Abstract
Bacterial microcompartments (MCPs) are the simplest organelles known. They function to enhance metabolic pathways by confining several related enzymes inside an all protein envelope called the shell. In this study, we investigated the factors that govern MCP assembly by performing scanning mutagenesis on the surface residues of PduA, a major shell protein of the MCP used for 1,2-propanediol degradation. Biochemical, genetic and structural analysis of 20 mutants allowed us to determine that PduA K26, N29 and R79 are crucial residues that stabilize the shell of the 1,2-propanediol MCP. In addition, we identify two PduA mutants (K37A and K55A) that impair MCP function most likely by altering the permeability of its protein shell. These are the first studies to examine the phenotypic effects of shell protein structural mutations in a microcompartment system. The findings reported here may be applicable to engineering protein containers with improved stability for biotechnology applications.
Keywords: microcompartment; carboxysome; B12; 1,2-propanediol; Salmonella
Introduction
Bacterial microcompartments (MCPs) are unique, highly sophisticated protein-based organelles used to optimize metabolic pathways that have toxic or volatile intermediates1; 2; 3; 4. MCPs are assembled from about 5,000–20,000 polypeptides of 10–20 different types, but there is no evidence for lipid or DNA components. Bioinformatics studies indicate that MCPs are present in about 17% of bacterial species and are involved in seven or more different metabolic processes of which global carbon fixation and pathogenesis in enteric bacteria are the most notable 1; 2; 5; 6; 7; 8. A typical MCP is 100–150 nm in diameter and consists of an enzyme system encapsulated within a protein shell 1; 2; 4; 9. Several lines of research indicate that compartmentalization of enzymes and substrates within MCPs is used to enhance process efficiency while sequestering toxic and/or volatile metabolic intermediates 2; 10; 11; 12. The enzyme systems of several MCPs have been well characterized biochemically and recent studies showed that resident enzymes are directed to the MCP lumen by short N-terminal targeting sequences 13; 14; 15. The overall architecture of the MCP shell is, at present, understood on the basis of high resolution crystal structures and model building 3; 16; 17; 18. The MCP shells are built from a conserved family of proteins that have bacterial microcompartment (BMC) domains as well as a conserved pentameric protein that forms the vertices 16; 17; 19. The BMC-domain proteins are hexagonal in shape (they can be hexamers or pseudohexameric trimers), and tile edge-to-edge to form extended protein sheets3; 17; 18; 20; 21. In addition, shape complementarity shared along the edges of different types of BMC-domain proteins allows them to tessellate into the functionally sophisticated protein shells required by MCPs. The flat protein sheets formed by BMC-domain proteins interact with the pentameric vertex proteins to develop the intricate 3D architecture which is icosahedral or polyhedral in shape 16.
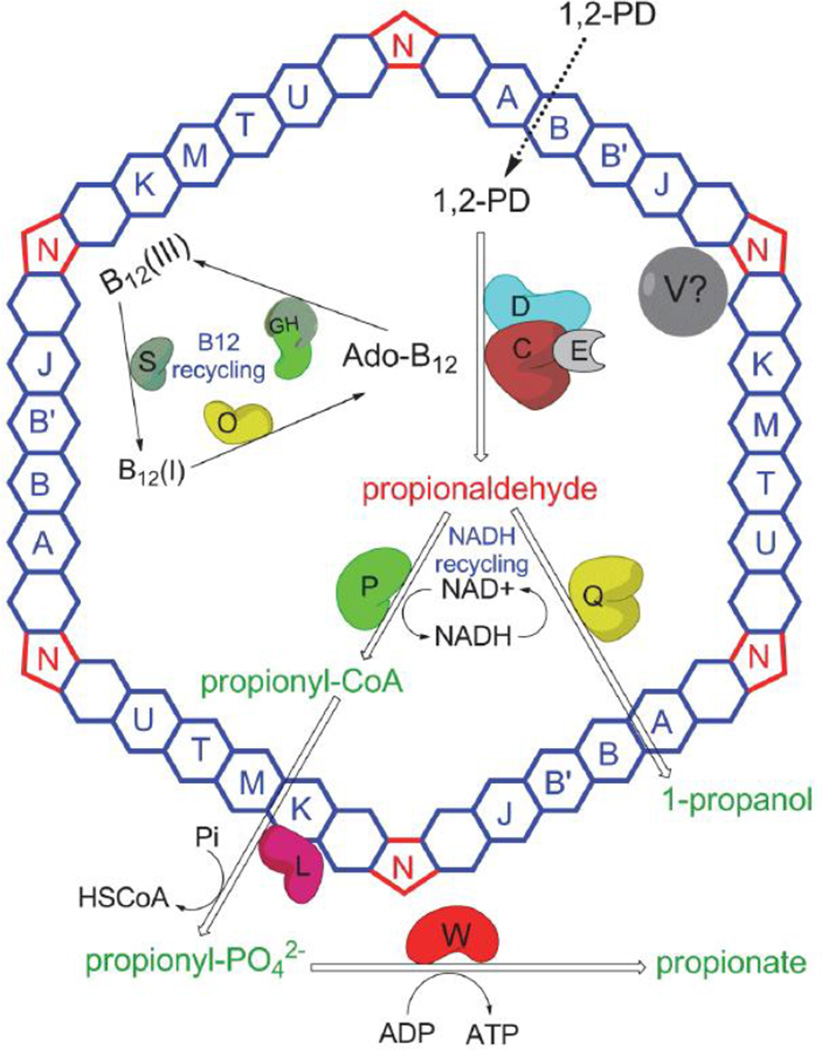
One of the best understood bacterial MCPs is the 1,2-propanediol (1,2-PD) utilization (Pdu) MCP of Salmonella enterica1; 22; 23 (Fig. 1). This MCP consists of enzymes involved in 1,2-PD utilization encapsulated inside a multi-protein shell that acts as a diffusion barrier to the cytotoxic/volatile pathway intermediate, propionaldehyde 11; 22; 23; 24; 25. The genes involved in 1,2-PD utilization (pdu) including pathway enzymes and MCP shell proteins are found in a large contiguous cluster of 23 genes 22; 26. The shell of the Pdu MCP is composed of nine different proteins: PduA, PduB, PduB’, PduJ, PduK, PduN, PduT, PduU and PduM 11; 23; 24. The PduA, PduB, PduB’, PduJ, PduK, PduT, and PduU are BMC-domain proteins proposed to amalgamate together to form the flat faces of the polyhedral MCP shell. The PduN protein is a homolog of pentameric proteins thought to form the vertices of MCPs, while PduM is an essential MCP structural protein whose exact role is uncertain 24; 27. The major shell proteins are PduA, PduB, PduB’, and PduJ, while PduK, PduN, PduT and PduU are present in low proportions23. Deletion of the pduBB’, pduJ, pduM or pduN genes severely impaired MCP formation and resulted in propionaldehyde toxicity during growth on 1,2-PD 24; 27. However, deletion of the pduK, pduT, or pduU genes did not greatly influence MCP structure or growth on 1,2-PD 24. On the other hand, a pduA deletion mutant formed larger than normal MCPs subject to an intermediate level of propionaldehyde toxicity 24.
Figure 1. Present understanding of the Pdu MCP.
The shell is made up of nine different polypeptides (PduA, PduB, PduB', PduJ, PduK, PduM, PduN, PduT, PduU). PduA, PduB, PduB’, PduJ, PduK, PduM, PduT, and PduU form the flat surfaces of the shell while PduN forms the pentameric vertices. The enzyme system for 1,2-PD metabolism is encapsulated within the shell: PduCDE (coenzyme B12-dependent diol dehydratase), PduP (propionaldehyde dehydrogenase), PduQ (1-propanol dehydrogenase) and PduGH, PduS and PduO (B12 recycling enzymes). The proposed function of the Pdu MCP is to sequester propionaldehyde and channel it to downstream enzymes thereby preventing toxicity and DNA damage. PduL (phosphotransacylase) and PduW (propionate kinase) were not observed in purified MCPs and are positioned outside the MCP shell. The exact function of PduV is not understood and its position is also not certain.
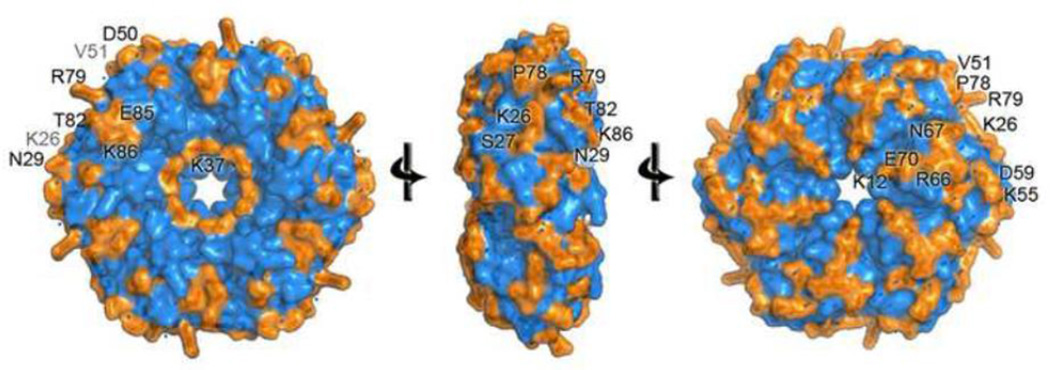
In this study, we investigated the assembly of the Pdu MCP by mutational analysis of the exposed residues in the hexameric PduA protein. All residues that are greater than 25% solvent accessible in the PduA hexamer were individually changed to alanine using site-directed chromosomal mutations (Table 1). These residues were not limited to residues involved in edge interactions between hexagonal tiles, but also included those exposed on the hexagonal faces (Fig. 2). The rationale behind selecting solvent accessible residues is that these residues are likely to participate in inter-protein interactions needed for the assembly and organization of the Pdu MCP. PduA was chosen since it is the only major Pdu shell protein (it is estimated to comprise 16% of the MCP shell) whose crystal structure is available 28. These are the first studies to investigate the key shell protein residues that drive the formation of a bacterial MCP. This information might be helpful for designing more stable MCPs for biotechnology applications.
Table 1.
