Abstract
Signaling initiated by hypoxia and insulin powerfully alters cellular metabolism. The protein stability of hypoxia-inducible factor-1 alpha (Hif-1α) and Hif-2α is regulated by three prolyl hydroxylase domain–containing protein isoforms (Phd1, Phd2 and Phd3). Insulin receptor substrate-2 (Irs2) is a critical mediator of the anabolic effects of insulin, and its decreased expression contributes to the pathophysiology of insulin resistance and diabetes1. Although Hif regulates many metabolic pathways2, it is unknown whether the Phd proteins regulate glucose and lipid metabolism in the liver. Here, we show that acute deletion of hepatic Phd3, also known as Egln3, improves insulin sensitivity and ameliorates diabetes by specifically stabilizing Hif-2α, which then increases Irs2 transcription and insulin-stimulated Akt activation. Hif-2α and Irs2 are both necessary for the improved insulin sensitivity, as knockdown of either molecule abrogates the beneficial effects of Phd3 knockout on glucose tolerance and insulin-stimulated Akt phosphorylation. Augmenting levels of Hif-2α through various combinations of Phd gene knockouts did not further improve hepatic metabolism and only added toxicity. Thus, isoform-specific inhibition of Phd3 could be exploited to treat type 2 diabetes without the toxicity that could occur with chronic inhibition of multiple Phd isoforms.
Under normoxia, PHD-containing proteins hydroxylate critical pro-line residues on Hif-1α and Hif-2α, regulating their stability3,4. To date, three oxygen-dependent prolyl hydroxylases5 have been identified (Phd1, Phd2 and Phd3), but their relative roles in regulating the protein levels of the Hif isoforms in normal tissues and general effects on metabolism are poorly defined. To address this issue, we created mice with all possible combinations of homozygously floxed alleles encoding the Phd isoforms (Phd1fl/fl, Phd2fl/fl, Phd3fl/fl, Phd1fl/fl; Phd2fl/fl, Phd1fl/fl; Phd3fl/fl, Phd2fl/fl; Phd3fl/fl and Phd1fl/fl; Phd2fl/fl; Phd3fl/fl) that have been previously described6. We injected 8-week-old male littermates of each individual homozygous floxed genotype with control adenoviral GFP (adGFP) or adenoviral Cre (adCre) by tail vein to achieve liver-specific deletion of an individual Phd gene or a combination of Phd genes7.
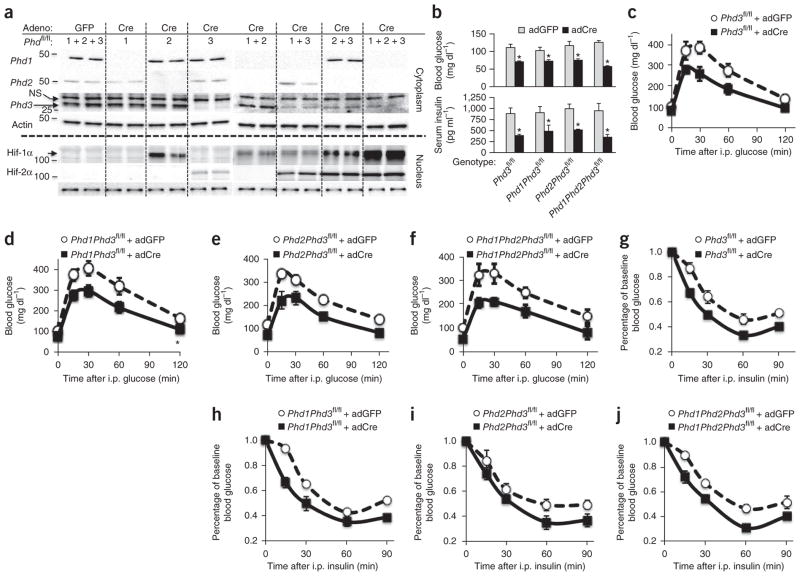
Injection of adCre resulted in an approximate 80–90% deletion of Phd1, Phd2 or Phd3 in the liver compared to control adGFP, as quantified in immunoblots (Fig. 1a). Knockout of Phd2 in the liver stabilized Hif-1α protein expression but not that of Hif-2α, and deletion of other Phd genes in addition to Phd2 further stabilized Hif-1α (Fig. 1a). Conversely, abrogation of Phd3 specifically stabilized Hif-2α expression over that of Hif-1α (Fig. 1a). Knockout of other Phd genes in combination with knockout of Phd3 enhanced Hif-2α expression in an additive fashion (Fig. 1a), which correlated with increased expression of known Hif-2α targets such as vascular endothelial growth factor (Vegfa) and erythropoietin (Epo, Supplementary Fig. 1). These data demonstrate both overlapping and isoform-specific regulation of Hif-1α and Hif-2α by the Phd proteins in the liver.
Figure 1.
Phd3 specifically regulates hepatic Hif-2α expression and glucose metabolism in vivo. (a) Western blots from nuclear and cytoplasmic lysates for the indicated proteins and genotypes. Phd1fl/fl, Phd2fl/fl and Phd3fl/fl mice treated with adGFP used as expression control. For the other genotypes, mice were treated with adCre by tail vein injection to achieve a liver-specific knockout. Each lane represents lysates from an individual mouse liver. Molecular weights in kDa are shown at left. Adeno, adenovirus; NS, nonspecific. (b) Fasting blood glucose and insulin levels from the indicated knockout animals. (c–j) Glucose tolerance tests (GTTs) (c–f) and insulin tolerance tests (g–j) of mice of the indicated genotypes and adenoviruses. Data are expressed as mean ± s.e.m. (n = 8 male mice per group).
