Abstract
Itk and Btk are nonreceptor tyrosine kinases of the Tec family that signal downstream of the T cell receptor (TCR) and B cell receptor (BCR), respectively. Despite their high sequence similarity and related signaling roles, Btk is a substantially more active kinase than Itk. We showed that substitution of six of the 619 amino acid residues of Itk with those of Btk was sufficient to completely switch the activities of Itk and Btk. The substitutions responsible for the swap in activity are all localized to the activation segment of the kinase domain. Nuclear magnetic resonance and hydrogen-deuterium exchange mass spectrometry analyses revealed that Itk and Btk had distinct protein dynamics in this region, which could explain the observed differences in catalytic efficiency between these kinases. Introducing Itk with enhanced activity into T cells led to enhanced and prolonged TCR signaling compared to that in cells with wild-type Itk. These findings imply that evolutionary pressures have led to Tec kinases having distinct enzymatic properties depending on the cellular context. We suggest that the weaker catalytic activities observed for T cell–specific kinases is one mechanism to regulate cellular activation and prevent aberrant immune responses.
Introduction
Finely tuned enzymatic activity controls cellular function at all levels. Of the numerous cellular activities controlled by enzymes, kinase-mediated phosphotransfer reactions are a key feature of intracellular signaling cascades, directing information flow and ultimately cellular responses to various stimuli. Kinase activity can be controlled by specific regulatory mechanisms, the amount of enzyme expressed in the cell, the localization pattern of the enzyme, or the intrinsic activity of the kinase in question (1). It is well known that dysregulation of kinase activity by mechanisms such as increasing kinase abundance or mutations that alter regulatory control is associated with numerous disease states (2–4).
Detailed knowledge of kinase structures and their associated regulatory mechanisms can lead to an understanding of disease at a molecular level, and can be harnessed to intentionally modulate enzymatic activity by mutation of specific amino acid residues. Indeed, hyperactivated or constitutively activated kinases serve as valuable tools to dissect specific signaling pathways, increasing our ability to fully understand cellular communication at the molecular level. High-resolution crystal structures of many protein kinases have been solved (5–7), and they reveal a common kinase domain fold consisting of two lobes (Fig. 1A); the smaller N-terminal lobe (N-lobe) is made up of five β strands and one helix referred to as the C-helix, whereas the larger C-terminal lobe (C-lobe) consists primarily of α-helices. The substrate adenosine triphosphate (ATP) binds in the catalytic cleft that is formed between the two lobes, whereas the protein or peptide substrate interacts primarily with the C-lobe. A large flexible loop lies between the N-lobe and C-lobe, and it is referred to as the activation segment. This activation segment is not always visible in electron density maps, which has made this region of the kinase domain seem somewhat enigmatic. Most, but not all, kinases have one or more residues in the activation segment that undergo phosphorylation as a prerequisite for activation (8). Phosphorylation on the activation loop switches the enzyme from an inactive to an active state by triggering concerted movements in different regions of the kinase to form a catalytically competent active site.
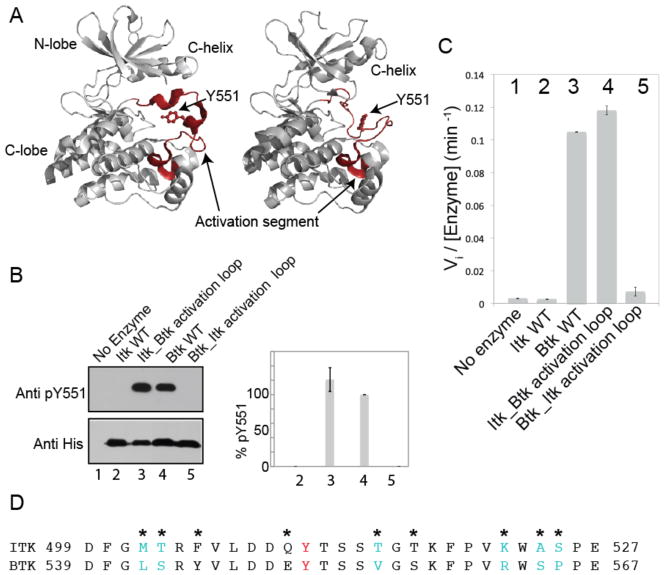
Fig. 1. Swapping the activation segments of Itk and Btk switches the catalytic activity profiles of these two related kinases.
(A) Structures of the kinase domain of Btk with the activation segment in multiple conformations [Protein Data Bank (PDB): 3GEN, 1K2P]. The N-terminal and C-terminal lobes of the kinase domain, the C-helix, and Tyr551 (Y551) on the activation loop are labeled, and the activation segment in each structure is highlighted in red and labeled. (B and C) The in vitro kinase activities of the indicated proteins were monitored by (B) Western blotting analysis for autophosphorylation on the activation loop at Tyr551 (Y551) or (C) phosphorylation of a peptide substrate (Peptide B) with 32P-ATP and determination of initial velocity (Vi). Pooled Western blotting data were quantified by normalizing the intensity of the band corresponding to pY551 of wild-type (WT) Btk (lane 4) to 100% and reporting the extent of phosphorylation of Y551 for the other proteins accordingly. Lane 1: negative control (no enzyme); lane 2: WT Itk; lane 3: the Itk_Btk activation loop; lane 4: WT Btk; lane 5: the Btk_Itk activation loop. Data in (B) and (C) are mean values ± SD from three independent experiments. The Western blot in (B) is representative of three independent experiments. (D) Amino acid sequences of the activation segments of Itk and Btk. The phosphorylation site, Y511 in Itk and Y551 in Btk is shown in red, and asterisks indicate sequence differences between the two kinases. Residues in cyan indicate the minimal sequence changes required to swap the activities of Itk and Btk [determined in (Fig. 2, A and B)].
Kinases that share high sequence similarity and belong to the same family are often assumed to share similar activity and regulatory features; however, there are indications that important differences exist between closely related kinases. One interesting example of such a difference emerges from early studies of Syk (spleen tyrosine kinase) and ZAP-70 (ζ chain–associated protein kinase of 70 kD), related tyrosine kinases that exhibit very different levels of regulation (9, 10). ZAP-70, which is found in mature T cells, exhibits much stricter control over its catalytic activity that does Syk, which is found in B cells (11).
Like ZAP-70 and Syk, the Tec family kinases interleukin-2 (IL-2)–inducible T cell kinase (Itk) and Bruton’s tyrosine kinase (Btk) are nonreceptor tyrosine kinases that function downstream of the T cell receptor (TCR) and B cell receptor (BCR), respectively (12),(13). We performed a direct comparison of the catalytic activities of Itk and Btk and found that Btk was the more active kinase, a finding that mirrors previous findings from analyses of ZAP-70 and Syk. In an effort to better understand how and why highly related kinase sequences exhibit such different levels of catalytic activity, we determined the molecular basis for the catalytic differences between Itk and Btk. We found that specific amino acids within the activation loop segments of Itk and Btk were wholly responsible for the observed differences in catalytic activity. Our findings demonstrate that activation loop dynamics, which are determined by just a few amino acids within the activation segment, directly control the catalytic efficiency of the kinase. Our findings led to construction of the first hyperactivated form of Itk, and incorporating this mutant into T cells enabled us to speculate on why specific kinase activities, or enzymatic activities in general, have evolved precise activity levels that suit the cellular environment in which they are found.