Solvent accessible surface area of residues in the PduA protein
| Residue | Surface area of free amino acid (Å2) |
Accessible surface area in hexamer (Å2) |
% Surface Area Accessible in hexamer |
|
|---|---|---|---|---|
| 1 | K12 | 321.4 | 90.9 | 28 |
| 2 | K26 | 321.4 | 112.2 | 35 |
| 3 | S27 | 232.4 | 64.9 | 28 |
| 4 | N29 | 270.2 | 135.8 | 50 |
| 5 | M31 | 302.6 | 109.8 | 36 |
| 6 | V33 | 265.7 | 71.7 | 27 |
| 7 | K37 | 321.4 | 91.6 | 28 |
| 8 | D50 | 260.2 | 82.4 | 32 |
| 9 | V51 | 268.6 | 73.4 | 27 |
| 10 | K55 | 321.4 | 96 | 30 |
| 11 | D59 | 269.1 | 80.7 | 30 |
| 12 | R66 | 359.6 | 145.1 | 40 |
| 13 | N67 | 273.7 | 140 | 51 |
| 14 | E70 | 293 | 94.8 | 32 |
| 15 | V74 | 262.1 | 67.2 | 26 |
| 16 | P78 | 254.3 | 81.9 | 32 |
| 17 | R79 | 361.1 | 172.1 | 48 |
| 18 | T82 | 258.4 | 101.8 | 39 |
| 18 | E85 | 279 | 83.2 | 30 |
| 20 | K86 | 321.4 | 177.7 | 55 |
Figure 2. Mutagenesis sites.
Each residue of PduA with a side chain more than 25% exposed to solvent was changed to alanine by mutagenesis. Changed residues are colored orange and labeled.
Results
Examination of the previously reported crystal structure of wild-type PduA protein with Swiss PDB viewer (http://www.biomedcentral.com/1471-2105/13/173) showed that twenty residues in the PduA hexamer are more than 25% solvent exposed (Table 1). Each of these residues was changed to alanine individually via chromosomal mutations. The mutations were verified by DNA sequencing and then we screened for mutations that impaired MCP function by growing strains on 1,2-PD at limiting and saturating B12 concentrations. These conditions were chosen because prior studies showed that mutational impairment of shell formation results in fast growth on 1,2-PD at limiting B12 and propionaldehyde toxicity at saturating B12 concentrations11; 24. The fast growth phenotype is understood to result from increased permeability or abrogation of the MCP shell leading to a higher availability of enzyme substrates and cofactors to the 1,2-PD degradative enzymes encased within the MCP 24 with a presumed cost of increased DNA damage 25. On the other hand, at saturating B12 propionaldehyde rapidly leaks from defective MCPs and growth arrest/inhibition due to toxicity ensues 11; 24; 25. Out of the 20 mutants examined in this study, five (PduA-K26A, PduA-N29A, PduA-K37A, PduA-K55A and PduAR79A) demonstrated a change in phenotype during growth on 1,2-PD indicative of an MCP defect (as further discussed below) while the rest behaved similarly to the wild-type.
Residue K26 of PduA is vital for MCP assembly
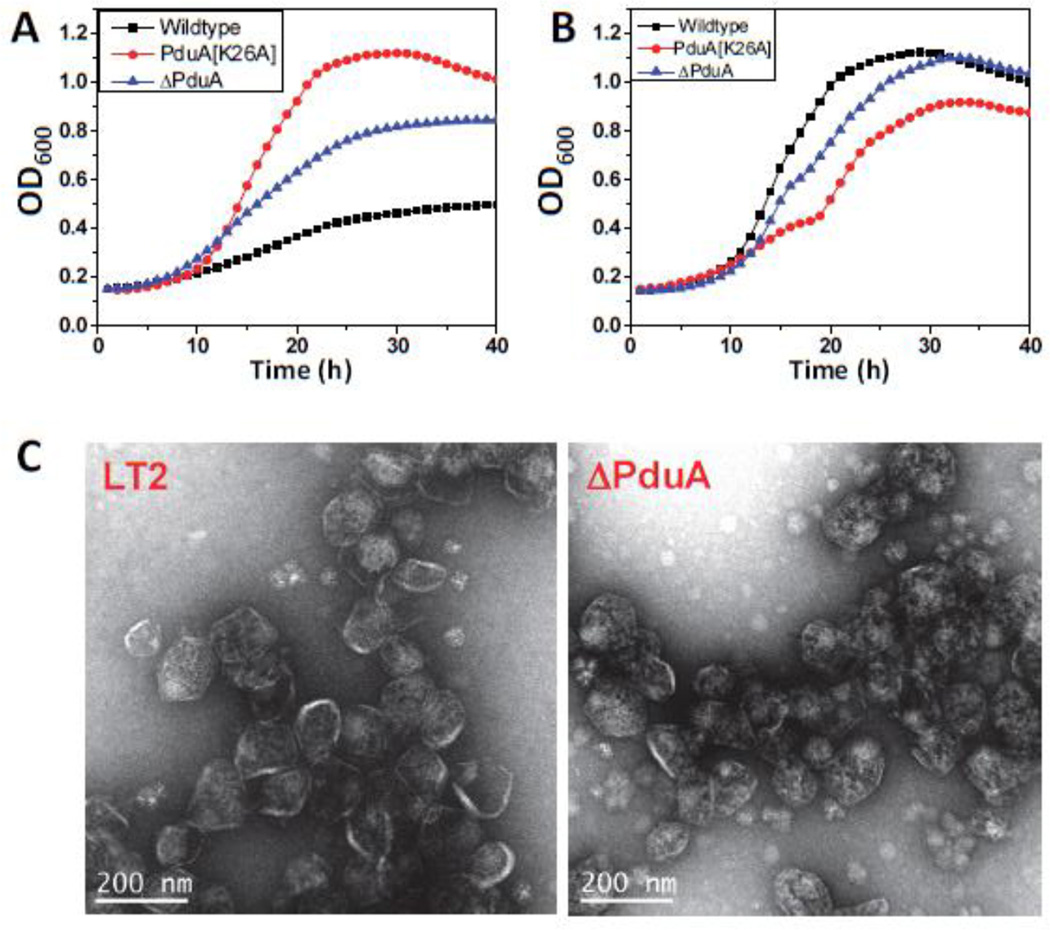
As controls, growth tests were performed on wild-type LT2 and a pduA deletion mutant. As expected, the ΔpduA mutant showed a phenotype indicative of MCPs with a broken or more porous shell, namely faster growth than wild-type at limiting vitamin B12 concentrations, and propionaldehyde toxicity at saturating B12 concentrations (Fig. 3). We note, however, that the period of growth inhibition (due to propionaldehyde toxicity) observed for the ΔpduA mutant was shorter than previously observed 11. In prior studies, a ΔpduA mutant underwent growth arrest for about 12 h 24. Here, growth of the ΔpduA mutant was only slightly inhibited between the 14 and 16 hour time points. Controls showed that growth arrest was reduced in this study due to the adsorption of propionaldehyde by plastic culture plates (Sinha and Bobik, unpublished results). In previous studies glass culture flasks were used 11; 24.
Figure 3. PduA-K26A phenotype.
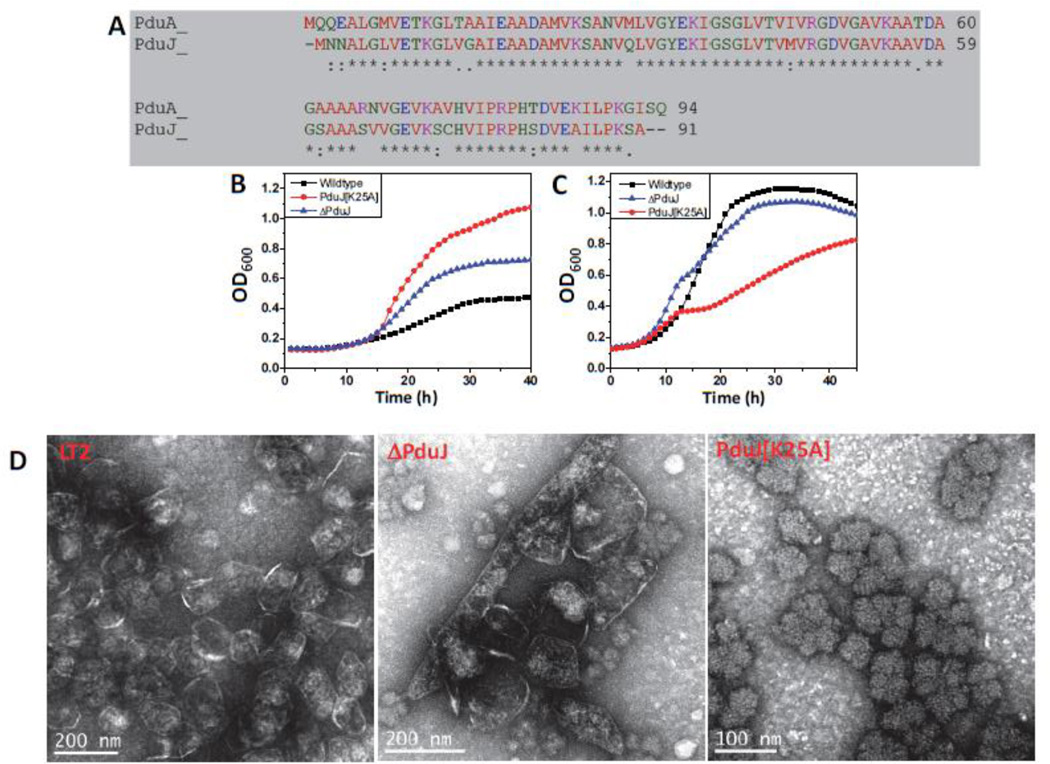
Respective strains were grown on minimal 1,2-PD medium with (A) limiting (20 nM) or (B) saturating (100 nM) CN-B12, respectively. The growth curves were repeated at least three times in duplicate and representative curves are shown. (C) Electron Micrographs of purified wild-type and ΔPduA MCPs. PduA-K26A strain did not yield any MCPs.
For the PduA-K26A strain, the growth rate on 1,2-PD at limiting B12 was much faster than wild-type, and, somewhat unexpectedly, substantially faster than that of the ΔpduA mutant as well (Fig. 3). This indicated that the PduA-K26A MCPs are more highly permeable than the ΔPduA MCPs, which are partially functional. Moreover, at higher B12 concentrations, the period of growth inhibition for the PduA-K26A mutant was longer than that of the ΔpduA mutant (it was observed from about hour 12 to hour 20 of the growth curve) suggesting a higher level of propionaldehyde escape into the cytoplasm (Fig. 3). Given the location of K26 in the crystal structure of PduA, at first we reasoned that changing K26 to A26 impaired shell formation due to the loss of edge contacts between adjacent PduA hexamers. This would lead to the formation of aberrant MCPs, which could account for the observed phenotypes. However, at this point it was unclear to us why the K26A point mutation in PduA produced a substantially more severe phenotype that did the PduA deletion mutant.