Phd3fl/fl animals treated with adCre to achieve a liver-specific knockout exhibited lower fasting glucose and fasting insulin levels (Fig. 1b) compared to adGFP-treated controls. Deletion of additional Phd genes further lowered fasting glucose and fasting insulin compared to littermate adGFP-treated control mice. The changes in blood glucose for all genotypes were stable over at least 30 d (Supplementary Fig. 2a). Phd3fl/fl, Phd1fl/fl; Phd3fl/fl, Phd2fl/fl; Phd3fl/fl and Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice treated with adCre demonstrated similarly higher glucose tolerance (Fig. 1c–f) and insulin tolerance (Fig. 1g–j) compared to their age-matched littermate controls treated with adGFP. These differences in glucose tolerance and insulin tolerance were statistically significant for the Phd3-containing genotypes, as determined by area-under-the-curve calculations (*P < 0.05 compared to adGFP controls, Supplementary Fig. 2b,c). However, Phd1fl/fl, Phd2fl/fl and Phd1fl/fl; Phd2fl/fl mice treated with adCre showed no improvements in fasting glucose, insulin, glucose tolerance or insulin tolerance compared to adGFP-treated controls (Supplementary Fig. 3a–d), despite detectable expression of Hif-1α in the knockout combinations lacking Phd2 (Fig. 1a). These data indicate that Phd3 is the dominant Phd isoform in the hepatic regulation of glucose metabolism, and that other Phd isoforms are not able to completely compensate for loss of Phd3, as has been suggested in other studies8.
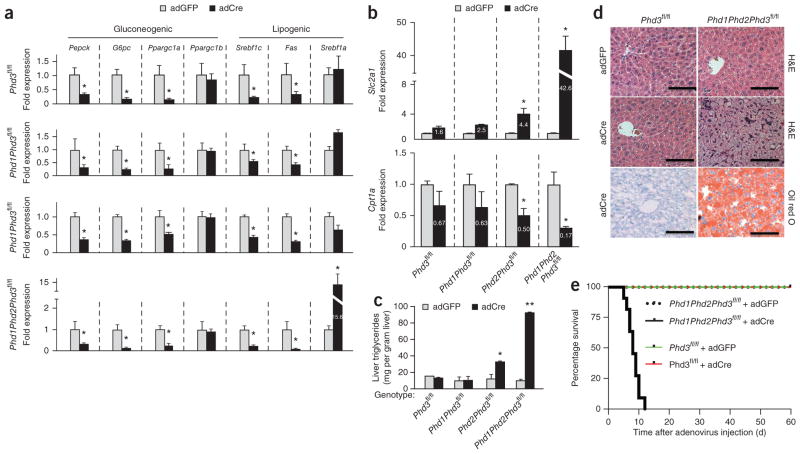
Abrogation of hepatic Phd3 through treatment with adCre led to lower expression of mRNAs encoding key gluconeogenic enzymes, Pck1, G6pc and Ppargc1a, but not Ppargc1b, compared with adGFP control treatment (Fig. 2a). The deletion of additional Phd genes in combination with Phd3 deletion did not further decrease gluconeogenic gene expression and, furthermore, the loss of Phd1 or Phd2 appeared to make no difference in gluconeogenic gene regulation compared to controls (Supplementary Fig. 4a).
Figure 2.
Worsened hepatotoxicity without improved metabolism in combination Phd knockout animals. (a) Quantitative PCR for relative mRNA levels of the indicated gluconeogenic and lipogenic genes in the livers in the indicated mice of the indicated genotypes and treatments (n = 8 male mice per group). (b) Gene expression data for Slc2a1 (top) and Cpt1a (bottom) in the livers of animals with the indicated genotypes. (c) Liver triglyceride measurements in control and knockout animals as indicated. Data are expressed as mean ± s.e.m. (n = 6 male mice per group). *P < 0.05 compared to adGFP controls and **P < 0.001 compared to adGFP controls. (d) H&E-stained and oil red O–stained sections of mouse livers of the indicated genotypes treated with adGFP or adCre. Scale bars, 100 μm. (e) Kaplan-Meier analysis of survival after adenoviral injection. (n = 8 male mice per treatment group). Log-rank analysis with P < 0.0001 for the Phd1Phd2Phd3fl/fl + adCre compared to Phd1Phd2Phd3fl/fl + adGFP controls.
Isoforms of sterol regulatory element-binding protein 1 (Srebf1) regulate nearly all aspects of lipid metabolism, with Srebf1c controlling lipogenesis and fatty acid synthase (Fas) levels and Srebf1a having broader control over fatty acid metabolism and cholesterol synthesis9. In mice with deletion of hepatic Phd3, expression of Srebf1c and Fas were lower than in adGFP control mice, whereas the expression of Srebpf1a was unchanged (Fig. 2a). Deletion of either Phd1 or Phd2 in addition to Phd3 further diminished Srebf1c and Fas expression when compared to their individual controls. Animals lacking all three Phd genes in the liver exhibited a 15-fold increase in the expression of Srebf1a, (Fig. 2a). This increase may be related to Phd1, as knockouts of Phd1 showed seven- and fivefold increases in Srebf1a and Srebf1c, respectively, that trended toward statistical significance compared to littermate controls (Supplementary Fig. 4b, P = 0.06).
Slc2a1 (also known as Glut1) is a hypoxia-inducible gene that increases intracellular glucose levels to be used by glycolysis and other metabolic pathways10. Phd2fl/fl; Phd3fl/fl and Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice treated with adCre exhibited a 4.5-fold and a 42-fold increase in Slc2a1 expression, respectively, compared to adGFP-treated mice. In addition, adCre treatment of Phd2fl/fl; Phd3fl/fl and Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice was associated with lower expression of a key regulator of mitochondrial β-oxidation, carnitine palmitoyl transferase-1a (Cpt1a) by 50% and 83%, respectively, compared to littermates treated with adGFP (Fig. 2b).