Results
Btk is a more active kinase than Itk
In our previous work on the Tec family kinases, we noted that the purified full-length kinase Btk has greater activity than full-length Itk in activity assays in vitro (14, 15). As a first step to identify the molecular basis for this unexpected difference in activity, we directly compared the catalytic activities of the kinase domains of Itk and Btk, and we found that the activity difference between these kinases was pronounced and persisted even for the isolated catalytic kinase domains (Fig. 1, B and C).
We turned our attention to the activation segment of the kinase domain, which is defined as the long flexible loop between a conserved “DFG” motif at the N-terminal end and an “APE” motif at the C-terminal end (Fig. 1, A and D). The “APE” motif is partially conserved in the Tec kinases and consists of an “SPE” and a “PPE” motif in Itk and Btk, respectively (Fig. 1D). Crystal structures of Btk show that the activation segment can undergo a major conformational change from a collapsed inactive conformation (Fig. 1A, left) to an extended conformation similar to that found in the active tyrosine kinase Lck (Fig. 1A, right) (16, 17).
Substitution of the activation segment of Itk with that of Btk activates Itk
To determine whether the activation segment was responsible for the activity differences between Btk and Itk, we swapped the activation segment sequences between the two proteins. The kinase domain of Itk containing the Btk activation segment sequence (the Itk_Btk activation loop mutant) and the kinase domain of Btk with the Itk activation segment (the Btk_Itk activation loop mutant) were expressed and purified from bacteria, and their activities were measured either by comparing the extents of phosphorylation of residues in the activation loops of each kinase [Tyr511 (Y511) for Itk and Tyr551 (Y551) for Btk] (Fig. 1B), or by monitoring the kinetics of phosphorylation (initial velocity, Vi) of a general tyrosine kinase peptide substrate, Peptide B (Fig. 1C) (15, 18). Comparison of the activities of these two swapped mutants with those of the kinase domains of wild-type Itk and Btk demonstrated that substitution of the activation segment of Itk with that of Btk activated the Itk kinase domain such that it had a level of activity similar to that of wild-type Btk, whereas substitution of the activation segment of Btk with that of Itk resulted in a Btk kinase domain that had poor activity, similar to that of wild-type Itk (Fig. 1, B and C). Thus, the activity difference between Itk and Btk is completely determined by the amino acid sequences of the activation segments of these enzymes.
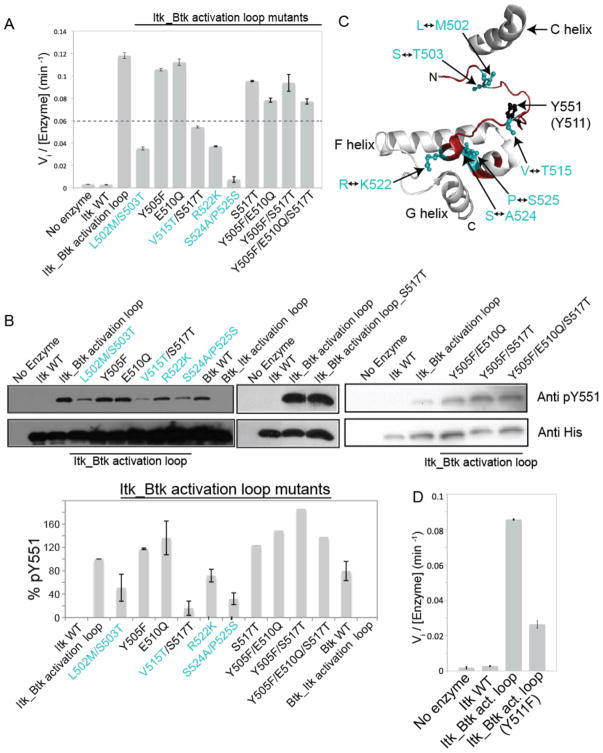
Comparison of the sequences of the kinase activation segments showed that there are nine amino acids that differ between Itk and Btk (Fig. 1D). To determine the relative contribution of each of these nine amino acids to the observed activity difference between Itk and Btk, we mutated each of these amino acids in the Itk_Btk activation loop mutant, individually or in pairs, to the corresponding Itk residues (Fig. 2, A and B). Mutation of the activation segment residues Tyr505 (Y505), Glu510 (E510), and Ser517 (S517) to the corresponding residues of Itk had no effect on the activity of Itk_Btk activation loop mutant (Fig. 2, A and B). However, mutation of the Btk activation segment residues at positions 502, 503, 515, 522, 524, and 525 to the corresponding residues in Itk reduced the kinase activity of the Itk_Btk activation loop mutant to less than half that of the original Itk_Btk activation loop protein (Fig. 2, A and B). In a complementary fashion, mutation of the Btk activation segment residues Leu542 (L542), Ser543 (S543), Val555 (V555), Arg562 (R562), Ser564 (S564), and Pro565 (P565) to the corresponding residues of Itk decreased the activity of Btk (fig. S1). Together, these findings indicate that swapping six residues between the activation segments of Itk and Btk is sufficient to completely switch the activity of these two kinases. Five of these six residues are located at the N-terminal and C-terminal anchor points of the activation segment of the kinase domain, whereas only one residue is located in the middle of the activation segment (Fig. 2C).
Fig. 2. Detailed mutational analysis of the kinase activation segment of Itk.
(A) The Itk_Btk activation loop mutant containing the entire activation segment of Btk was mutated back to Itk by modifying the indicated residues. The numbering used to indicate mutations is that of Itk. Proteins were then used in at least two experiments to determine Vi by monitoring phosphorylation of a peptide substrate (Peptide B) with 32P-ATP. Those single or double mutants whose Vi values were below half that of the Itk_Btk activation loop mutant were considered important for conferring Itk with an activity similar to that of Btk. (B) An extended set of mutants including those examined in (A) were analyzed by Western blotting to determine the extent of phosphorylation of Tyr551 (pY551) as a measurement of their overall activity. Quantification of the relative abundance of pY551 from two experiments is shown in the bar graph. The abundance of pY551 for the Itk_Btk activation loop mutant was set to 100%, and the abundances of pY551 for all of the other proteins were calculated as percentages of that of the Itk_Btk activation loop mutant. Four mutants (Itk_Btk activation loop S517T single mutant, Y505F/E510Q double mutant, Y505F/S517T double mutant and Y505F/E510Q/S517T triple mutant) were tested only once by Western blotting and were not pursued further because the difference in activity between these mutants and wild-type Btk were small when determined both by Western blotting and measurement of Vi (see A). Western blots are representative of two experiments. (C) The structure of the activation segment of Btk (PDB: 1K2P) illustrates the location of the minimal set of residues that must be swapped between Itk and Btk to confer Itk with the activity of Btk. As in (A) and (B), residue numbering is that of Itk. The phosphorylation sites in the activation segments of Itk (Y511) and Btk (Y551) are shown. The N-terminal and C-terminal ends of the activation segment (shown in red) are labeled, and the C, F, and G helices are included for reference. (D) Phosphorylation of Itk Tyr511 is required for the full activity of the Itk_Btk activation loop. The recombinant proteins WT Itk, Itk_Btk activation loop, and the same Itk_Btk activation loop construct with an additional Y511F mutation were used in three experiments to measure Vi by monitoring phosphorylation of a peptide substrate (Peptide B) with 32P-ATP. Mean Vi values ± SD were determined from three independent experiments.