Next, we used protocols developed in our laboratory 34 to purify Pdu MCPs from wild-type Salmonella and the ΔpduA and PduA-K26A mutant strains and measure their DDH activity. The DDH activity of the purified wild-type Pdu MCP in vitro was 28.3±.3 µmol min−1mg−1 while that of the ΔPduA MCP was 41.1±2 µmol min−1mg−1 (Table 4). The higher specific activity of the ΔPduA MCPs suggest easy access of substrates and cofactors to the diol dehydratase, which is expected for MCPs whose shells have altered permeability properties. We also attempted to purify MCPs from the PduA-K26A mutant, but several attempts failed. Even increasing the culture volumes to four times that used for the wild-type and reducing the temperature of purification steps resulted in no detectable amount of the PduA-K26A MCPs. These results indicated that the PduA-K26A protein destabilized or disrupted the entire MCP assembly. This is in contrast to the pduA deletion mutant, where the slightly enlarged MCPs that are formed can be readily purified and retain partial functionality based on phenotypic tests 24 (Fig 3C). Hence, results suggested that PduA-K26A might be disrupting the Pdu MCP through aberrant interactions with other MCP components.
Table 4.
DDH activity and yield of Mutant MCPs*
| PduA Mutant | Specific Activity (µmol min−1mg−1) |
Yield (% wt) | |
|---|---|---|---|
| 1 | Wild-type | 28.3±0.3 | 100±5 |
| 2 | PduA-K26A | ND | ND |
| 3 | PduA[N29A] | 39.3±0.9 | 75±4 |
| 4 | PduA[K37A] | 27.8±0.6 | 92±3 |
| 5 | PduA[K55A] | 30.4±0.8 | 97±7 |
| 6 | PduA[R79A] | 40.1±2 | 70±3 |
| 7 | PduJ[K25A] | 37.9±1.3 | 66±3 |
| 9 | ΔPduA | 41.1±2 | 72±1 |
| 10 | ΔPduJ | 36.6±1 | 83±6 |
The DDH activities are the mean of three independent observations.
Dominance test
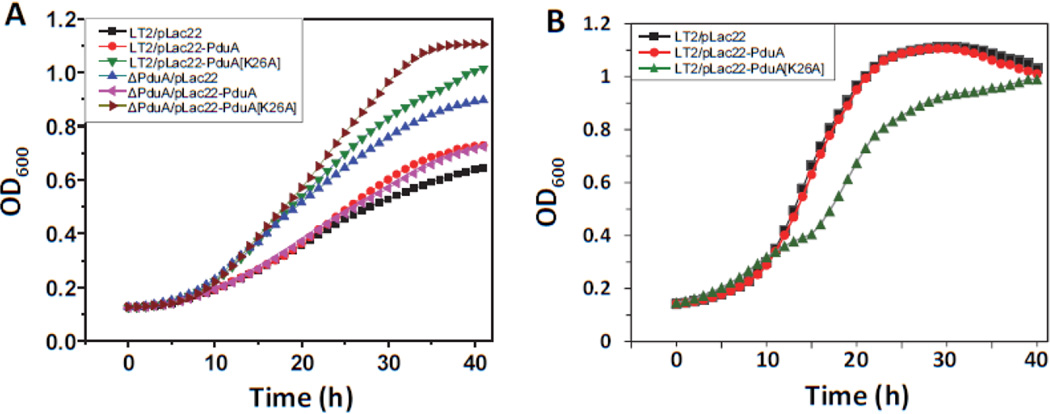
If PduA-K26A disrupted the Pdu MCP due to aberrant interactions with other MCP components, the phenotypes described above should be dominant or partially dominant. To test this, we cloned PduA-K26A into vector pLac22 and examined the effects of PduA-K26A production on 1,2-PD catabolism. As controls, we used plasmid pLac22-pduA (produces native PduA) and pLac22 vector alone. At limiting B12, the production of native PduA protein restored the wild-type phenotype to a pduA deletion, but had little effect on the wild-type strain showing that the pduA deletion is recessive (Fig 4A). In contrast, production of PduA-K26A aggravated the phenotype of the pduA deletion and also increased the growth rate of the wild-type indicating impaired MCP function (Fig. 4A). In addition, production of PduA-K26A during growth on 1,2-PD at saturating B12 levels, resulted in growth arrest from about 10–16 hours whereas production of native PduA had little effect (Fig. 4B). These results show that the phenotypes described above for the PduA-K26A mutant are dominant. This suggested that aberrant interactions between PduA-K26A and other MCP components blocked MCP assembly, which would explain why the K26A phenotype is more severe than that of the pduA deletion mutant. Nevertheless, at this stage, the molecular details underlying the PduA-K26A phenotypes were unclear.
Figure 4. PduA-K26A dominance test.
Production of PduA-K26A from a plasmid in an otherwise wild-type strain resulted in phenotypes indicative of a broken MCP: fast growth at limiting B12 (A) and temporary growth inhibition at saturating B12 (B). This indicated that the PduA-K26A phenotype was dominant. Respective strains were grown on minimal 1,2-PD medium with (A) limiting (20 nM) or (B) saturating (100 nM) CN-B12, respectively. The growth curves were repeated at least three times in duplicate and representative curves are shown.
Crystallographic structure of the PduA-K26A mutant
To obtain atomic level insights into the effects of changing K26 of PduA to A26, we determined the structures for 2 crystal forms of H6-PduA-K26A. Coordinates of the mutant protein hexamer assemblies from crystal forms 1 and 2 were superimposed on the wild-type PduA hexamer structure with RMSDs of 1.0 Å and 0.9–1.2 Å, respectively. Hence, the H6-PduA-K26A protein had no major conformational differences compared to wild-type PduA. Interestingly, unlike the wild-type PduA protein, the hexamers of the K26A mutant did not form extended protein sheets in crystals. Instead, for both crystal forms, some of the hexamer edges were exposed to solvent while others made incidental (non-biologically relevant) crystal contacts. The apparent disruption in the K26A mutant of natural edge-to-edge interactions offers a possible explanation for the observed phenotypes, as further described in the discussion.
A PduJ-K25A mutant is impaired for Pdu MCP assembly and function
PduJ is a major component of the Pdu MCP and is estimated to constitute 22% of the shell 23. PduA and PduJ share 83% amino acid sequence identity (Fig. 5A). Residue K25 of PduJ, corresponds to the K26 edge lysine of PduA. To test the role of PduJ K25 in MCP function and assembly, we generated a PduJ-K25A by chromosomal mutation and compared its phenotypes to wild-type Salmonella and a pduJ deletion mutant. At limiting B12 concentrations, a ΔpduJ mutant showed a higher growth rate compared to the wild-type as expected 24 (Fig. 5B). Expectations were again met as the PduJ-K25A mutant showed a higher growth rate than both the wild-type and the ΔpduJ strain, a situation similar to that of the PduA-K26A mutant. Furthermore, at saturating B12 concentrations, the ΔpduJ strain showed only a slight growth arrest due to propionaldehyde toxicity during the 1,2-PD catabolism, while PduJ-K25A mutant showed extended period of growth arrest (Fig. 5C). Thus, as was seen for the corresponding pduA K26A mutant, the phenotype of the PduJ-K25A mutant was more severe than that of pduJ deletion mutant.
Figure 5. PduJ-K25A phenotype.
(A) Salmonella enterica PduA and PduJ sequence alignment. (B and C) Respective strains were grown on minimal 1,2-PD medium with (B) limiting (20 nM) or (C) saturating (100 nM) CN-B12, respectively. The growth curves were repeated at least three times in duplicate and representative curves are shown. (D) Electron Micrographs of purified wild type and ΔPduA MCPs. The PduA-K26A strain did not yield any MCPs.
We also purified MCPs from the ΔpduJ and the PduJ-K25A mutants. For ΔpduJ, electron microscopy showed elongated MCPs that were surrounded by a protein shell (as observed previously in thin sections of whole cells24). In contrast, the PduJ-K25A strain produced amorphous aggregates with no clear indication of a shell (Fig. 5D). Furthermore, analyses of purified MCPs showed that the DDH activities of the wild-type, ΔpduJ and PduJ-K25A mutants were 28.3±0.3, 36.6±1 and 37.9±1.3 µmol min−1mg−1, respectively. These results supported the growth tests that indicated PduJ-K25A impairs the assembly of Pdu MCP. More importantly, in conjunction with studies on the PduA-K26A mutant, these findings support a conserved role for edge lysines in MCP assembly for different shell protein paralogs.