Germline knockouts of multiple Phd genes11 and transgenic animals expressing activated Hif-1α or Hif-2α develop hepatic steatosis (ref. 12). Phd2fl/fl; Phd3fl/fl and Phd1; Phd2; Phd3fl/fl mice treated with adCre exhibited higher hepatic triglyceride levels (Fig. 2c), and a grossly fatty liver (Supplementary Fig. 5a) confirmed by steatosis seen at microscopic level by H&E and oil red O staining (Fig. 2d), compared to littermate controls treated with adGFP. The other single or combination Phd knockout animals showed no alterations in liver triglycerides (Fig. 2c,d and Supplementary Figs. 5b,c and 6). The marked fat accumulation in the livers of Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice treated with adCre (Fig. 2c) may be caused by a combination of increased glucose uptake through increased Slc2a1 expression, and increased lipogenesis caused by increased Srebf1a expression and decreased β-oxidation evidenced through low Cpt1a levels.
The hepatic dysfunction in Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice treated with adCre also manifested as progressive hypoglycemia (Supplementary Fig. 5d) that culminated with hypoglycemic seizures and death by 12 d after adenoviral treatment (Fig. 2e). There was no such toxicity in Phd1fl/fl; Phd2fl/fl; Phd3fl/fl mice treated with adGFP. Thus, the simultaneous deletion of multiple Phd genes causes marked hepatotoxicity without additional metabolic benefits over those observed in mice with only Phd3 knockout.
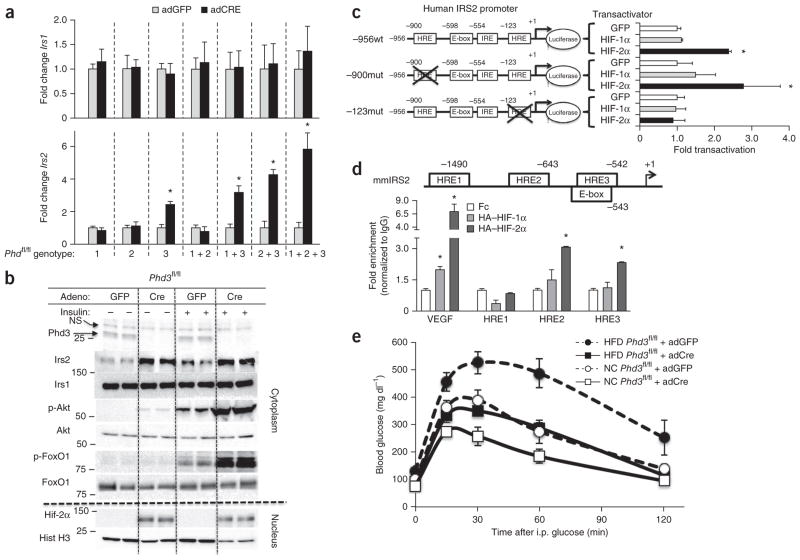
Loss of Phd3 altered expression of Irs1 and Irs2, which are critical components of insulin signaling7. Irs1 expression was unchanged by any Phd abrogation, but Irs2 mRNA levels (Fig. 3a) were higher in mice containing a Phd3 deletion, either alone or in combination with other Phd genes, compared to their littermate adGFP-treated controls. For instance, adCre treatment of Phd3fl/fl mice increased Irs2 protein expression by threefold compared to adGFP control treatment (Fig. 3b). This enhanced Irs2 expression led to a threefold increase in the insulin-stimulated phosphorylation of protein kinase B (Akt) and the transcription factor FoxO1 compared to insulin-stimulated adGFP-treated mice. Akt and FoxO1 are critical regulators of gluconeogenesis13, and this may be an important step by which Phd3 and Hif-2α improve glucose homeostasis.
Figure 3.
A Hif-2α –mediated increase in Irs2 expression in mice lacking hepatic Phd3 improves insulin action and reverses diabetes. (a) Quantitative PCR of Irs1 and Irs2 from livers of the indicated genotype and adenovirus treatments. Bars represent mean ± s.e.m. (n = 8 age-matched male mice, *P < 0.05). (b) Insulin-stimulated liver lysates from Phd3fl/fl mice infected with adGFP or adCre; molecular weights in kDa are shown at left. Each lane represents a lysate from a different mouse. p-Akt, phosphorylated Akt; p-FoxO1, phosphorylated FoxO1; Hist, histone. NS, nonspecific. (c) Luciferase reporter assay in Fao hepatoma cells with human IRS2 promoter–luciferase constructs containing wild-type sequence (−956wt) or mutations of distal (−900mut) or proximal (−123mut) HREs with HIF-1α, HIF-2α or GFP controls. (d) In vivo ChIP assay. Eight-week-old male C57/BL6 mice were treated with HA–HIF-1, HA–HIF-2 or control Fc control adenoviruses; liver lysates subjected to a ChIP assay to detect the enrichment HIF binding on the indicated HREs within the mouse Irs2 promoter (mmIRS2) are shown. (e) Glucose tolerance tests on Phd3fl/fl mice that were fed either a HFD or normal chow (NC) then treated with adGFP or adCre to induce deletion of Phd3. Data are expressed as mean ± s.e.m. (n = 8 mice per group).
To determine whether the induction of Irs2 by Hif-2α occurs directly or indirectly, we created luciferase reporter constructs for the human IRS2 promoter and mutated the two hypoxia response element (HRE) consensus sites at nucleotide positions −900 and −123 from the +1 initiation site14 (Supplementary Fig. 7a). We transfected these constructs into Fao hepatocytes along with an expression vector encoding GFP or constitutively active human HIF-1α or HIF-2α (ref. 15). HIF-2α, but not HIF-1α, specifically activated the wild-type human IRS2 promoter (Fig. 3c). A point mutation of the HRE at −900 did not alter the activation of the human IRS2 promoter by HIF-2α, but this activation was abolished by the mutation of the HRE at −123 (Fig. 3c).