Activation of Itk by substituting its residues with those of Btk only partially depends on phosphorylation at Tyr511
Activation loop phosphorylation is a well-studied activation mechanism for many kinases, including those of the Tec family (19, 20). To determine the extent to which enhancement of the activation of Itk by substitution of its activation segment residues with those of Btk required phosphorylation on the activation loop of Itk, we mutated Tyr511 to phenylalanine and assessed the kinase activity of the mutant protein by monitoring phosphorylation of the Peptide B substrate. The Y511F mutation of Itk in the context of activating Btk residues resulted in decreased kinase activity, but not to the level of activity of wild-type Itk (Fig. 2D). This finding suggests that the activating effect of the activation segment residues of Btk extended beyond increasing the accessibility of Tyr511 for phosphorylation. Based on our previous work with various Itk mutations (14), we suggest that the activation segment residues of Btk might serve to stabilize the active conformation of the kinase, either by stabilizing the conserved regulatory spine (21), the C helix, or both to a greater extent than do the corresponding activation segment residues of Itk.
Introduction of the activation segment residues of Btk into full-length Itk activates Itk
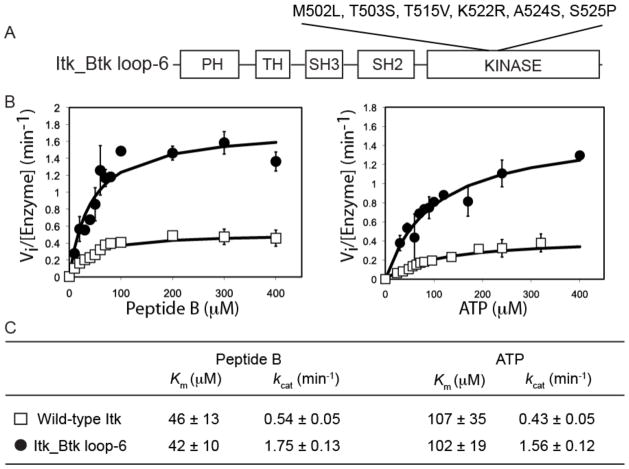
We next introduced the six activation segment residues of Btk that we identified in the earlier kinase domain studies (M502L, T503S, T515V, K522R, A524S, and S525P) into full-length Itk to determine whether these mutations also activated the full-length enzyme (Fig. 3A). This full-length Itk mutant is hereafter referred to as the Itk_Btk loop-6 mutant to indicate the minimum set of activation loop mutations required to activate the kinase activity of Itk. We expressed the Itk_Btk loop-6 mutant protein and purified it from Sf9 cells, and then we compared its activity to that of full-length, wild-type Itk. The Peptide B and ATP substrate curves indicated that the Itk_Btk loop-6 protein was more active than wild-type Itk (Fig. 3B). The Km values (an indicator of substrate affinity) of the hyperactive Itk_Btk loop-6 mutant protein for Peptide B and ATP were similar to those of wild-type Itk (Fig. 3C). The increased activity of the Itk_Btk loop-6 protein came entirely from an increase in kcat (a measure of catalytic rate); the value of kcat for the hyperactive Itk mutant was ~3.5- to 4-fold greater than that of wild-type Itk, and was the same as that of full-length Btk (15, 22). Thus, introduction of six activation segment residues of Btk into full-length Itk activated the mutant Itk similarly to full-length Btk by altering the catalytic rate of the Itk_Btk loop-6 protein, and not by increasing its affinity for its substrates. To our knowledge, Itk_Btk loop-6 is the first hyperactive form of full-length Itk.
Fig. 3. Kinetic analysis of full-length Itk and the full-length Itk_Btk loop-6 protein.
(A) Substitution of the indicated six amino acid residues in the activation segment of Itk with those of Btk was required to create the full-length Itk_Btk loop-6 mutant. The N-terminal regulatory domains of full-length Itk or Itk_Btk loop-6 proteins consists of an N-terminal pleckstrin homology (PH) domain, followed by a Tec homology (TH) domain, and Src homology (SH) domains SH3 and SH2. (B) Peptide B and ATP substrate curves comparing the in vitro phosphorylation kinetics of wild-type Itk (open squares) and the Itk_Btk loop-6 mutant (filled circles). The magnitude of the activity increase for full-length Itk_Btk loop-6 compared to that of full-length WT Itk was less than that observed for the corresponding WT and mutant isolated kinase domain fragments (Fig. 1B), whereas the overall activity (Vi value) was higher for the full-length proteins as compared to the Vi of the isolated kinase domain fragments. (C) Kinetic constants for the WT Itk and Itk_Btk loop-6 proteins were derived from two independent experiments. Error bars represent SD from the mean.