PduA-N29A and PduA-R79A mutants have impaired shell function
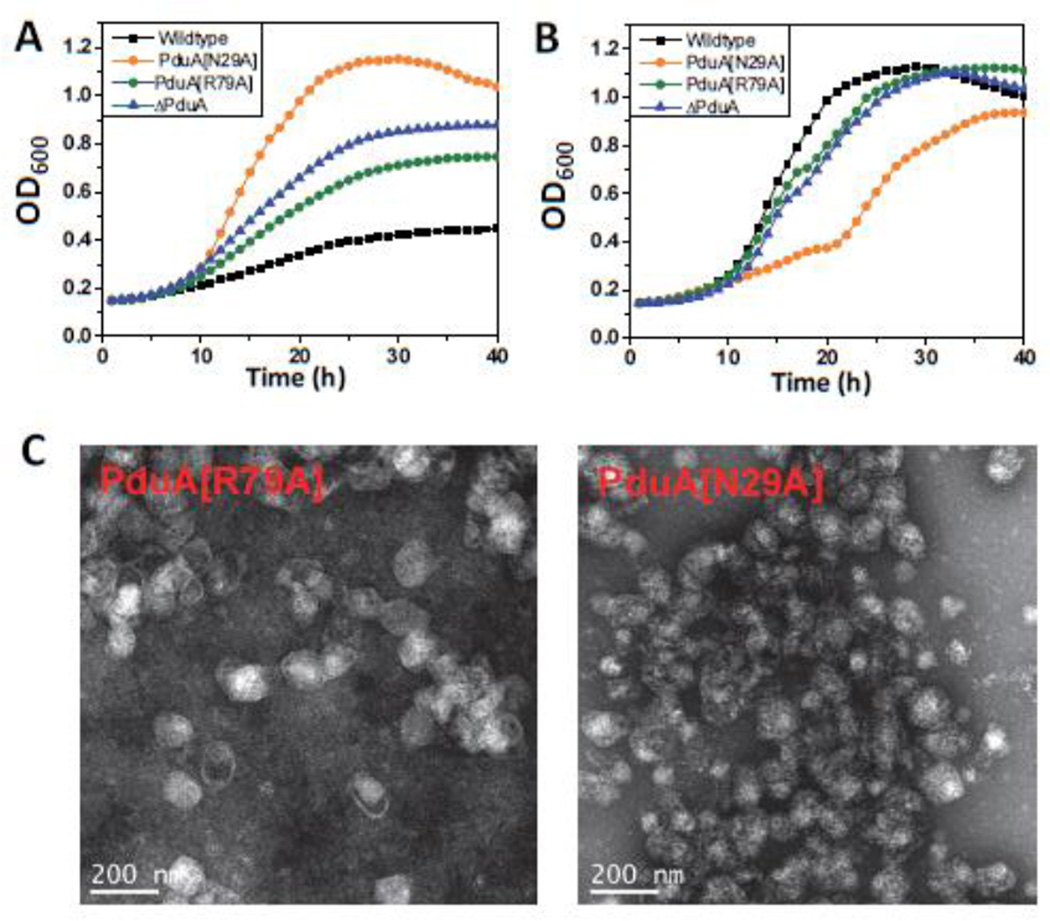
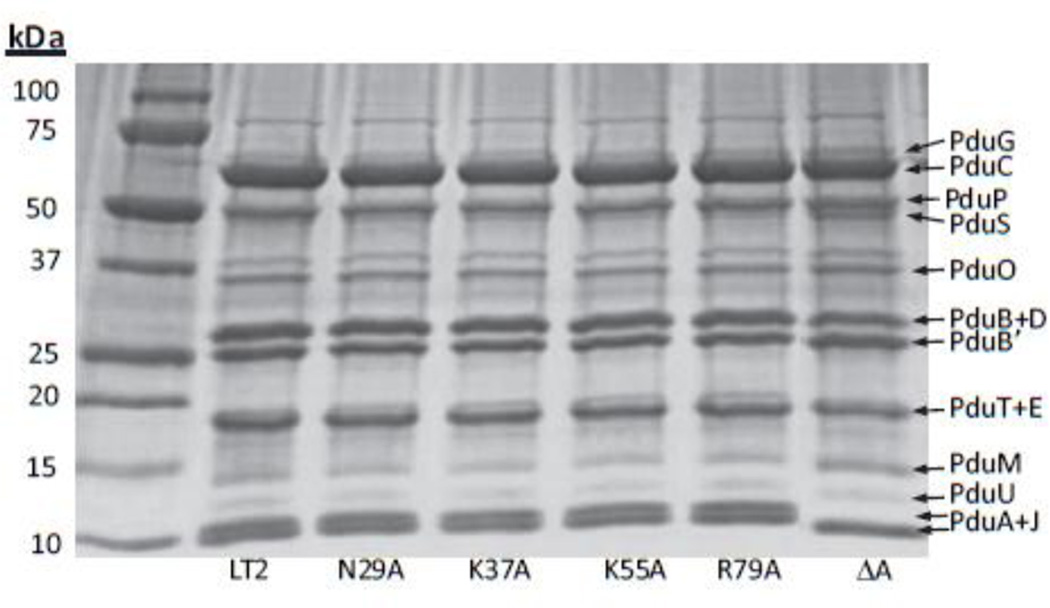
Growth tests showed that mutations in two other residues at the hexamer perimeter, PduA-N29A and PduA-R79A, also produced phenotypes indicative of impaired shell function. Both mutants grew faster on 1,2-PD than the wild-type at limiting B12 concentrations (Fig. 6A). At saturating B12 concentrations, both were subject to propionaldehyde toxicity, with PduA-N29A showing a more severe phenotype (Fig. 6B). Upon purification, the yields of PduA-N29A and PduA-R79A MCPs were ~75% and ~70% of the wild-type MCPs, respectively, suggesting that these mutant MCPs are unstable during purification (Table 4). Electron microscopy of the purified MCPs showed that PduA-R79A mutant MCPs are similar in appearance to wild-type, while the shells of PduA-N29A mutant MCPs have a more wrinkled appearance (Fig. 6C). We also measured the DDH activities of purified PduA-N29A and PduA-R79A MCPs. The activities were 39.3±0.9 and 40.1±2 µmol min−1mg−1, respectively, which are much higher compared to the wild-type Pdu MCPs (specific activity 28.3±0.3 µmol min−1mg−1). The SDS-PAGE of MCPs purified from the respective strains showed that PduA-R79A and PduA-N29A were incorporated into the MCPs at levels similar to wild-type (Fig. 7). This indicated that these mutants had no dramatic effects on protein folding. However, even though the PduA-R79A and PduA-N29A were stably incorporated into the MCP, the phenotypic tests described above clearly showed that these MCPs were nonfunctional, highly porous and unable to regulate the inward and outward movement of metabolites. Cumulatively, these results highlight the key roles of the R79 and N29 of PduA in forming well-packed protein arrangements in the shell of the Pdu MCP. The atomic basis for these critical edge interactions are detailed subsequently.
Figure 6. PduA-N29A and PduA-R79A phenotype.
Respective strains were grown on minimal 1,2-PD medium with (A) limiting (20 nM) or (B) saturating (100 nM) CN-B12, respectively. The growth curves were repeated at least three times in duplicate and representative curves are shown. (C) Electron Micrographs of purified PduA-N29A and PduA-R79A MCPs.
Figure 7. SDS-PAGE gel of purified MCPs.
Ten µg Pdu MCPs purified from wild-type Salmonella or respective mutants were loaded onto a 10–20% SDS-PAGE gel. The respective lanes of the mutants are indicated on the gel.
PduA-K37A and PduA-K55A mutants grow slowly on 1,2-PD
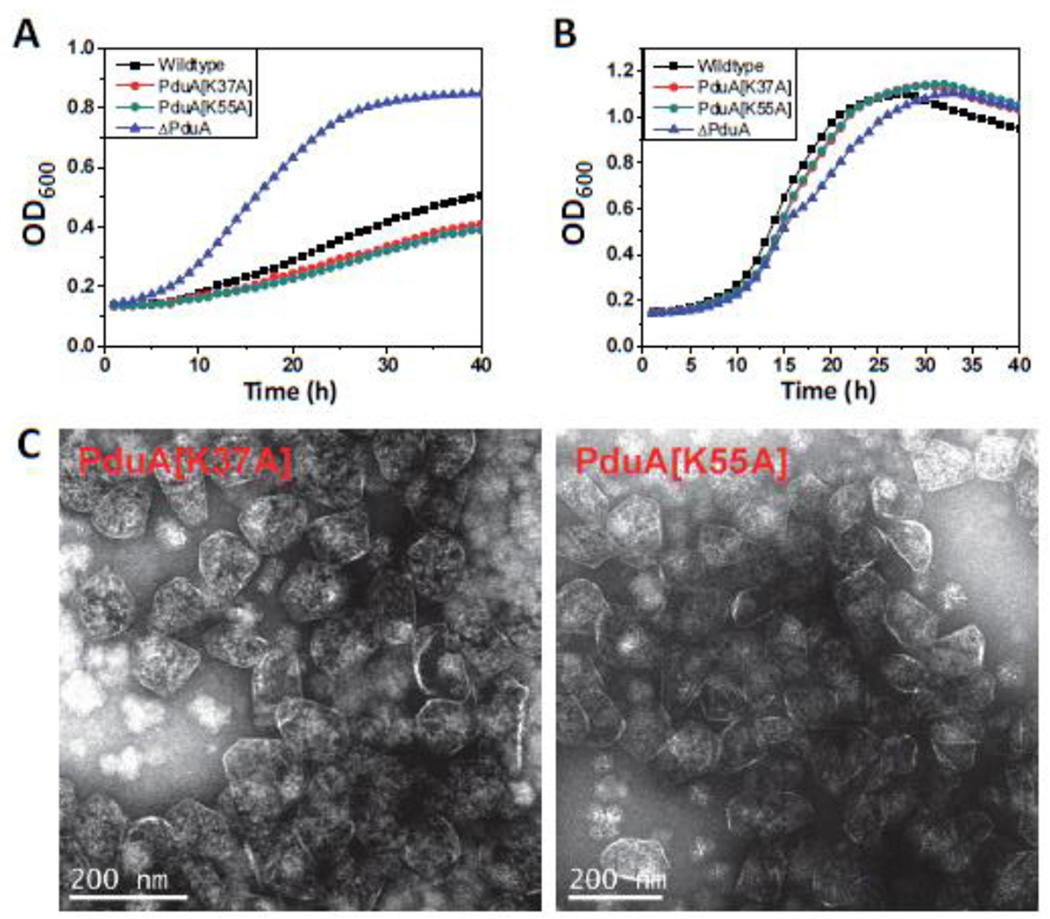
Two of the chromosomal mutants constructed in this study (PduA-K37A and PduA-K55A) showed slow growth on 1,2-PD compared to the wild-type at limiting B12 concentrations (Fig. 8). However, at saturating B12 concentrations no noticeable difference was observed. For both the mutants, the DDH activity of the purified mutant MCPs (28.3±0.3 µmol min−1mg−1 and 27.8±0.6 µmol min−1mg−1 for PduA-K37A and PduA-K55A, respectively) was close to the wild-type. There was no change in the yield of the mutant MCPs nor in the SDS-PAGE pattern of the purified MCPs (Fig. 7). The slow growth phenotypes were modest but reproducible. The doubling times calculated from semi-log plots where doubling time = 0.693/(2.303) (slope of the linear region of the plot) for wild type, PduA-K37A, PduA-K55A and ΔpduA are 18.2±1.6h, 23.2±1.2h, 24.5±41.7h and 10.9±0.3h, respectively. The results shown are the mean of four independent experiments. This indicated that the mutants grew slower compared to the wild type. We also fitted the growth curve to , where y (OD600) is a function of the time t. t50 is the time required for OD600 to reach halfway between starting and maximal values. The Slope describes the steepness of the curve, with a higher value denoting a shallow curve. For data fitting, the portion of the curves where the OD600 dropped after reaching maxima was ignored. The wild-type has a t50 and Slope of 20.2±0.7h and 8.4±0.3, respectively. Both the mutants showed higher t50 (23.5±1.1h and 24±1.2 h for PduA-K37A and PduA-K55A respectively) and Slope (9.3±0.1 and 9.4±0.1 for PduA-K37A and PduA-K55A, respectively) values compared to the wild-type further supporting a significantly slower growth rate at limiting B12 concentrations. The ΔpduA strain has the lowest t50 and Slope indicating fastest growth rate. The t50 and Slope values are listed in Table 5. A reasonable explanation for these mutants is that they impair the activity of 1,2-PD degradative enzymes found within the MCP by restricting the flow of 1,2-PD or required cofactors into the MCP.