We confirmed the specificity of Irs2 induction by Hif-2α with in vivo chromatin immunoprecipitation (ChIP) assays from intact livers of mice infected with adenoviruses encoding hemagglutinin-tagged constitutively active human HIF-1α, constitutively active human HIF-2α or Fc control. We found three major HRE sites within the mouse Irs2 promoter (mmIRS2), and the HREs at −643 and −543 displayed enrichment of HIF-2α binding over HIF-1α (Fig. 3d). Although the HRE at −643 in the mmIRS2 promoter is not conserved in humans (Supplementary Fig. 7b), the HRE at −543 is embedded within a conserved E-box that is essential for IRS2 activation16. These data demonstrate that Hif-2α activates Irs2 in an isoform-specific manner through a conserved site within the Irs2 promoter.
To test the therapeutic efficacy of Phd3 deletion, we treated Phd3fl/fl mice with a high-fat diet (HFD) or normal chow for 6 weeks and then acutely deleted Phd3 through administration of adCre. Both adGFP and adCre treatment groups gained 25–30% more weight compared to normal chow, but there was no difference in weight gain between adGFP and adCre animal on either diet (Supplementary Fig. 8a). The HFD treatment induced hyperglycemia, hyperinsulinemia and glucose intolerance, which is consistent with diabetes, in the control adGFP animals (Fig. 3e). The acute loss of Phd3 from adCre in HFD cohort of animals decreased fasting blood glucose by 30% (Supplementary Fig. 8b) and fasting serum insulin by 50% compared to the diabetic adGFP controls (Supplementary Fig. 8c). Notably, the diabetic phenotype induced by the HFD was completely ameliorated by the loss of Phd3, as evidenced by glucose tolerance returning to normal levels with adCre treatment (Fig. 3e, quantified in Supplementary Fig. 8d).
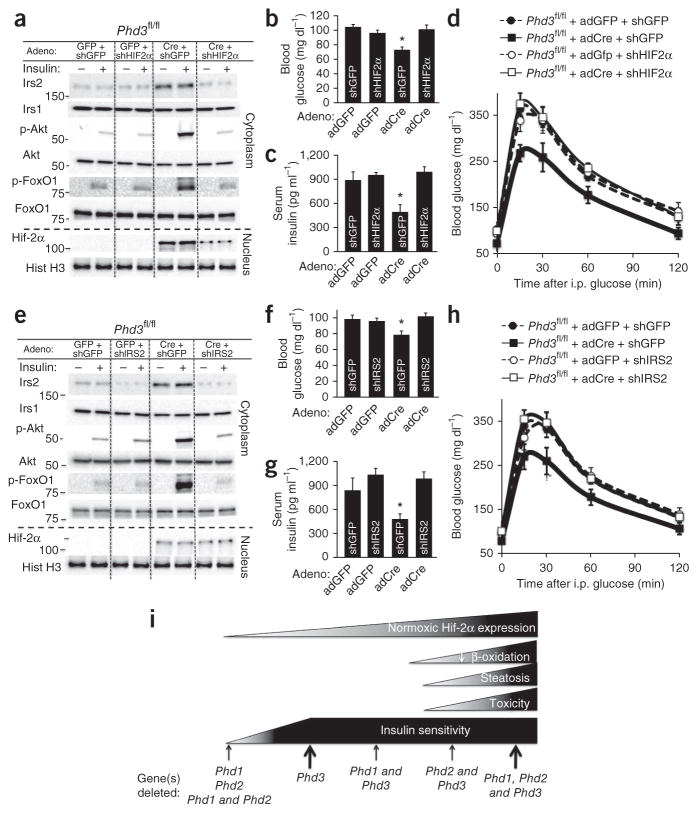
To determine whether either Hif-2α or Irs2 is necessary for the metabolic phenotype of Phd3 knockout animals, we constructed adenoviral shRNAs against both molecules17 and injected a mixture of adenoviruses containing adCre or control adGFP) along with an shRNA adenovirus at a constant titer. Phd3fl/fl mice treated with adCre and a control shRNA adenovirus against GFP (shGFP) showed an induction of Hif-2α and Irs2 expression, whereas Phd3fl/fl mice treated with both adCre and an shRNA adenovirus against Hif-2α (shHIF2) displayed a 70% decrease in Hif-2α and Irs2 expression compared to adGFP controls (Fig. 4a). The reduced expression of Hif-2α decreased the activation of Akt and FoxO1 (Fig. 4a) and consequently reversed the improvements in fasting blood glucose (Fig. 4b), insulin levels (Fig. 4c) and glucose tolerance (Fig. 4d, quantified in Supplementary Fig. 8e) in Phd3 knockout animals. In a similar vein, when Irs2 was knocked down in Phd3 knockout mice by an shRNA adenovirus targeting Irs2 (shIRS2. Fig. 4e), insulin-stimulated Akt and FoxO1 activation decreased, and fasting blood glucose (Fig. 4f), insulin levels (Fig. 4g) and glucose tolerance (Fig. 4h, quantified in Supplementary Fig. 8f) returned to levels seen in wild-type mice.
Figure 4.