Hyperactive Itk is more active than wild-type Itk in T cells
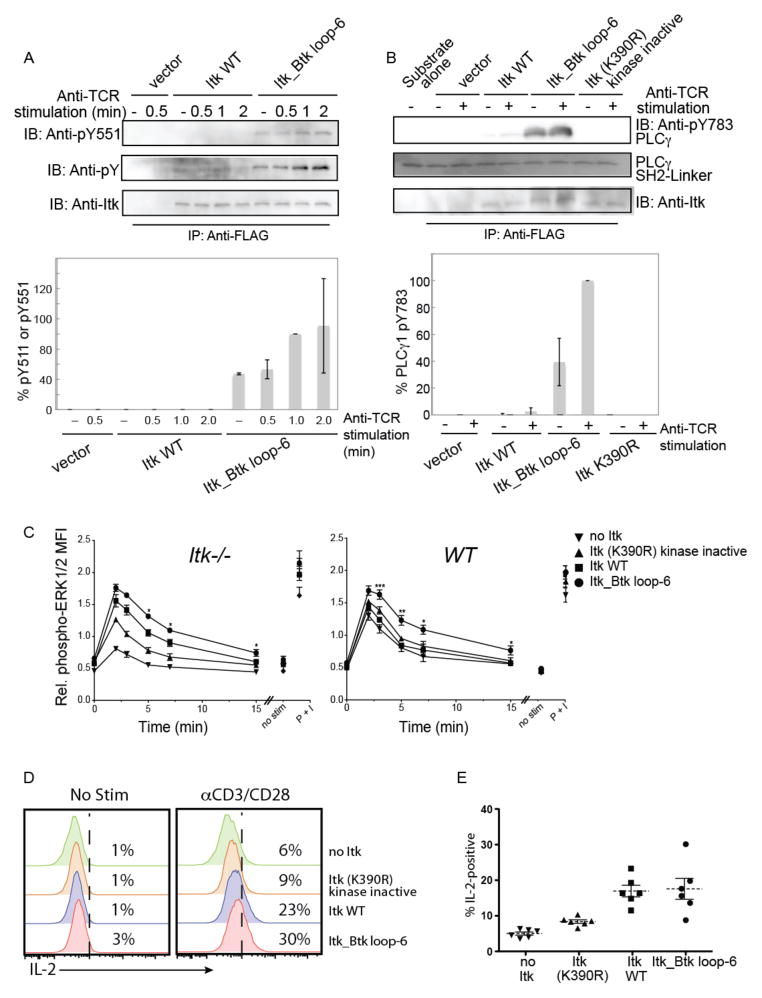
To determine whether the Itk_Btk loop-6 mutant was also more active than wild-type Itk in a cellular setting, we transfected Jurkat cells with plasmids encoding either FLAG-tagged wild-type Itk or the Itk_Btk loop-6 mutant. We stimulated the cells with anti-CD3 antibody to activate the TCR, and then monitored Itk activity by Western blotting analysis of the abundance of phosphorylated Tyr511 (pY511), or by means of an in vitro kinase assay with immunoprecipitated Itk enzyme and phosphoylipase C γ1 (PLC-γ1) as the substrate. Cells expressing the full-length Itk_Btk loop-6 mutant exhibited enhanced phosphorylation of the activation loop residue Tyr511 compared to that in cells expressing wild-type Itk (Fig. 4A). In addition, immunoprecipitated Itk_Btk loop-6 from stimulated Jurkat T cells generated more phosphorylated PLC-γ1 than did wild-type Itk (Fig. 4B). Interestingly, cells expressing Itk_Btk loop-6 mutant had higher basal activity as compared to cells expressing wild-type Itk. In the absence of TCR stimulation, Itk_Btk loop-6 had higher phosphorylation on the activation loop residue Tyr511 and phosphorylated PLC-γ1 to a greater extent as compared to wild-type Itk (Fig. 4, A and B). Thus, substitution of just six activation segment residues among the 619 amino acids of full-length Itk with the corresponding residues of Btk was sufficient to activate Itk in Jurkat cells.
Fig. 4. Hyperactive Itk enhances phosphorylation of the activation loop and PLC-γ1 in Jurkat cells, and enhances and prolongs TCR signaling.
(A) Jurkat cells were transfected with plasmids encoding FLAG-tagged WT Itk or the Itk_Btk loop-6 mutant. Twenty four hours after transfection, cells were left unstimulated or were stimulated with C305 (anti-TCR) antibody for the indicated times. Cells were lysed, subjected to immunoprecipitation with anti-FLAG antibody, and the extent of phosphorylation on the activation loop was determined by Western blotting analysis of samples for pY511 (top blots). Data are representative of two independent experiments. Band intensities from both experiments were quantified by densitometric analysis. The abundance of pY511 in the Itk_Btk loop-6 mutant after 1 min of TCR stimulation was normalized to 100%, and the relative amounts of pY511 for the other proteins at all time points were determined. (B) WT Itk or the Itk_Btk loop mutant were immunoprecipitated from unstimulated or TCR-stimulated Jurkat cells and their activities were monitored by an in vitro kinase assay with the SH2-linker region of PLC-γ1 as a substrate. The abundance of PLC-γ1 pY783 was determined by Western blotting analysis with a specific antibody. Data are representative of two independent experiments. Band intensities from both experiments were quantified by densitometric analysis. The abundance of PLC-γ1 pY783 in the reaction containing the Itk_Btk loop-6 mutant (normalized for total Itk) after TCR stimulation was normalized to 100%, and the relative amounts of PLC-γ1 pY783 from the reactions with the other proteins were determined. (C) Itk−/− or WT OT-II TCR transgenic CD4+ T cells were infected with control retrovirus (no Itk) or with retroviruses expressing the indicated Itk constructs. Thy1.1+ cells were isolated and were left untreated or were incubated with anti-CD3 antibody (αCD3) to stimulate the TCR for 2, 3, 5, 7, or 15 min. Cells were fixed and permeabilized, and incubated with antibodies specific for CD4, Thy1.1, and pERK1/2. Graphs show the normalized mean fluorescence intensities (MFIs) ± SEM for pERK1/2 in gated CD4+ Thy1.1+ cells (see fig. S2B). Values were normalized within each experiment to the average MFI of pERK1/2 staining for all samples in that experiment, and data are means ± SEM from 4 to 6 independent experiments. Statistically significant differences between cells expressing hyperactive Itk_Btk loop-6 compared to cells expressing WT Itk are indicated: *P < 0.05; **P < 0.01; ***P < 0.001. Non-stimulated cells (no stim) were not incubated with αCD3. As a positive control for the activation of ERK1/2, cells were stimulated with PMA and ionomycin (P+I) for 4 min. (D and E) The Itk_Btk loop-6 construct does not promote increased IL-2 production after TCR stimulation of primary T cells. Itk−/− OT-II TCR transgenic CD4+ T cells were infected with control retrovirus (no Itk) or with retroviruses expressing the indicated Itk proteins. Infected cells were left untreated or were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies (αCD3/CD28) for 5 to 6 hours before being incubated with fluorescently tagged antibodies specific for CD4, TCRβ, Thy1.1, and IL-2. (D) Histograms show representative flow cytometry plots of IL-2 staining in gated CD4+, TCRβ+, Thy1.1+ cells. (E) The graph shows a compilation of flow cytometry data from six independent experiments showing the percentages of cells under each condition that were positive for IL-2. Dashed horizontal lines indicate the means and bars represent the SEM. The percentages of cells expressing WT Itk or the Itk_Btk loop-6 mutant that produced IL-2 were significantly greater than those of cells expressing kinase-deficient Itk or those that had no Itk (P < 0.05). There was no statistically significant difference in the amounts of IL-2 produced by cells expressing WT Itk and those expressing they hyperactive Itk_Btk loop-6 mutant.