Figure 8. PduA-K37A and PduA-K55A phenotype.
Respective strains were grown on minimal 1,2-PD medium with (A) limiting (20 nM) or (B) saturating (100 nM) CN-B12 respectively. The growth curves were repeated at least three times in duplicate and representative curves are shown. (C) Electron Micrographs of purified PduA-K37A and PduA-K55A MCPs.
Table 5.
t50 and Slopes for PduA[K37A] and PduA[K55A] growth
| Strain | t50 | Slope | Doubling Time (td) |
|---|---|---|---|
| Wild-type | 20.2±0.7 | 8.4±0.3 | 18.2±1.6 |
| PduA[K37A] | 23.5±1.1 | 9.3±0.1 | 23.2±1.2 |
| PduA[K55A] | 24±1.2 | 9.4±0.2 | 24.5±41.7 |
| ΔPduA | 15.4±1.5 | 4.8±0.2 | 10.9±0.3 |
Values reported are the mean of four independent experiments
Discussion
Our present understanding of the MCP shell models it as being comprised of sheets of hexagonally-shaped BMC-domain protein oligomers that tile edge-to-edge to form polyhedral facets, along with pentameric proteins of the CsoS4/CcmL family placed at the vertices. Edge complementarity between different types of BMC domain proteins is thought to allow the formation of mixed sheets with diversified function. This model is supported by several structural studies 3; 18. What is not well-understood are the key interactions that hold the various BMC-domain proteins (and the pentamers) together, stabilizing the shell and perhaps determining how its components are assembled and organized. Here, we used a rational approach of alanine scanning mutagenesis of exposed side chains to identify residues of the PduA protein that are instrumental in stabilizing the overall MCP architecture. Our studies identified three crucial edge residues important for inter-protein interactions. The most important among them are the edge lysines (corresponding to PduA K26 and PduJ K25) of the BMC domain proteins. Two other residues located along the edge of PduA (N29 and R79) were also critical for MCP assembly and function. Moreover, we also identified two residues (K37 and K55) that impaired growth on 1,2-PD, tentatively by altering the permeability properties of the MCP shell. However, the other mutants tested (Table 1) had no substantial difference in phenotype or MCP activity in our tests. It was somewhat surprising to us that most of the mutants we constructed had no obvious phenotype. Prior studies indicated that shell proteins are involved in the recruitment of enzymes to the lumen and exterior surfaces of the Pdu MCP 14; 15; 49; hence, surface mutations might be expected to result in abnormal enzyme content. This was not observed in the present study. Thus, our results tentatively suggest enzyme recruitment to bacterial MCPs requires multivalent protein-protein interactions.
Prior crystallographic studies proposed the potential importance of the PduA K26 edge lysine in protein sheet formation 28. However, it was also observed in the wild-type PduA crystal structure that the surface area buried at the interface between two hexamers where two lysines interact is ~1230 Å2, which is substantially less than that of typical dimeric interactions; the interface between carboxysome shell protein hexamers is ~1700 Å2 28. Hence, although the edge lysines in PduA (and in corresponding shell proteins from other types of MCPs) were implicated in inter-protein interactions, there was no experimental investigation of their actual contribution in vitro or in vivo. In this article, we demonstrated that residue K26 of PduA is essential to the assembly and function of the Pdu MCP using a combination of genetic, physiological, biochemical, microscopic and structural tools.
Prior studies also found that the edge lysines corresponding to PduA K26 are widely conserved among different types of BMC domain proteins 17; 20; 50 suggesting that these residues may be of importance to sheet formation in a variety of different MCP shells (Fig. S1). In this study, we report that changing edge lysines to alanine in two different shell proteins (PduA and PduJ) produces phenotypes indicative of a key role in MCP formation. Both PduA-K26A and PduJK25A mutants showed a faster growth rate compared to the wild-type at low B12 concentrations and were subject to propionaldehyde toxicity during growth on 1,2-PD, indicating a major defect in shell structure or assembly. Furthermore, we found that mutating the corresponding edge lysine of PduB resulted in similar phenotypes (data not shown). Thus, studies reported here indicate that the critical role of edge lysines in MCP shell formation is conserved among different BMC domain proteins.
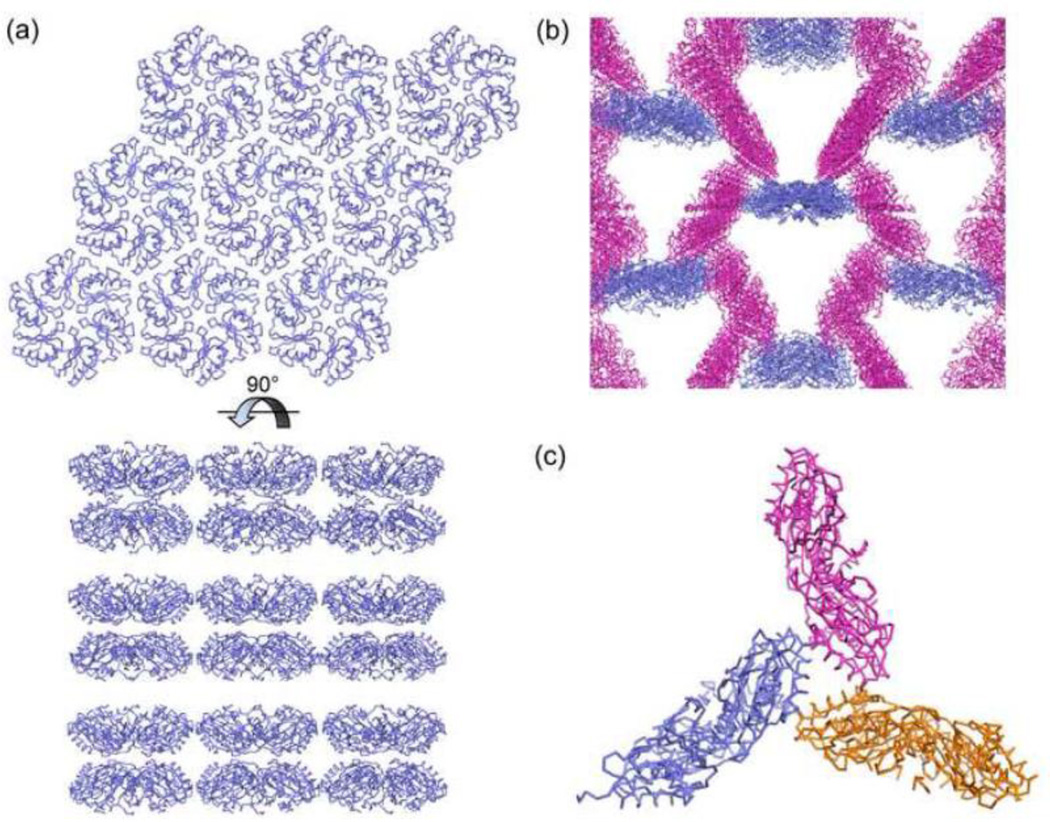
To obtain an atomic-level understanding of the phenotypes of the PduA-K26A mutant we determined its crystal structure and compared it to that of the wild-type PduA protein. In contrast to the wild-type PduA protein, the PduA-K26A mutant did not form extended protein sheets in crystals (Fig. 9A). Instead, one crystal form revealed two interacting edges of PduA-K26A that resulted in the formation of extended one-dimensional strips (Fig. 9B). Moreover, in both crystal forms, some subunits contacted adjacent hexamers edge-to-face rather that edge-to-edge (Fig. 9C). These finding are consistent with the idea that PduA-K26A is impaired for sheet formation which could reasonably account for the observed phenotypes. In addition, the dominant phenotype of the PduA-K26A mutant (which shows that it interferes with MCP assembly in some way) could also be explained by impaired edge contacts. PduA-K26A might be incorporated into the MCP (even in competition with wild-type PduA) but form weak edge contacts with neighboring shell proteins resulting in an unstable MCP. Alternatively, PduA-K26A might be recruited to the MCP by interaction with cargo proteins and essentially block closure of the shell due to weak or nonspecific edge contacts with adjacent BMC domain proteins. Thus, overall, structural data support the idea that residue K26 of PduA has a key role in stabilizing the protein sheets that form the shell of the Pdu MCP and might also play a role in determining shell protein edge-to-edge binding specificity.
Figure 9. Extended macromolecular structure of (A) PduA protein and (B) PduA-K26A.
(A) Wild-type PduA forms extended molecular layers in crystals. (B) Crystal form 1 of PduA-K26A contains one dimensional strips whose edges interact with the faces of other hexamers. (C) The edge-on-face interactions between three hexamers in crystal form 2 of PduA-K26A.