Both Hif-2 and Irs2 are required for improved metabolism in mice with a liver-specific knockout of Phd3. (a) Insulin signaling studies on mice lacking hepatic Phd3 with an additional shRNA knockdown of Hif2a. Total adenovirus amount for each group was the same (5 × 109 PFU bolus) but was divided equally between the two indicated adenoviruses (see Online Methods). Each vertical lane represents a different mouse liver of the indicated treatments. Molecular weights in kDa are shown at left. (b,c) Fasting blood glucose (b) and insulin levels (c) from the indicated adenoviral treatments (n = 6 male mice per treatment group; data are mean ± s.e.m.; *P < 0.05). (d) GTTs performed on Phd3fl/fl mice treated with the indicated adenovirus combinations. Data are expressed as mean ± s.e.m. (n = 8 male mice per treatment group). (e) Insulin signaling studies on mice lacking hepatic Phd3 with an additional shRNA knockdown of Irs2. (f,g) Fasting blood glucose (f) and insulin levels (g) from the indicated adenoviral treatments (n = 6 male mice per treatment group; data are mean ± s.e.m.; *P < 0.05). (h) GTTs performed on Phd3fl/fl mice treated with the indicated adenovirus combinations. (i) Proposed model of metabolic improvements and toxicity in Phd knockout animals.
In this study, we demonstrate a direct link between oxygen sensing and insulin signaling through Phd3, which specifically stabilizes Hif-2α and augments Irs2 expression. Phd3 appears to be a critical and specific regulator of Hif-2α in vivo. These data are distinct from in vitro5 and global knockout8 data, which do not demonstrate such a relationship between Phd3 and Hif-2α, probably owing to chronic compensatory mechanisms that are invoked in germline knockout models.
We show that Hif-2α directly promotes the transcription of Irs2 in both the human and mouse Irs2 promoter constructs (Fig. 3c,d and Supplementary Fig. 7a,b). Despite potential pleiotropic effects of Phd3 deletion, the mechanism of metabolic improvements appears to require both Hif2a and Irs2, as knockdown of either molecule with shRNA adenovirus was sufficient to reverse the improvements in fasting glucose metabolism in liver-specific knockouts of Phd3. Irs2 may be the major mediator of these metabolic improvements since Irs2 has previously been shown to be critically important for metabolic homeostasis in the fasted state18 and is necessary and sufficient to prevent type 2 diabetes19.
As summarized in Figure 4i, the loss of Phd3 alone stabilizes Hif-2α at a low level that is capable of improving glucose tolerance without discernible toxicity. The deletion of additional Phd alleles increases Hif-2α expression; however, it also causes hepatic steatosis without any further metabolic improvements (Fig. 2d and Supplementary Fig. 6). It is not known whether this increased toxicity is a dose-dependent effect of Hif-2α, an Hif- independent effect of Phd deletion or some combination of these scenarios. However, when taken at face value our data suggest that there may be a therapeutic window of Hif-2α expression that maximizes metabolic improvement while minimizing toxicity that can be achieved by inhibiting Phd3 in an isoform-specific fashion.
Notably, we demonstrate that long-term Phd3 inhibition causes minimal toxicity, and this is corroborated by germline knockouts of Phd3 that do not demonstrate any obvious deleterious phenotype8,20. Pan–prolyl hydroxylase inhibitors are being tested for a wide variety of maladies, including anemia21, bone fractures22 and wound healing23, although the toxicity displayed in the pan-Phd knockouts of our study might caution against the chronic use of such nonspecific inhibitors. However, the isoform-specific inhibition of Phd3 could represent a new therapeutic approach to treat diabetes with a mechanism distinct from those of other available treatments and with potentially lower toxicity.
ONLINE METHODS
Animals and breeding strategy
All animals were housed on a 12-h light-dark cycle and fed a standard rodent chow unless otherwise indicated. All procedures involving mice were performed in accordance with the NIH guidelines for use and care of live animals and were approved by the Stanford University Institutional Animal Care and Use Committee. All mice in this study were of a mixed C57BL/6-FVB genetic background; therefore, all studies used only littermate homozygous floxed mice with adGFP as controls. The breeding strategy in this study involved the crossing of triple-heterozygous (Phd1fl/+, Phd2fl/+ and Phd3fl/+) mice to generate all the possible genotypes and has been described previously6. At least three different breeders were used to generate each floxed genotype, and the phenotypes were checked among the litters of all breeders. Genotyping analysis of these Phdfl/fl conditional mice has been previously described20. After this initial generation of each genotypes, the individual Phd phenotypes were maintained as single or multiple homozygous floxed alleles (for example, Phd1fl/fl or Phd1fl/fl; Phd2fl/fl; Phd3fl/fl), and experiments involving adenoviral treatments and diet were only compared among male littermate controls to account for subtle differences in genetic background. No female mice were used in the metabolic analyses in this manuscript. For the adenovirus injections studies, 8-week-old male C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). A sample size of six to eight male mice per treatment was deemed appropriate based on the magnitude of the phenotype in preliminary studies. All animals within an experimental cohort were included in the final analysis, although there were no preset inclusion or exclusion criteria. The mice in this study were not randomized to their treatments and were selected based purely on availability. No blinding was done for the investigators performing these studies.
Generation and use of adenoviral constructs
Purified adCre and adGFP was obtained from University of Iowa Gene Transfer Vector Core. Human HIF-1 and HIF-2 adenoviruses for the in vivo ChIP experiment were produced from cDNA constructs obtained through Addgene. HIF-1 contains proline-to-alanine mutations at P402 and P564 (Addgene #18955), and HIF-2 contains proline-to-alanine mutations at P405 and P531 (Addgene #18956) (ref. 15). These constructs were cloned into the pShuttle-IRES-GFP2 vector (Agilent, Palo Alto, CA) to generate adenoviruses through the AdEasy system24. The shHIF2, shIRS2 and Fc control adenoviruses were a gift from C.J.K. and colleagues25. All viruses were purified on a CsCl gradient then dialyzed into 20 mM Tris (pH 8.0), 25 mM NaCl + 5% glycerol solution. For adCre-mediated deletion, 1 × 109 PFU of adCre was injected as a 100-μl bolus. For hemagglutinin-tagged HIF-1 and hemagglutinin-tagged HIF-2 overexpression, 1 × 108 PFU was injected as a 100-μl bolus. For shRNA studies, a 50/50 mixture of adenoviruses was used, with the total of 5 × 109 PFU injected as a 200-μl bolus into the tail vein.