To test whether the Itk_Btk loop-6 mutant led to enhanced signaling in the physiological setting of primary T cells, we used retroviral transduction to introduce Itk constructs into murine Itk-deficient (Itk−/−)CD4+ T cells. To ensure the production of equivalent amounts of the various Itk constructs, we used a retroviral vector that expresses enhanced green fluorescent protein (eGFP) from an internal ribosome entry site (IRES) on the same mRNA as that encoding Itk; in all experiments, only GFP+ cells with comparable magnitudes of GFP fluorescence were included in the analysis. Furthermore, to confirm the equivalent abundances of the various Itk constructs, we sorted GFP+ cells by flow cytometry and determined Itk protein abundance by Western blotting analysis (fig. S2A). Transduced cells were stimulated by crosslinking TCR-associated CD3 with an activating antibody, and then were analyzed for the phosphorylation (and thus activation) of the mitogen-activated protein kinases (MAPKs) extracellular signal–regulated kinase 1 (ERK1) and ERK2 (collectively known as ERK1/2), which are downstream of PLC-γ1 activation in T cells. In this system, cells with the Itk_Btk loop-6 mutant exhibited enhanced ERK1/2 phosphorylation compared to that in cells with wild-type Itk (Fig. 4C and fig. S2B). In addition to the enhanced magnitude of phosphorylation of ERK1/2 at the peak time of the response (2 to 3 min after stimulation), cells expressing Itk_Btk loop-6 also exhibited prolonged signaling that lasted up to 15 min after stimulation. We also observed that the Itk_Btk loop-6 mutant enhanced TCR signaling in wild-type CD4+ T cells, even in the presence of endogenous Itk (Fig. 4C). In a long-term assay of TCR signaling, the measurement of IL-2 production 5 to 6 hours after stimulation, we detected no enhancement in the response of Itk−/− CD4+ T cells expressing the Itk_Btk loop-6 mutant compared to that in cells expressing wild-type Itk (Fig. 4, D and E). This latter result may be accounted for by the low abundance of Itk protein in the retrovirally transduced Itk−/− T cells (fig. S2A).
Tyr511 is more readily phosphorylated in Itk containing the Btk activation segment
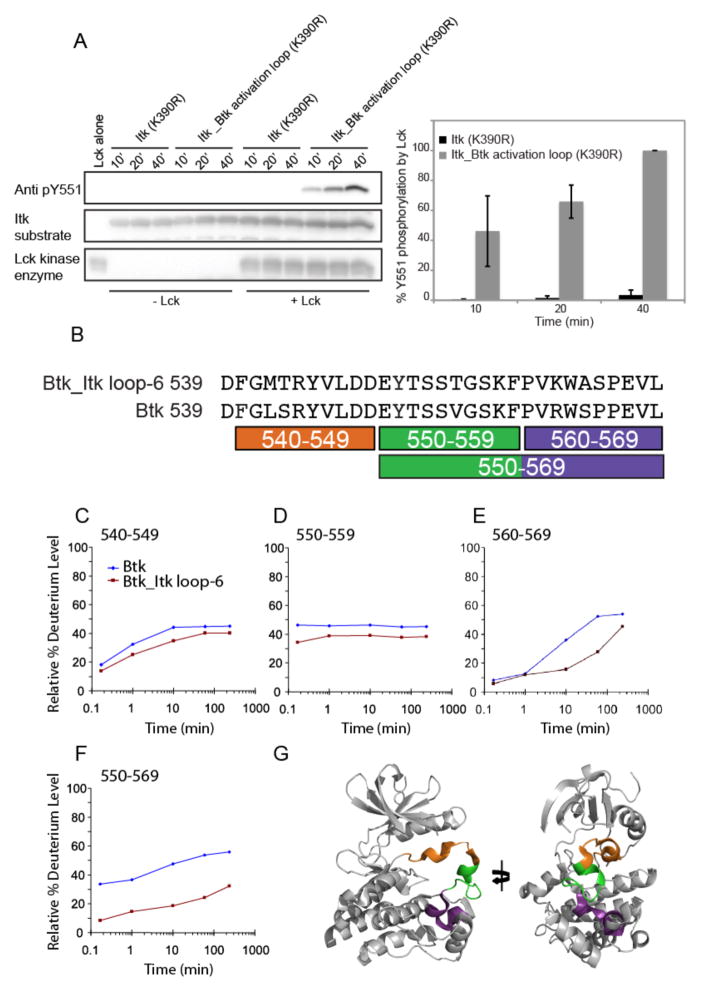
Having successfully swapped the catalytic activities of Itk and Btk by swapping specific activation segment residues between the two enzymes, we next wished to determine how a small subset of the total amino acid residues in these kinases could confer such different activity profiles. We reasoned that the increased flexibility, accessibility, or both of the activation segment could lead to increased phosphorylation of Btk Tyr551 (or Itk Tyr511), providing one mechanism to explain the enhanced activity of Btk and the Itk_Btk activation loop mutant compared to that of wild-type Itk. To test this, we purified catalytically inactive (K390R) versions of the kinase domains of wild-type Itk and of the Itk_Btk activation loop mutant, and subjected them to in vitro phosphorylation by purified Lck, the upstream Src family kinase member that phosphorylates Itk on Tyr511 in T cells (19). The abundance of pTyr511 in reactions containing the Itk_Btk activation loop mutant (K390R) was greater than that in reactions containing Itk (K390R) (Fig. 5A), consistent with the hypothesis that the Btk activation loop, particularly Tyr511, is more accessible to Lck, whereas in the Itk loop, this residue is more occluded.
Fig. 5. Tyr511 of Itk is more accessible for phosphorylation by Lck when in the context of the Itk_Btk activation loop.
(A) Kinase-deficient (K390R) mutants of Itk or the hyperactive Itk_Btk activation loop proteins were subjected to phosphorylation by Lck in an in vitro kinase assay. Phosphorylation of Itk Tyr511 was monitored at three time points (10, 20, and 40 min) by Western blotting analysis with an anti-pY511 antibody. The Western blot is representative of three independent experiments. Densitometric data from all three experiments were quantified and are presented in the bar graph on the right. To normalize for exposure times between Western blots from different experiments, we set the abundance of pY551 in the Itk_Btk activation loop (K390R) mutant after 40 min of stimulation to 100% in each independent experiment. The abundances of the phosphorylated Itk (K390R) protein (all three time points) and Itk_Btk activation loop (K390R) protein (at the 10- and 20-min time points) are shown relative to that for the Itk_Btk activation loop (K390R) protein at the 40-min time point. Data summarized from three independent experiments with error bars representing SD from the mean. (B) Pepsin digestions of the WT Btk and Btk_Itk loop-6 mutant proteins produced coincident peptides, which are indicated under the sequences that covered the entire activation segments of both proteins. (C to F) Deuterium exchange was measured for the WT Btk and Btk_Itk loop-6 mutant proteins (see the full dataset in fig. S3) and data were plotted as the relative percentage of deuterium incorporation versus time for the peptides identified in (B) for Btk (blue diamonds) and Btk_Itk loop-6 (red squares). (G) The locations of the individual peptides, color-coded as in (B), are shown on the structure of the kinase domain of Btk, with the activation segment in the collapsed inactive conformation (PDB: 3GEN).
