Neuman K. C. (2011) Single-molecule measurements of topoisomerase activity with magnetic tweezers. Methods Mol. Biol. 778, 229–241. [DOI] [PubMed] [Google Scholar]
- Seol Y.; Zhang H.; Pommier Y.; Neuman K. C. (2012) A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 109, 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay M. A.; He C.-Y.; Chen Z.-Y. (2010) A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 28, 1287–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii A.; Frank-Kamenetskii M. D. (2013) Strong bending of the DNA double helix. Nucleic Acids Res. 41, 6785–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrhofer P., Schleef M., and Jechlinger W. (2009) Use of Minicircle Plasmids for Gene Therapy. in Gene Therapy of Cancer, pp 87–104, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Vandermeulen G.; Marie C.; Scherman D.; Préat V. (2011) New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. 19, 1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni G.; Di Martino E.; Di Mauro E.; Caserta M. (1989) Regulation of the function of eukaryotic DNA topoisomerase I: Topological conditions for inactivity. Proc. Natl. Acad. Sci. U.S.A. 86, 3080–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porecha R. H.; Stivers J. T. (2008) Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl. Acad. Sci. U.S.A. 105, 10791–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko L. S., Gall A. A., and Lyubchenko Y. L. (2012) Mica Functionalization for Imaging of DNA and Protein-DNA Complexes with Atomic Force Microscopy. In Methods in Molecular Biology, pp 295–312, Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.