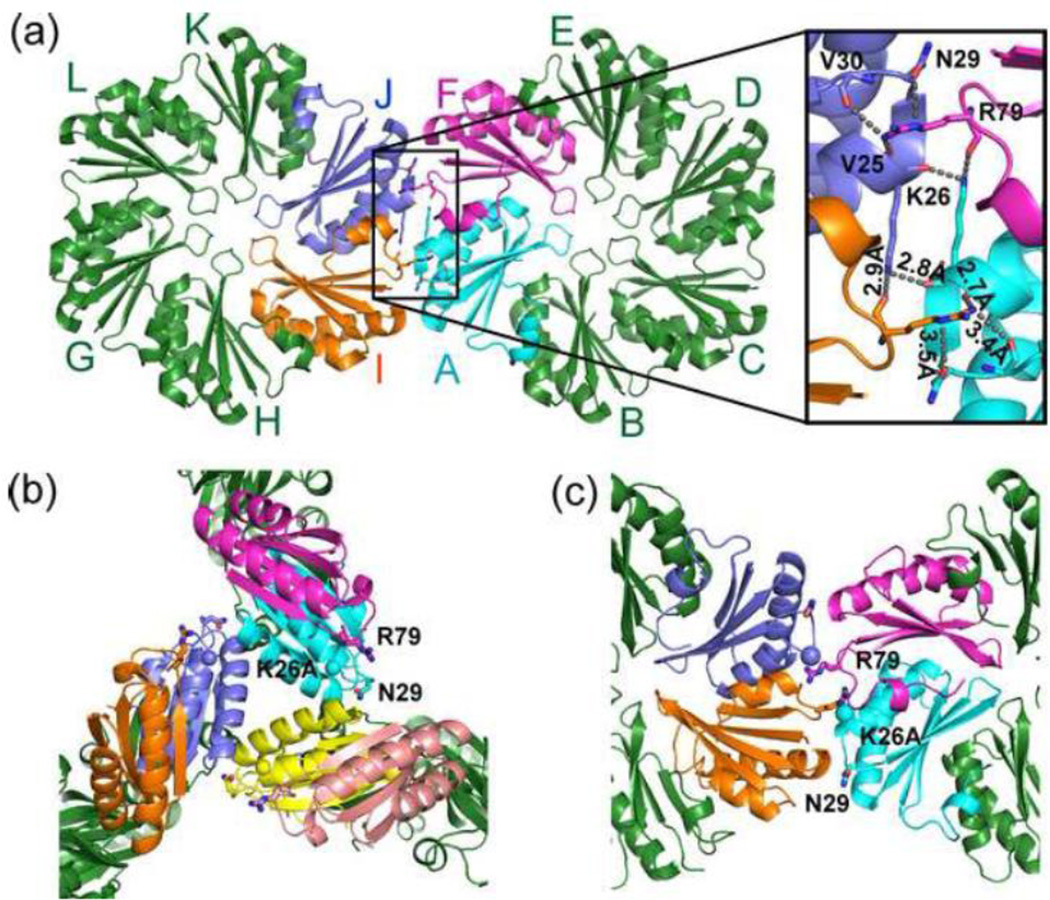
Our scanning mutagenesis of the surface of PduA also indicated important roles for PduA N29 and R79 in shell formation. This is supported by growth tests as well as the low yields and high DDH activity of the purified PduA-N29A and PduA-R79A MCPs. To better understand the atomic details of how residues K26, N29 and R79 of PduA stabilize the MCP shell, we reexamined our previously reported wild-type PduA structure 28. This analysis found that the ammonium nitrogen of K26 from one subunit forms hydrogen bonds to carbonyl oxygens in two different subunits: R79 from an adjacent subunit in the same hexamer, and K26 from a subunit in the adjacent hexamer (Fig. 10A). We also observed that the guanidino ω-nitrogen atom of R79 forms hydrogen bonds with the backbone carbonyl-oxygens of residues V25 and V30 further stabilizing the interface. Moreover, the guanidino ε-nitrogen atom of R79 forms a hydrogen bond with the amide oxygen of the N29 side chain from the adjacent hexamer. These interactions are reciprocal across every two-fold interface between adjacent hexamers of wild-type PduA. The details of the interactions are shown in Table 6. It is particularly notable that this interaction hub connects four monomers (two from each hexamer), thus strongly fortifying the edge. Another interesting observation about this interface is that the key amino acids side chains interdigitate. For example, R79 from one subunit is sandwiched between K26 and N29 from another subunit (Fig. 10A). This sandwiched side chain interaction creates an interface with high surface complementarity, which stabilizes the entire MCP assembly. These stabilizing interactions explain the critical roles played by K26, N29 and R79 in the stability of the Pdu MCP. On the other hand, the crystal structures of PduA-K26A revealed drastically altered lateral interfaces between hexamers. Edge-to-face interactions were observed in both crystal forms with the loss of the interactions described above (Fig. 10B). Furthermore, for the edge-to-edge interactions observed in PduA-K26A crystal form 1, N29 and R79 no longer made the inter-hexamer contacts observed in the wild-type PduA crystal structure (Fig. 10C).
Figure 10. PduA inter-hexamer interactions.
(A) The interface is stabilized by ten sets of interactions contributed by 4 different monomers. As indicated in the figure, interacting subunits A (cyan) and F (magenta) are from one hexamer and interacting subunits I (orange) and J (light blue) are from another hexamer. The important interactions are listed in Table 6. (B) PduA-K26A crystal structures contain new edge-to-face interactions not present in wild-type PduA. (C) Crystal form 1 of PduA-K26A also has drastically altered edge-to-edge interactions between hexamers. The β-carbon of the alanine mutation is highlighted as a sphere in B and C.
Table 6.
Lys 26, Asn 29 and Arg 79 Interactions at the interface
| Interacting Atom (Chain/Residue) |
Interacting Atom (Chain/Residue) |
Distance | Interaction type | |
|---|---|---|---|---|
| * | O (J/Lys26) | NZ (A/Lys26) | 2.8 Å | Hydrogen Bonding |
| * | NZ (J/Lys26) | O (A/LysK26) | 2.8 Å | Hydrogen Bonding |
| * | NZ (J/Lys26) | O (I/Arg79) | 2.9 Å | Hydrogen Bonding |
| * | NZ (A/Lys26) | O (F/Arg79) | 2.9 Å | Hydrogen Bonding |
| * | OD1 (J/Asn29) | NE(F/Arg79) | 3.5 Å | Hydrogen Bonding |
| * | OD1 (A/Asn29) | NE(I/Arg79) | 3.5 Å | Hydrogen Bonding |
| O (J/Val30) | NH1 (F/Arg79) | 3.4 Å | Hydrogen Bonding | |
| O (A/Val30) | NH1 (I/Arg79) | 3.4 Å | Hydrogen Bonding | |
| O (J/Val25) | NH1 (F/Arg79) | 2.7 Å | Hydrogen Bonding | |
| O (A/Val25) | NH1 (I/Arg79) | 2.7 Å | Hydrogen Bonding |
These interactions were determined as described previously1
We also examined the conservation of K26, N29 and R79 among diverse MCP shell proteins. K26 and R79 are conserved across several hexameric BMC-domain proteins from varied MCPs (Fig. S1A). In trimeric/pseudo-hexameric BMC domain proteins like PduB or EutL, the conserved arginine is also present, but not in PduT. The N29 residue is found in all BMC-domain proteins of the Pdu MCP, but is not conserved in the BMC domain protein of other MCPs (Figs. S1A and S1B). This conserved asparagine is replaced by arginine in the CcmK1, CcmK2 and CcmK4 proteins from the β-carboxysome and the EutM protein of the ethanolamine utilization (Eut) MCP and by glutamate in the CsoS1A and CsoS1C shell proteins of the α-carboxysome. This suggests that the K-N-R sandwich is a conserved feature that defines edge complementarity at the Pdu MCP hexameric interfaces, but this feature is only partially conserved among other BMC-domain proteins.
It is interesting to note that although PduA-N29A has phenotypes consistent with a complete block in MCP formation (similar to PduA-K26A) it still forms MCPs with an SDS-PAGE profile indistinguishable from wild-type, and PduA-N29A itself is present in the purified MCPs (Fig. 7). This tentatively suggests that this residue might play a specific role in controlling the permeability of the MCP to propionaldehyde as well as enzymatic substrate and cofactors. Alternatively, the PduA-N29A phenotype could be due to weaker edge contacts or perhaps altered binding specificity, resulting in aberrant contacts to other shell proteins. The exact nature of that role will require further experimentation.
During these studies we also observed that the solubility of PduA was greatly increased by changing the K26 edge lysine to alanine. The solubility of the wild-type PduA protein is about 1 mg/mL, whereas we were able to concentrate PduA-K26A to 10 mg/mL or more for crystallographic studies. Similar increases in solubility resulted from changing the corresponding edge lysines in PduB (data not shown). Presumably, the relative insolubility of shell proteins results from sheet formation, and disruption of the sheets by mutation therefore dramatically increases their solubility. These observations have important implications for purification and study of BMC domain proteins. In general, due to their inherent property of sheet formation, it is difficult to obtain high yields of BMC-domain proteins during overexpression and purification. Alteration of edge lysines by mutation could solve this problem, allowing the production of large amounts of protein for structural and biochemical studies.
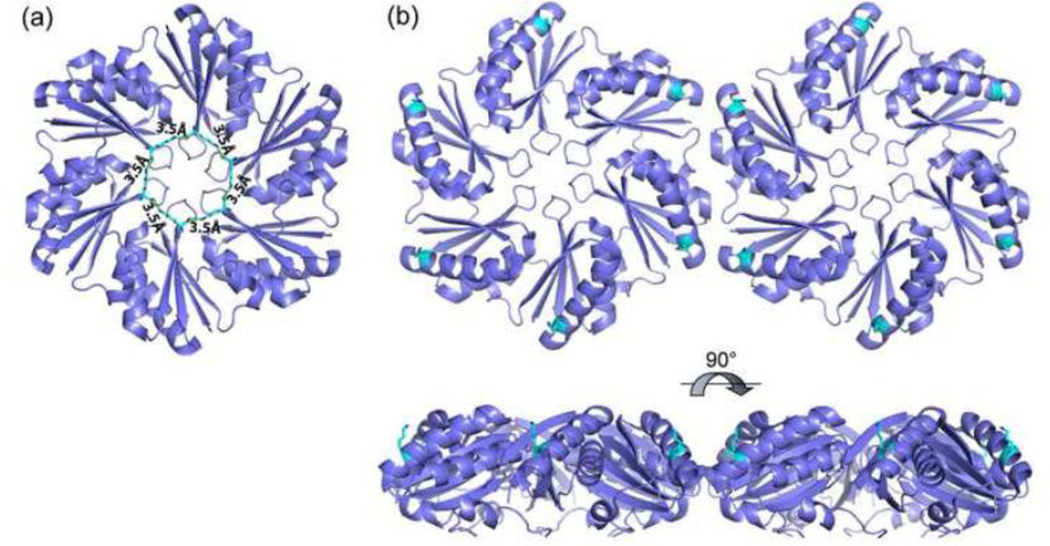
Two mutants identified in this study, PduA-K37A and PduA-K55A grew more slowly than wild-type on 1,2-PD. This suggests that the lysine residues at positions 37 and 55 are involved in the dynamics of metabolite transport across the shell since a reduced flux of enzyme substrates or cofactors across the shell would be expected to inhibit growth on 1,2-PD. K37 is present near the central pore and six copies of the side chain (one from each monomer forming the hexamer) form a rigid ring-like structure stabilized by hydrogen bonds between the ammonium nitrogen and the main chain carbonyl oxygen of the lysine from another subunit (Fig.11A). It was suggested in our prior work that the PduA central pore acts as the conduit for 1,2-PD transport across the shell 28. The K37A mutation most likely disturbed this rigid architecture and disrupted the pore function. The case of K55 is slightly different as this residue is present near the vertex of the PduA hexamer (Fig.11B). K55 might affect pore structure from a distance or be directly involved in transport via previously unrecognized openings near where the vertices of three BMC hexamers meet. Alternatively, the PduA K37A and K55A mutants might impair the recruitment of MCP enzymes some of which have been shown to associate with the shell 14; 15; 49. No major changes in protein content were seen by SDS-PAGE (Fig. 7) but this method does not resolve all the enzymes of the Pdu MCP. Further work will be needed to address these varied possibilities.