The activation loop segment of Btk is more solvent-accessible than that of Itk
Based on the differential extents of tyrosine phosphorylation within the Itk and Btk activation loops (Fig. 5A), we next directly tested whether the activation segment of Btk was more solvent-accessible than that of Itk in experiments with hydrogen-deuterium exchange mass spectrometry (HDX-MS). To avoid unwanted heterogeneity in the sample because of Btk autophosphorylation of the activation segment, we performed the HDX-MS analysis on kinase-inactivated Btk mutants (Btk K430R, which corresponds to position 390 in Itk). In addition, because of the sample demands of biophysical techniques such as HDX-MS and nuclear magnetic resonance (NMR) spectroscopy, we used the Btk kinase domain and the mutant Btk kinase domain containing residues from the activation loop segment of Itk (Btk_Itk loop-6) for ease of protein production (see the Materials and Methods for details). The lack of a bacterial expression system to produce large amounts of soluble Itk kinase domain prevented us from performing similar biophysical analysis on the Itk system.
The complete peptide map, as well as the raw deuterium uptake curves, for all of the peptides derived from Btk kinase domain and the Btk_Itk loop-6 mutant were determined (fig. S3, A to E). Comparison of the deuterium uptake curves of peptides derived from the activation segment of wild-type Btk or the Btk_Itk loop-6 mutant (Fig. 5, B to F) showed that the activation segment of wild-type Btk incorporated deuterium more readily than did that of the Btk_Itk loop-6 mutant. This suggests that the activation segment of wild-type Btk is more flexible and therefore more easily deuterated than is the corresponding segment of the Btk_Itk loop-6 mutant. These results are consistent with our observation that the activation loop tyrosine (Tyr511 for Itk and Tyr551 for Btk) was more readily phosphorylated by Lck in the context of the Btk activation segment residues (Fig. 5A). Peptides derived from the C-terminal third of the activation segments of Btk and Btk_Itk loop-6 (Fig. 5, D to G) showed the most marked differences in deuterium uptake. This finding is consistent with biochemical results that showed that mutation of residues at the C-terminal end of the activation segment of Itk or Btk resulted in the most substantial changes in kinase activity (Fig. 2 and fig. S1). Peptides derived from the N-terminal third as well as the middle third of the activation segment (Fig. 5, C and D) showed minor differences in deuterium uptake.
Distinct dynamics characterize the activation segments of Itk and Btk
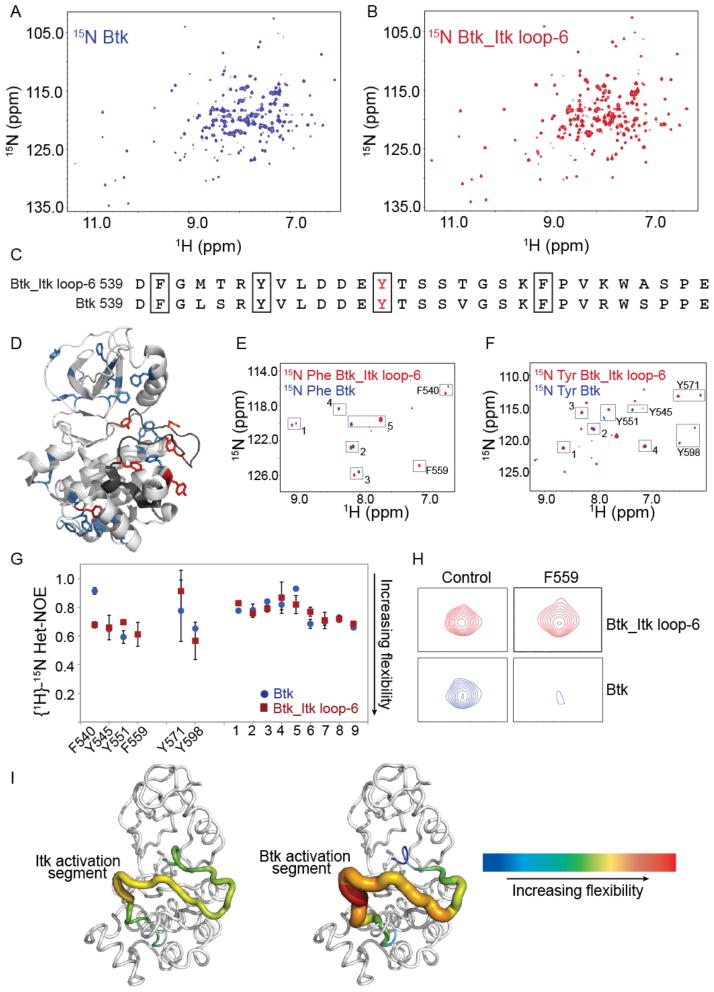
To complement the dynamic picture that emerged from HDX-MS analysis, we next analyzed the two kinase domains (Btk and Btk_Itk loop-6) by NMR spectroscopy. The resonance frequencies of every hydrogen (1H) that is directly attached to a nitrogen (15N) is measured using the Heteronuclear Single Quantum Correlation (HSQC) spectrum, allowing a direct measure of each amide N-H group in the protein. Given the large size of the kinase domain, we used a TROSY (Transverse Relaxation-Optimized Spectroscopy) version of the HSQC to improve spectral quality. Comparison of the 1H-15N TROSY HSQC spectrum of the uniformly labeled kinase domain of Btk (Fig. 6A) with that of the Btk_Itk loop-6 mutant (Fig. 6B) showed differences in both chemical shifts and linewidths. The linewidths observed for the Btk_Itk loop-6 sample were quite uniform compared to those of Btk. In contrast, the linewidths for the Btk sample are variable across the spectrum. The linewidth differences among the Btk amide resonances indicate variations in dynamics throughout the kinase domain of Btk that depended on the amino acid sequence of the activation segment. We pursued a residue-selective isotopic labeling approach to obtain a partial set of sequence-specific chemical shift assignments for the kinase domain of Btk. Phenylalanine and tyrosine were selected for specific labeling based on their lower propensity for isotope scrambling (metabolic interconversion to other amino acids during protein expression in bacteria), as well as because of the relatively low number of these residues in the kinase domain. Comparison of the activation segments of wild-type Btk and the Btk_Itk loop-6 mutant showed that the residues Phe450 (F540), Tyr545 (Y545), Tyr551 (Y551), and Phe559 (F559) of Btk were common to both proteins, and hence could serve as effective probes to monitor the dynamics of the activation segment in wild-type Btk and the Btk_Itk loop mutant (Fig. 6, C and D). Chemical shift assignments for the specifically 15N-labeled residues in Btk and Btk_Itk loop-6 (Fig. 6, E and F) were obtained by site-specific alanine mutagenesis (figs. S4 and S5).
Fig. 6. The C-terminus of the kinase activation segment of Btk is more dynamic than that of Itk.