Quantitative RT-PCR analysis
Total RNA was isolated from mouse tissues using an RNeasy Mini Kit (QIAGEN, Valencia, CA). cDNA was prepared from 1 μg of RNA using the QuantiTect Kit (QIAGEN, Valencia, CA) with random hexamer primers, according to manufacturer’s instructions. The resulting cDNA was diluted 100-fold, and a 2.5-μl aliquot was used in a 10-μl PCR reaction (SYBR Green, ABI) containing primers at a concentration of 300 nM each. PCR reactions were run in triplicate and quantified in the ABI 7900HT Sequence Detection System. Cycle threshold (Ct) values were normalized to TATA box–binding protein (TBP) expression, and results were expressed as a fold change of mRNA compared to the indicated control mice. Primers were optimized over exon-exon junctions whenever possible and were optimized to a calculated Tm of 60 °C. Primer sequences are as follows: Irs1 forward 5′-TCC CAA ACA GAA GGA GGA TG-3′ and Irs1 reverse 5′-CAT TCC GAG GAG AGC TTT TG-3′, Irs2 forward 5′-GTA GTT CAG GTC GCC TCT GC-3′ and Irs2 reverse 5′-TTG GGA CCA CCA CTC CTA AG-3′, Pgc1a forward 5′-GTC AAC AGC AAA AGC CAC AA-3′ and Pgc1a reverse 5′-TCT GGG GTC AGA GGA AGA GA-3′, Pgc1b forward 5′-TCC TGT AAA AGC CCG GAG TAT-3 ′ and Pgc1b reverse 5′-GCT CTG GTA GGG GCA GTG A-3′, Srebpf1a forward 5′-GAA CTG GAC ACA GCG GTT TT-3′ and Srebpf1a reverse 5′-GGC CAG AGA AGC AGA AGA GA-3′, Srebpf1c forward 5′-GAG CCA TGG ATT GCA CAT TT-3′ and Srebpf1c reverse 5′-CTC AGG AGA GTT GGC ACC TG-3′, Fas forward 5′-GAG GAC ACT CAA GTG GCT GA-3′ and Fas reverse 5′-GTG AGG TTG CTG TCG TCT GT-3′, Cpt1 forward 5′-CCA ATC ATC TGG GTG CTG G-3′ and Cpt1 reverse 5′-AAG AGA CCC CGT AGC CAT CA-3′, Slc2a1 forward 5′-CCA TGT ATG TGG GAG AGG TGT-3′ and Slc2a1 reverse 5′-TTG CCC ATG ATG GAG TCT AAG-3′, Epo forward 5′-CAT CTG CGA CAG TCG AGT TCT G-3′ and Epo reverse 5′-CAC AAC CCA TCG TGA CAT TTT C-3′, Vegfa forward 5′-CCA CGT CAG AGA GCA ACA TCA- ′ and Vegfa reverse 5′-TCA TTC TCT CTA TGT GCT GGC TTT-3′ and Tbp forward 5′-ACC CTT CAC CAA TGA CTC CTA TG-3′ and Tbp reverse 5′-TGA CTG CAG CAA ATC GCT TGG-3′.
Metabolic studies
Only male mice that were littermate and weight matched were used in metabolic analyses. For GTTs, mice were fasted overnight, and then blood samples were obtained at 0, 15, 30, 60 and 120 min after intraperitoneal (i.p.) injection of 2 g per kg body weight dextrose. ITTs were performed by injecting 0.5 U per kg body weight insulin (Novolin, Novo Nordisk) i.p. into mice after a 4-hr fast, followed by blood collection at 0, 15, 30 and 60 min after injection. Blood glucose values were determined using a One Touch II glucose monitor (Lifescan, Milipitas, CA). Serum insulin levels were measured by ELISA using mouse insulin as a standard (Crystal Chem, Chicago, IL). Triglyceride levels in liver were measured using a kit from Sigma-Aldrich. For HFD, mice were fed a chow that consisted of 60% fat (D14292, Research Diets) for a total of 6 weeks, and normal chow was a standard 17% fat diet (D12451, Research Diets). Metabolic studies on these HFD-fed mice were carried out in the same manner as above.
In vivo insulin signaling
Following an overnight fast, mice were anesthetized with 2,2,2-tribromoethanol in PBS (Avertin) and injected with 5 U per kg body weight of regular human insulin (Novolin, Novo Nordisk, Denmark) through tail vein. Five minutes after the insulin bolus, mice were euthanized and tissues were removed and frozen in liquid nitrogen. Immunoblot analyses of insulin signaling molecules were performed using tissue homogenates prepared in a tissue homogenization buffer that contained 25 mM Tris–HCl (pH 7.4), 10 mM Na3VO4, 100 mM NaF, 50 mM Na4P2O7, 10 mM EGTA, 10 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, 1% Nonidet–P40 and 0.1% SDS supplemented with the cOmplete Protease Inhibitor Cocktail (Roche). All protein-expression data were quantified by Chemi–Doc XRS+ system (Bio-Rad).