Figure 11. Structural positions of K37 and K55 in PduA.
(A) Six PduA K37 residues form a ring-like structure stabilized by hydrogen bonds. This ring may be important for the function of the central pore which is thought to act as a conduit for 1,2-PD. (B) K55 is located near the vertices of PduA hexamers.
This study highlights important aspects of the PduA shell protein and the Pdu shell more generally. A pduA deletion mutant is able to synthesize partially competent microcompartments and perform the MCP functions to a reduced extent. In the absence of the entire PduA gene, its task may be taken up by other shell proteins. However, a single amino acid change in PduA (K26A) abolished sheet propagation and completely stalled MCP formation, presumably by preventing the formation of a stable shell. These observations suggest that the MCP structure evolved from, and relies upon, an intricate set of interactions between just a few key residues at the perimeter of the hexameric building blocks. These insights might allow engineering MCPs with enhanced stability.
Materials and Methods
Chemicals and reagents
Antibiotics, vitamin B12 (CN-B12, B12), NAD+, NADH, NADP+, and NADPH were from Sigma Chemical Company (St. Louis, MO). Isopropyl β-D-1-thiogalactopyranoside (IPTG) was from Diagnostic Chemicals Limited (Charlotteville PEI, Canada). KOD DNA polymerase was from Novagen (Cambridge, MA). Restriction enzymes and T4 ligase were from New England Biolabs (Beverly, MA). Bacterial protein extraction reagent (B-PER II) was from Pierce (Rockford, IL). Other chemicals were from Fisher Scientific (Pittsburgh, PA).
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 2. The rich media used were lysogeny broth (LB) also known as Luria-Bertani/Lennox medium (Difco, Detroit, MI) and Terrific Broth (TB) (MP Biomedicals, Solon, OH). The minimal medium used was no-carbon-E (NCE) medium29.
Table 2.
Bacterial strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BE293 | *LT2/pKD46 | Lab Collection |
| BE287 | LT2/pLac22 | Lab Collection |
| BE1349 | LT2/pLac22-pduA | This Study |
| CS728 | LT2, ΔpduA:PblaP3-sacB-cat/pKD46 | This Study |
| CS732 | LT2, ΔpduA689 | This Study |
| SS40 | LT2, pduA690 [K26A] | This Study |
| SS41 | LT2, pduA691 [K37A] | This Study |
| SS42 | LT2, pduA692 [K55A] | This Study |
| SS64 | LT2, pduA693 [N29A] | This Study |
| SS84 | LT2, pduA694 [R79A] | This Study |
| SS106 | LT2, pduJ695 [K25A] | This Study |
| SS110 | LT2, ΔpduJ696 | This Study |
| SS141 | LT2, ΔpduA689/plac22 | This Study |
| SS142 | LT2, ΔpduA689/plac22-pduA | This Study |
| SS144 | LT2, ΔpduA689/plac22-pduA [K26A] | This Study |
| SS145 | LT2/plac22-pduA [K26A] | This Study |
| BE1722 | E. coli BL21DE3 RIL/pTA925-H6-PduA-K26A | This Study |
LT2 = S. enterica serovar Typhimurium LT2
Construction of chromosomal pduA mutations
A scarless chromosomal pduA deletion and point mutants were constructed by a modified PCR-based recombineering method30. Briefly, the sacB-cat cassette of pDS132 31; 32 along with an upstream bla P3 promoter of pBR32233 was subcloned into pET-41a (Novagen) between SphI and XhoI, generating pCS693. Primers pduA-DSCF (TATAGTCCCAACTATCGGAACACTCCATGCGAGGTCTTTGCTCTCCCTTATGCGACTCCT) and pduA-DSCR (TTCCACCAGCTCATTGCTGCTCATTGGCTAATTCCCTTCCCTTTCGGGCTTTGTTTAGCA) were used to amplify the 2.4 kb PblaP3-sacB-cat from pCS693. Strain BE293 which expresses the lambda red recombinase from pKD46 30 was transformed with the gel-purified PCR product and AmpR, CamR transformants were selected at 30 °C. An AmpR, CamR strain (CS728) was then transformed by electroporation with a mutagenic oligo or PCR product. To generate a scarless pduA deletion the following oligo was used: AACTATCGGAACACTCCATGCGAGGTCTTTGAAGGGAATTAGCCAATGAGCAGCAATGAG. For the construction of pduA missense mutations, mutagenic DNA was generated by fusion PCR. The primers used are listed in Supplementary Table 1. In the first PCR reaction, primers F-PduA-Flank and respective R-Fusion were used with LT2 genomic DNA as template. In the second PCR reaction primers R-PduA-Flank and respective F-Fusion were used LT2 genomic DNA as template. In the third and final PCR reaction, primers F-PduA-Flank and R-PduA-Flank were used with PCR1 and PCR2 as templates in the same reaction. The final PCR product was transformed into CS728 and SucS transformants were selected on TYE plates (10 g of Bacto-tryptone, 5 g of yeast extract, 15 g of agar in 1 L of the medium) with 5% sucrose and without NaCl31 at 37 °C and screened for chloramphenicol sensitivity. The pduA mutations were verified by DNA sequencing of PCR-amplified genomic DNA.
Bacterial growth assay
Growth studies were performed using a Synergy HT Microplate reader (BioTek, Winooski, VT) as previously described, with the following modifications: Cells were grown overnight in LB followed by transfer of 10 µl of this culture to 10 ml of LB supplemented with 0.6% 1,2-PD. After 6 h, cells were pelleted and washed three times with NCE medium containing 0.6% 1,2-PD and 1 mM MgS04. The washed cells were suspended in NCE medium (supplemented with 0.6% 1,2-PD; 0.3 mM each of valine, isoleucine, leucine, and threonine; 50 µM ferric citrate; and 20 nM CN-B12 or 100 nM CN-B12) to an OD600 of 0.15 and allowed to grow at 37°C with constant shaking in a microplate reader as described24. The growth curves were repeated at least three times in duplicate and representative curves are shown.
Pdu MCP purification
Pdu MCPs were isolated as described with some minor modifications 34. 20 ml of LB was inoculated with 200 µl of overnight LB culture and grown for 6 h till an OD600 of 0.6–0.8 was reached. Next, 4 ml of the 6 h culture was used to inoculate 400 ml of NCE medium, supplemented with 1 mM MgSO4, 0.5% succinate, and 0.6% 1,2-PD in a 1 L baffled Erlenmeyer flask. Cultures were incubated at 37°C with shaking at 275 rpm until an optical density at 600 nm (OD600) of 1.0 to 1.2 was reached. Cells were harvested by centrifugation and washed twice with 40 ml of buffer A (50 mM Tris-HCl pH 8.0, 500 mM KCl, 12.5 mM MgCl2, 1.5% 1,2-PD). Cells (~1 g wet weight) were resuspended in a mixture of 10 ml of buffer A and 15 ml of BPER-II supplemented with 5 mM β-mercaptoethanol, 0.4 mM protease inhibitor AEBSF, 25 mg of lysozyme, and 2 mg of DNase I. Vortexing was avoided for resuspension. The resultant mixture was incubated at room temperature with 70 rpm shaking for 30 min followed by incubation on ice for 5 min. In the case of low-yield mutant MCPs, the entire purification was performed at 4°C. Cell debris were removed by centrifugation twice at 12,000 × g for 5 min at 4°C, and then the MCPs were spun down at 20,000 × g for 20 min at 4°C. The pellet was washed once with a mixture of 4 ml of buffer A and 6 ml of BPER-II containing 0.4 mM AEBSF and then was resuspended in 0.5 ml of buffer B (50 mM Tris-HCl pH 8.0, 50 mM KCl, 5 mM MgCl2, 1% 1,2-PD) containing 0.4 mM AEBSF. Remaining cell debris were removed by centrifugation at 12,000 × g for 1 min at 4°C repeated three times. Purified MCPs were stored at 4°C until used.
Electron microscopy
Electron microscopy was carried out as previously described with slight modifications 34. Immediately after purification, MCPs were diluted to a concentration of 0.1 mg/ml in buffer B and 20 µl was applied to formvar coated copper grids (400 mesh). Grids were washed twice with ultrapure water containing 0.1% 1,2 PD. Uranyl acetate (2%) containing 0.1% 1,2-PD was applied to the grids and incubated for 1 min. The uranyl acetate was removed by absorbing the excess liquid with Whatman 40 filter paper. The grids were dried in the dark and viewed under a JEOL 2100 scanning transmission EM (JEOL USA, Inc., Peabody, MA) 34.
DDH activity assay
Diol dehydratase (DDH) assays were performed using a coupled NADH-dependent alcohol dehydrogenase reaction as previously described 26. The propionaldehyde produced by DDH is converted to 1-propaniol by alcohol dehydrogenase concomitant with the oxidation of NADH to NAD+. The absorbance at 340 nm was followed and rates were quantified using ε340 = 6.22 mM−1 cm−1.