(A and B) Comparison of the 1H-15N TROSY HSQC spectra of the uniformly 15N labeled (A) kinase domain of WT Btk and (B) kinase domain of the Btk_Itk loop-6 mutant. Data is representative of at least three independent experiments. (C) Comparison of the sequences of the activation segments of WT Btk and the Btk_Itk loop-6 mutant. (D) Structure of the kinase domain of Btk (PDB: 1K2P) showing the tyrosine and phenylalanine residues throughout the protein. Six tyrosine and phenylalanine backbone amide resonances have been assigned (shown in red), whereas the remainder of the tyrosines and phenylalanines (in blue) have not been assigned. (E and F) 1H-15N TROSY HSQC spectra for the kinase domains of WT Btk and the Btk_Itk loop-6 mutant specifically labeled with (E) 15N-Phe and (F) 15N-Tyr. Peaks that were assigned are labeled, and the unassigned peaks in the spectra that correspond to either phenylalanine or tyrosine resonances are numbered. (G) Steady-state {1H}-15N heteronuclear NOE data for WT Btk (blue circles) and the Btk_Itk loop-6 mutant (red squares) are plotted for assigned and unassigned tyrosines and phenylalanines. F540, Y545, Y551, and F559 reside in the activation loop segment of each kinase. (H) Btk F559 shows extensive line broadening in the context of WT Btk as compared to the Btk_Itk loop-6 mutant. (I) Dynamical differences based on Het-NOE values for the activation loop segments of Itk (left) and Btk (right) are plotted onto the structure of WT Btk (PDB: 1K2P). Increasing flexibility is indicated by the color and width of the structural trace.
We next measured the steady-state {1H}-15N heteronuclear NOE for the backbone amides of each of the phenylalanine and tyrosine resonances. The backbone {1H}-15N heteronuclear NOE (Nuclear Overhauser Effect) provides information about the motion of individual N-H bond vectors. Amide bonds undergoing motions that are faster than the overall tumbling of the molecule show a decreased NOE while those undergoing slower motions are characterized by higher NOE values. In spectra of the selectively labeled proteins, nine resonances were identified as corresponding to phenylalanine or tyrosine, but were not specifically assigned. The NOE values for these resonances did not differ between wild-type Btk and the Btk_Itk loop-6 mutant (Fig. 6G). Tyr571 (Y571) and Tyr598 (Y598) were assigned previously (23), and also showed no difference within error between the two samples (Fig. 6G). Focusing our attention on activation segment dynamics, the NOE values measured for Phe540 (F540), Tyr545 (Y545), Tyr551 (Y551), and Phe559 (F559) within the kinase domain of the Btk_Itk loop-6 mutant were all between 0.6 and 0.7 (Fig. 6G), consistent with uniform flexibility across the activation segment containing the six Itk residues. These NOE values were consistent with the range of NOE values expected for residues in loop regions of folded proteins (24).
The activation segment of the kinase domain of Btk exhibited distinct dynamics. The NOE value measured for Phe540 (F540) at the N-terminus of the activation loop was ~0.9, indicating rigidity on the nanosecond timescale compared to the corresponding residue within the activation loop of the Btk_Itk loop-6 mutant (Fig. 6G). Phe540 is one of five kinase regulatory spine residues (14), and the greater rigidity observed for this residue at the N-terminal end of the activation segment of Btk may be indicative of a higher degree of pre-organization in the regulatory spine of the more active Btk compared to that of the less active Btk_Itk loop-6 mutant. There was no difference in the flexibility of Tyr545 between Btk and the Btk_Itk loop-6 mutant, and Tyr551 (the site of phosphorylation) was only slightly more flexible in Btk than in the Btk_Itk loop-6 mutant. In contrast to all of the other resonances examined by selective labeling, the peak corresponding to Phe559 (F559) at the C-terminal end of the activation segment in Btk showed extensive line-broadening in the HSQC spectrum (compare to the peak for the corresponding residue in the Itk loop, Fig. 6H) prohibiting NOE measurement for this residue. Nevertheless, the extent of line broadening clearly indicates that Phe559 of Btk underwent conformational exchange on the microsecond, or greater, timescale (25). Thus, the N-terminal and C-terminal regions of the activation loop in Btk are characterized by very different types of motions. The N-terminus of the Btk loop (Phe540) showed restricted motion on the nanosecond timescale, whereas the C-terminus of the same loop (Phe559) underwent motions extending to the microsecond timescale. The slower conformational fluctuations on the microsecond timescale for the C-terminal end of the Btk loop sequence may facilitate phosphorylation of the activation segment tyrosine by providing greater access to Tyr551 or, as has been suggested for a number of other enzyme systems (26, 27), the microsecond timescale motions observed for the Btk activation segment may contribute to catalysis.
The extent of loop dynamics based on measured NOE values and linewidth comparisons for the activation segments of Btk and Btk-Itk loop-6 were visualized on the structure of Btk (Fig. 6I). The variation in flexibility from the N-terminus to the C-terminus of the activation loop of Btk (Fig. 6I, right) was more substantial than that of the Itk loop (Fig. 6I, left). The flexibility of the C-terminus of the Btk activation loop, in particular, was consistent with the biochemical and HDX-MS analyses, which pointed to the C-terminal end of the activation segment as being the primary region responsible for the differences in activity and dynamics between Btk and Itk.
Discussion
Despite similarities in sequence and structure between closely related kinases, it is becoming increasingly clear that there are functional and regulatory differences between kinases even within the same family. For the Tec family kinases Itk and Btk, which share 58% sequence identity and 76% similarity in the isolated kinase domains, we have observed distinct extents of activity and identified a specific and small set of amino acid residues that control the catalytic efficiencies of each kinase. Swapping just six residues in the activation segments of Itk and Btk completely switched the kinetic profile of these kinases; the Itk_Btk loop-6 mutant was kinetically indistinguishable from wild-type Btk, whereas the Btk_Itk loop-6 mutant exhibited the poor kinetics of wild-type Itk (Fig. 1, B and C). We found that the basis for the switch in kinase activity derives from differences between the dynamics of the activation loop segments of Itk and Btk.
The sequence differences between Itk and Btk in their activation segments are subtle (Fig. 1D). The one exception is Pro565 (P565) at the C-terminus of the activation segment of Btk; the corresponding residue in Itk is a serine. The nature of the prolyl amino acid does not intuitively point to the greater flexibility that we have observed for the C-terminus of the Btk activation loop. The role of this proline in stabilizing the active conformation of Btk may stem from improved packing of the side chain of Pro565 of Btk with the F-helix kinase core, which enables better coupling and coordinated movements of the activation segment with other regulatory regions of the kinase. In contrast, the side chain of Ser525 of Itk is less likely to contribute to the hydrophobic interactions that stabilize the kinase core.