Nuclear-cytoplasmic purification
This procedure has been described previously26, but we provide a detailed version here. Buffer A consisted of 10 mM Tris-HCl (pH 7.8), 1.5 mM MgCl2, 10 mM KCl, cOmplete Protease Inhibitor Cocktail (Roche), Pepstatin (Roche), 0.5 mM dithiothreitol (DTT), 0.4 mM PMSF and 1.0 mM Na3VO4. Buffer C consists of 20 mM Tris-HCl (pH 7.8), 1.5 mM MgCl2, 420 mM KCl, 20% glycerol, Complete Inhibitor Cocktail (Roche), Pepstatin (Roche), 0.5 mM DTT, 0.4 mM PMSF and 1.0 mM Na3VO4. Buffer D consisted of 20 mM Tris-HCl (pH 7.8), 1.5 mM MgCl2, 100 mM KCl, 20% glycerol and 0.2 mM EDTA. 250 mg of freshly isolated liver was homogenized in ice-cold Buffer A. The cells were further lysed by passing through a 26-gauge needle. The homogenate was then centrifuged for 5 min at 4,500g at 4 °C. The cytoplasmic fraction was obtained by recentrifugation of the supernatant for 10 min at 20,000g. The pelleted nuclei were resuspended in 500–750 μl of Buffer C for 30 min at 4 °C with gentle agitation. The extracted nuclei were centrifuged at 10,000g for 30 min at 4 °C and then dialyzed in a 3,500–molecular weight cut off (MWCO) dialysis cassette MWCO for 2 h with two buffer exchanges in 1 L of Buffer D (otherwise, the extracts form an insoluble precipitate in Laemmli buffer). The final nuclear lysate was obtained after centrifugation at 10,000g for 10 min at 4 °C.
General western blotting and antibodies
Rabbit polyclonal antibodies to HIF-1 (#ab2185), HIF-2 (#ab199) and PHD1 (#ab108980) were purchased from Abcam and used at a concentration of 1:500. Rabbit polyclonal antibody to PHD3 was purchased from Novus (NB100-139) and was used at a concentration of 1:500 (ref. 8). Rabbit monoclonal antibodies against PHD2 (#4835), IRS1 (#3407), p-Akt (S473, #4060), actin (#8456) and histone H3 (#4499) were purchased from Cell Signaling Technologies and used at a dilution of 1:1,000. Rabbit polyclonal antibodies against Irs2 (#4502) antibody (IRS-2), Akt (#9272), p-FoxO1 (#9456) and FoxO1 (#9454) were purchased from Cell Signaling Technology and used at a concentration of 1:1,000. Antibodies were diluted in Superblock TBS solution (Pierce) and maintained at 4 °C. Western blotting was run using precast gradient gels (4–15%) from Bio-Rad, and protein ranges of the relevant ranges were cut out and blotted with the individual anitbodies to save on reagent costs. Whenever possible, 50 μg of protein were loaded per well.
Immunohistochemistry
Livers were dissected and fixed in 10% formalin using standard technique, and then dehydrated in ethanol followed by paraffin embedding and sectioning for H&E analysis by Histo-Tec Laboratory (Hayward, CA). For oil red O stains, fresh liver chunks were embedded in OCT and then cryosectioned for oil red O analysis. Images were taken with a Leica DM6000 B microscope. Scale bars were taken using the native Leica imaging software. These data are representative of observations of at least six animals per group.
In vivo chromatin immunoprecipitation
Liver tissue (3–4 g) was cross-linked with 1% formaldehyde for 10 min and with 0.5 M glycine for 5 min, followed by centrifugation. The pellets were rinsed in 1× PBS, homogenized, centrifuged and resuspended in cell lysis buffer and protease inhibitors, incubated on ice for 15 min and centrifuged. Pellets were then resuspended in nuclear lysis buffer + protease inhibitor, sonicated 5 × 30 s with power of 5 W and centrifuged. 10 μL of sonicated sample was removed and combined with 400 uL elution buffer + 16 μL 5M NaCl and incubated at 65 °C overnight. The next day, 950 μL ethanol was added, followed by precipitation overnight at −20 °C and chromatin quantification using the QIAquick PCR purification kit (QIAGEN, Valencia, CA). The immunoprecipitation was performed with 100 μg of sonicated chromatin incubated with 18 μg of anti-hemagglutinin antibody (Abcam, Cambridge, MA) or 18 μg of normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA). Following overnight incubation, complexes were incubated with 25-μL Dynabeads (1:1 mix protein A/G) for 2 h at room temperature (Invitrogen, Grand Island, NY), followed by a series of washes and elution from beads. 16 μL of 5M NaCl was added for overnight incubation at 65 °C, followed by addition of 100% ethanol and −20 °C precipitation overnight. Samples were run through the QIAquick PCR Purification Kit, and quantitative RT-PCR was performed as described above, using a titration of pooled input samples as a standard curve and samples in duplicate (at a minimum). Signals were normalized to IgG, with Vegf serving as a positive control and m-5Ve as a negative control. Significance was determined using a two-tailed Student’s t-test. The mmIRS-2 HRE1 primers used were 5′-GCC CCA AAC CGT GTT CAC-3′ and 5′-AAA GGC CAC GTA GAT AGA GAA ATT CA-3′. The mmIRS-2 HRE2 primers used were 5′-CGA CGG ACA GCG AGA CGG AC-3′ and 5′-AAC GCA GCC CGG TGT CGG-3′. The mmIRS–2 HRE3 primers used were 5′-CCG CCG CAC AGT GAG TAA C-3′ and 5′-GCA GAG TCA CGT GTT GTT TTG C-3′. The mmVEGF HRE primers (+ control) were 5′-CGC GTC CTC CCT CAC GC-3′ and 5′-CTC GGC CAT CAC GGG G-3′. The m–5Ve (−control) primers were 5′-GGG GGA TAA TGA TTG CAA AA-3′ and 5′-GCG TGG ACA GAG ATC TAG GC-3′.