Protein methods
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using Bio-Rad Ready gels and Bio-Rad Mini-Protean Tetra electrophoresis cells according to the manufacturer’s instructions (Bio-Rad, Hercules, CA). Coomassie Brilliant Blue R-250 was used to stain proteins following electrophoresis and protein was measured using Bio-Rad protein assay reagent (Bio-Rad), which is based on the method of Bradford, with bovine serum albumin as the standard35
Cloning expression and protein purification of H6-PduA-K26A
The gene for production of His6-PduA-K26A, was generated by amplifying the pduA allele of strain SS40 using genomic DNA as template and the following primers: GCCGCCAGATCTATGCATCACCATCATCACCACCAACAAGAAGCA CTAGGA and GCCGCC AAGCTT TCATTGGCTAATTCCCTTCGGTAA as forward and reverse primers, respectively. The resulting PCR amplicons were cloned into T7 expression vector pTA925 36 followed by electroporation into E. coli BL21DE3 RIL (Stratagene). The insert was verified by DNA sequencing and the resulting strain (BE1722) was used for production of the H6-PduA-K26A protein.
The His6-PduA-K26A production strain (BE1722) was grown at 37° C with shaking at 240 rpm in 2 L baffled flasks containing 1 L of LB broth with 25 µg/mL kanamycin and 10 µg/mL chloramphenicol. At an OD600 of 0.6–0.8, the growth temperature was reduced to 30° C and 0.5 mM IPTG was added. Cells were grown an additional 6 hours, pelleted by centrifugation using a Sorvall RC-5C Plus centrifuge and Sorvall SLC-4000 rotor at 5,000 rpm and 4°C. Cells were washed with 50 mM Tris HCl pH 9, 200 mM NaCl and pelleted centrifugation using a Sorvall SS34 rotor at 7,000 rpm and 4°C, and stored at −80 °C until used.
For purification of H6-PduA-K26A, all procedures were carried out at 4 °C or on ice. Approximately 8 g of cells were thawed and resuspended 1:3 (w/v) in 50 mM Tris HCl, pH 9.0 containing 200 mM NaCl, 0.01 mg/mL DNase, and Roche complete protease inhibitor. Cells were broken with an Emulsiflex (Avestin, Inc., Ottawa, ON, Canada) as described 37. Lysates were clarified by centrifugation at 32,000×g for 30 min and filtered through a 0.45 µm syringe filter (Millipore). H6-PduA-K26A was purified from the filtered cell lysate using and AKTA purifier. Buffer A contained 50 mM Tris HCl pH 9 and 200 mM NaCl. Buffer B was similar to buffer A but also included 500 mM imidazole. A 5 mL HisTrap HP column (GE Healthcare) was equilibrated with 4% buffer B, and then about 35 mL of clarified cell lysate (~1 g protein) was loaded using a 50 mL Superloop (GE Healthcare). The column was eluted with 15 column volumes (CV) 4% buffer B, followed by 10 CV 15% buffer B and a 15 CV linear gradient from 4 to 100% buffer B. Proteins were followed by monitoring A280. Fractions were evaluated by SDS-PAGE then H6-PduA-K26A was concentrated to two- to three-fold and exchanged into 30 mM Tris HCl pH 9.0, 50 mM NaCl and 1% glycerol using a Hi-trap desalting column (GE healthcare). H6-PduA-K26A was concentrated using a 30 kDa centrifugal filter (Millipore) to give a protein concentration of 10 mg/mL determined by the BCA assay (Pierce). Concentrated H6-PduA-K26A was frozen in liquid N2 and stored at −80° C before being used to set crystal trays.
Crystallography
The hanging drop vapor diffusion method was used for crystallization of H6-PduA-K26A. Crystallization was performed at the UCLA Crystallization Facility using 96-well plates and commercially available sparse matrix screens set using a Mosquito liquid handling device (TTP LabTech). The crystals used for structure determination of crystal form 1 were obtained directly from Mosquito drops set in the Qiagen AmSO4 suite well C4 (2.2 M ammonium sulfate and 0.2 M potassium formate). Crystal form 2 was obtained following optimization of well B3 from the same suite. Seventy nanoliters of protein was mixed with 140 nL of well solution containing 1.8 M ammonium sulfate and 0.125 M cesium sulfate. Crystals took 3–7 days to grow.
Data collection
Diffraction data were collected at the APS-NECAT beamline 24-ID-C on a DECTRIS-PILATUS 6M detector. Single crystals were mounted with CrystalCap HT Cryoloops (Hampton Research, Aliso Viejo, CA). H6-PduA-K26A crystals were cryo-protected in mother liquor containing 33% glycerol and flash frozen in liquid nitrogen prior to data collection at 100K.
Data processing, phasing and Refinement
Data for crystal form 1 were processed using Denzo/Scalepack 38. Data for crystal form 2 were processed using XDS/XSCALE 39. Data quality statistics are reported in Table 3 40; 41. It was noted that the merging R-factor increased substantially in the higher resolution ranges. We explored whether crystal decay might be responsible and whether this could be mitigated by retaining only a subset of the diffraction data collected in the early stages of X-ray exposure. We found that stronger signal to noise, even at higher resolution, was obtained by retaining all the observed diffraction frames for processing 42. The structure of H6-PduA-K26A (crystal forms 1 and 2) was solved by molecular replacement using the program Phaser 43 and the PduA hexamer28 (PDB ID 3ngk) as a search model. There are nine PduA subunits in the asymmetric unit of crystal form 1; that is one and a half hexamers. Crystal form 2 contained seven PduA subunits in the asymmetric unit, divided into a one half-hexamer and two one-third hexamers. The H6-PduA-K26A models were refined to 1.9 Å and 2.4 Å resolution, respectively. In the Ramachandran plot of the crystal form 1 structure 92% of the residues in the model were found in the most favored regions, 7.5% in allowed regions and 0.5% in generously allowed regions by PROCHECK 44. The respective Ramachandran plot distribution for crystal form 2 was 93.3%, 6.7%, and 0.0%. Final iterative rounds of model building and refinement were carried out using Coot 45 and either REFMAC and BUSTER 46 (crystal form 1) or PHENIX 47 (crystal form 2). Refinement parameterization included non-crystallographic symmetry restraints and translation libration screw-motion (TLS) refinement with one TLS group per chain in the asymmetric unit 48. Data collection and refinement statistics are presented in Table 3.
Table 3.
Data collection and refinement statistics (molecular replacement)
| H6-PduA K26A (crystal form 1) |
H6-PduA K26A (crystal form 2) |
|
|---|---|---|
| Data collection | ||
| Space group | C2 | F23 |
| Cell dimensions | ||
| a, b, c (Å) | 183.2, 105.4, 67.3 | 235.3, 235.3, 235.3 |
| α, β ,γ (°) | 90.0, 94.8, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 1.94 (2.01–1.94)* | 2.4 (2.486–2.4) |
| Rmerge | 0.077 (0.523) | 0.076 (0.981) |
| Rpim | 0.059(0.373) | 0.017 (0.234) |
| I / σI | 15.6 (2.2) | 33.9 (4.2) |
| Completeness (%) | 96.4 (97.2) | 100.0 (100.0) |
| Redundancy | 4.1 (4.0) | 20.5 (20.6) |
| Wilson B-factor | 32.5 Å2 | 52.3 Å2 |
| Refinement | ||
| Resolution (Å) | 1.94 | 2.4 |
| No. reflections | 89773 | 42124 |
| Rwork / Rfree | 0.194/0.209 | 0.195/0.219 |
| No. atoms | ||
| Protein | 5662 | 4305 |
| Ligand/ion | 81 | 47 |
| Water | 260 | 62 |
| B-factors | ||
| Protein | 47.5 | 62.5 |
| Ligand/ion | 63.6 | 83.8 |
| Water | 47.4 | 48.7 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.002 |
| Bond angles (°) | 1.2 | 0.58 |
One crystal for each structure was used for data collection and structure determination.
Highest-resolution shell is shown in parentheses.
Supplementary Material
Highlights.
Microcompartment shells are made of proteins that tile edge-to-edge.
A triad of amino acids is required for stable edge contacts between shell proteins.
A small number of key interactions drives microcompartment stability.
Acknowledgements
This work was supported by grant AI081146 from the National Institutes of Health to TOY and TAB. DEM was supported by a National Institutes of Health Biotechnology Training Program award (grant no. T32GM067555). The authors thank members of the Bobik and Yeates laboratories for helpful discussions. We thank Michael Collazo for help with setting crystal trays, and M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov and F. Murphy at NECAT beamline 24-ID of the Argonne National Laboratory APS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The coordinates of the refined models and the merged structure factors have been deposited in the Protein Data Bank with PDB codes 4P2S for crystal form 1 and 4PPD for crystal form 2.
References
- 1.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rae BD, Long BM, Badger MR, Price GD. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 2013;77:357–379. doi: 10.1128/MMBR.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr. Opin. Struct. Biol. 2011;21:223–231. doi: 10.1016/j.sbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu. Rev. Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 5.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 7.Abdul-Rahman F, Petit E, Blanchard JL. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J Phylogen Evolution Biol. 2013;1 [Google Scholar]
- 8.Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 2013;22:179–195. doi: 10.1002/pro.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 10.Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J. Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan C, Bobik TA. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J. Bacteriol. 2011;193:5623–5628. doi: 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan C, Cheng S, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14995–5000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 17.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 18.Kinney JN, Axen SD, Kerfeld CA. Comparative analysis of carboxysome shell proteins. Photosynth Res. 2011;109:21–32. doi: 10.1007/s11120-011-9624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci. 2013;22:660–665. doi: 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the b-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Twodimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci. 2009;18:2629–2635. doi: 10.1002/pro.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J. Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella . J. Bacteriol. 2011;193:1385–1392. doi: 10.1128/JB.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium : three genes for the propanediol dehydratase. J. Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha S, Cheng S, Fan C, Bobik TA. The PduM protein is a structural component of the microcompartments involved in coenzyme B12-dependent 1,2- propanediol degradation by Salmonella enterica. J. Bacteriol. 2012;194:1912–1918. doi: 10.1128/JB.06529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J. Biol. Chem. 2010;285:37838–37846. doi: 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J. Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51:246–55. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brosius J, Cate RL, Perlmutter AP. Precise location of two promoters for the b-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J. Biol. Chem. 1982;257:9205–9210. [PubMed] [Google Scholar]
- 34.Cheng S, Fan C, Sinha S, Bobik TA. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One. 2012;7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Johnson CL, Pechonick E, Park SD, Havemann GD, Leal NA, Bobik TA. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
- 42.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 48.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 49.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol. Cell. 2010;38:305–315. doi: 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.