Cell culture and IRS2 promoter studies
The human IRS2 promoter14 from −963 to +1 was synthesized directly into the BamHI- and HindIII-flanked region of the pUC57 vector (Genscript, NY, USA). All mutant promoters were synthesized in a similar fashion. For the specific sequences of the mutations, see Supplementary Figure 8. The BamHI- and HindIII-flanked fragments for each of the promoter constructs were excised and then ligated into the compatible BglII- and HindIII-flanked sites in the pGL3 basic vector (Promega). Fao hepatoma cells (a gift from the laboratory of S. Biddinger) were cultured in RPMI + 10% FBS + 12.5 mM HEPES and 1 mM pyruvate and were rigorously tested to be mycoplasma negative. Cells were transfected using TransIT2020 (Mirus) using the manufacturer’s standard protocol. For instance, a six-well plate was transfected with 2.5 μg of luciferase construct, 2.25 μg of HIF or control DNA and 0.25 μg of Renilla luciferase along with 15 μl of TransIT-2020. The assay was scaled down accordingly for 12- or 24-well plates. The transfection was incubated for 36 h, and then the cells were serum-starved for 12 h before luciferase measurements. Luciferase data were measured with a Tecan Infinite M1000 luminometer and normalized to Renilla signal.
Statistical analyses
Data are presented as ± s.e.m. Student’s t-test was used for statistical analysis between two groups, whereas statistical significance between multiple treatment groups was determined by analysis of variance and Tukey’s t-test using Prism 6.0 for Macintosh. The data met assumptions of a normal distribution as determined by statistical software, and variance was estimated with s.d. and s.e.m. as reported in the manuscript. Kaplan-Meier analysis was performed using Prism 6.0 for Macintosh.
Supplementary Material
Acknowledgments
We thank K. Takeda and G.-H. Fong (University of Connecticut) for their generous gift of the Phd1fl/fl, Phd2fl/fl and Phd3fl/fl mice. We thank J. Boucher (Joslin Diabetes Center, Boston, MA) for sharing qPCR primer sequences and his critical reading of the manuscript. We thank S. Biddinger (Boston Children’s Hospital, Boston, MA) for providing the Fao hepatoma cells. C.M.T. was supported by Radiological Society of North America Research Resident grants 1018 and 1111. E.C.F. and E.L.L. were supported by US National Cancer Institute Training Grant CA121940. C.W. was supported by a training grant from the Canadian Institutes of Health and Research. A.N.D. was supported by a T32 training grant in Comparative Animal Medicine at Stanford University. A.J.K. was supported by grant P20 GM104936 from the US National Institute of General Medical Sciences (NIGMS). Fellowship support was from the NIGMS Stanford Medical Scientist Training Program grant T32 GM007365 (K.W.), Stanford Medical Science Training Program funding (K.W. and L.M.M.), Molecular and Cellular Immunobiology Program training grant 5T32AI07290 (L.M.M.), and US National Institutes of Health (NIH) R01HL074267, R01NS064517 and R01CA158528 (C.J.K.). A.J.G. was supported by NIH grants CA67166 and CA88480 and the Sidney Frank Foundation.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
C.M.T., E.C.F., A.J.K., E.L.L., K.W. and L.M.M. designed and performed experiments and analyzed data. C.W. and A.N.D. generated the knockout animals and contributed to design of all animal experiments. J.Y. and C.J.K. generated and purified the adenoviruses and contributed to experimental design of all adenovirus experiments. C.M.T. and A.J.G. wrote the manuscript and oversaw all aspects of this project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 2.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 3.Ivan M, et al. HIF-α targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 4.Min JH, et al. Structure of an HIF-1α–pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 5.Appelhoff RJ, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 6.Rankin EB, et al. The HIF signaling pathway in osteoblasts directly modulates erythropoiesis through the production of EPO. Cell. 2012;149:63–74. doi: 10.1016/j.cell.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniguchi CM, Ueki K, Kahn R. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J Clin Invest. 2005;115:718–727. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Minamishima YA, et al. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol. 2009;29:5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 11.Minamishima YA, et al. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol. 2009;29:5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim WY, et al. Failure to prolyl hydroxylate hypoxia-inducible factor α phenocopies VHL inactivation in vivo. EMBO J. 2006;25:4650–4662. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 14.Vassen L, Wegrzyn W, Klein-Hitpass L. Human insulin receptor substrate-2: gene organization and promoter characterization. Diabetes. 1999;48:1877–1880. doi: 10.2337/diabetes.48.9.1877. [DOI] [PubMed] [Google Scholar]
- 15.Kim WY, et al. Failure to prolyl hydroxylate hypoxia-inducible factor α phenocopies VHL inactivation in vivo. EMBO J. 2006;25:4650–4662. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa Y, et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med. 2006;12:107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- 17.Saito T, et al. Transcriptional regulation of endochondral ossification by HIF-2α during skeletal growth and osteoarthritis development. Nat Med. 2010;16:678–686. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 18.Kubota N, et al. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 2008;8:49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Canettieri G, et al. Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling. Cell Metab. 2005;2:331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, et al. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood. 2008;111:3229–3235. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamishima YA, Kaelin WG., Jr Reactivation of hepatic EPO synthesis in mice after PHD loss. Science. 2010;329:407. doi: 10.1126/science.1192811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. Prolyl hydroxylase inhibitors increase neoangiogenesis and callus formation following femur fracture in mice. J Orthop Res. 2009;27:1298–1305. doi: 10.1002/jor.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
- 24.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 25.Wei K, et al. A liver Hif-2α–Irs2 pathway sensitizes hepatic insulin signaling and is modulated by Vegf inhibition. Nat Med. 2013 doi: 10.1038/nm.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin EB, et al. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease–associated vascular tumors in mice. Mol Cell Biol. 2005;25:3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.