The localization of the functional differences between Itk and Btk to the dynamically variant activation segment is reminiscent of the tendency for kinase activation segments to be hot spots for oncogenic mutations (28). HDX studies of a mutant form of the tyrosine kinase c-Kit that is associated with gastrointestinal stromal tumors indicate that the D816H mutation, at the N-terminus of the activation segment, increases solvent-accessibility at the C-terminal end of the activation segment (29). Similar to the sequence changes that we introduced into Itk to produce the activated Itk_Btk loop-6 mutant, a change in activation loop dynamics induced by the D816H mutation of c-Kit correlates with a higher rate of autophosphorylation compared to that of the wild-type kinase (29). Indeed, it is likely that many disease-causing mutations fundamentally alter protein dynamics, which leads to functional deregulation. Several mutations that cause X-linked agammaglobulinemia (XLA) have been identified in the activation segment of Btk (30). Additionally, the COSMIC (catalogue of somatic mutations in cancer) database lists several mutations in the activation segments of Btk and Itk. Future studies will test the effects of these mutations on kinase dynamics. A clear understanding of how conformational dynamics in specific regions within the kinase scaffold controls catalytic function should lend itself to development strategies for new inhibitors.
The C-terminal end of the activation segment is important for the binding, alignment, or both of the substrate in other systems (31). Activation loop swapping between Itk and Btk altered the amino acid residues at the C-terminal end of the activation segment but they affected only kcat; the Km values measured for each enzyme and substrate were the same. We previously demonstrated that protein substrate selection in the Tec kinases occurs through a docking interaction that is mediated by helix G of the kinase domain, which is a region separate from, but adjacent to, the C-terminal end of the activation segment (23, 32). This docking interaction may be the primary determinant for substrate selection in the Tec kinases, or else the docking of substrate onto the G helix may allosterically alter the flexibility of the activation segment in a manner that promotes catalytic activity. It is also possible that the sequence of the activation segment might affect Km for substrates other than the model peptide substrate used here. Regardless, the amino acid differences between the activation segments of Itk and Btk have a clear effect on kcat, which strongly suggests that evolutionary pressures have resulted in sequence differences that altered loop dynamics and reduced the catalytic activity of Itk, which is found in T cells, compared to that of the kinase Btk, which is found in B cells.
T cells and B cells have remarkably similar signaling machinery downstream of the TCR and BCR, respectively (33, 34). Stimulation of TCRs and BCRs leads to the activation of Src family kinases, which phosphorylate receptor-associated immunoreceptor tyrosine-based activation motifs (ITAMs). The phosphorylated ITAM sites then recruit ZAP-70 in T cells or Syk in B cells, which are also activated by Src family kinases. The kinases ZAP-70 and Syk then phosphorylate the adaptor protein Src homology 2 (SH2) domain–containing leukocyte phosphoprotein of 76 kD (SLP-76) in T cells and SLP-65 in B cells, which in turn recruit the Tec kinases Itk in T cells and Btk in B cells, respectively. Our finding that Btk is a more efficient kinase than Itk parallels previous findings that Syk outperforms ZAP-70 and is under less regulatory control (9–11). We speculate that the weaker catalytic activity and more stringent regulation of the T cell kinases (ZAP-70 and Itk) compared to their B cell counterparts (Syk and Btk) might serve a regulatory role in T cell signaling that is not needed in B cells.
T cell activation is a tightly controlled process, which is necessary to avoid autoimmunity by preventing aberrant responses to weakly recognized self-antigens. Our data indicate that primary T cells expressing even a small amount of the Itk_Btk loop-6 mutant have enhanced and prolonged short-term responses to TCR stimulation. One outcome of this hyperactive Itk may be to lower the threshold amount of TCR stimulation required to promote T cell activation, thereby predisposing the system to autoimmune disease. Thus, a poorly active Itk in wild-type T cells may constitute an important mechanism for dampening responses to low abundance or weakly-recognized self-antigens. Indeed, in unrelated work aimed at developing an inhibitor screen, Douhan et al. showed that DT40-Btk−/− cells expressing a full-length Btk that contains the kinase domain of Itk in place of the kinase domain of Btk requires a four- to six-fold higher concentration of agonist antibody to elicit a calcium signaling response compared to the same cells expressing wild-type Btk (35). Moreover, the magnitude of the calcium response is lower in cells expressing the chimeric Btk molecule as compared to wild-type Btk (35), further supporting our notion that the weakly catalytic kinase domain of wild-type Itk is fine-tuned to maintain appropriate T cell responses.
It is also likely that additional mechanisms regulate the magnitude and duration of T cell responses. In spite of there being enhanced short-term signaling in T cells expressing the Itk_Btk loop-6 mutant, we did not observe an overall increase in IL-2 production. This latter response requires a complex series of events, including the activation and de novo synthesis of transcription factors, changes in chromatin structure, and reorganization of the actin cytoskeleton. Given this complexity, there are undoubtedly multiple checkpoints in place to regulate the throughput of this pathway. Nonetheless, it is evident that a small number of amino acid changes in the activation segment of the kinase domain of Itk are capable of altering the dynamic behavior of this loop, with consequences for enzyme activity in contexts ranging from the isolated protein to intact primary T lymphocytes.
Materials and Methods
Constructs
The baculoviral constructs for full-length mouse Itk and Btk have been described previously (15). The bacterial constructs for the isolated kinase domains (without the SH2-kinase linker) of mouse Lck (amino acid residues 230 to 509), Itk (356 to 619), and Btk (396 to 659) have been described elsewhere (14, 36). Full-length wild-type, kinase inactive (K390R), and hyperactive Itk (Itk_Btk loop-6) were also cloned into the pIRES2-EGFP vector (Clontech) for mammalian cell expression. The full length Itk_Btk loop-6 construct contains six mutations in the activation segment: M502L, T503S, T515V, K522R, A524S, and S525P. The bacterial construct for the C-terminal, SH2-linker construct of PLC-γ1 has been described elsewhere (37). All mutations were introduced with a site-directed mutagenesis (SDM) kit (Stratagene). All constructs were verified by sequencing at the Iowa State University DNA synthesis and sequencing facility.
Protein expression and purification
Baculoviral constructs were expressed and purified from Sf9 or High five cells as described previously (15). The bacterial constructs encoding the His-tagged kinase domains of Itk and Btk were expressed and purified from ArcticExpress cells (Stratagene) as described previously (36). None of the kinases was co-expressed with the Src family kinase Lck to avoid differential extents of phosphorylation of the activation loop in the in vitro kinase assays. For the HDX-MS analysis, the proteins were further purified by size exclusion chromatography (Hiload Superdex 26/60 75 pg, GE Healthcare). The final buffer consisted of 20 mM Tris (pH 8.0), 150 mM sodium chloride, 10% glycerol. The expression and purification of the C-terminal SH2-linker fragment of PLC-γ1 has been described elsewhere (37).
Kinase assays and Western blotting
In vitro kinase assays were performed by incubating the isolated kinase domain of Itk or Btk in kinase assay buffer [50 mM Hepes (pH 7.0), 10 mM MgCl2, 1 mM dithiothreitol (DTT), bovine serum albumin (BSA, 1 mg/ml), 1 mM Pefabloc, 200 μM ATP] at room temperature for 1 hour. The samples were boiled, resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Western blotting with an antibody against Btk phosphorylated at Tyr551 (anti-Btk pY551, BD Biosciences), anti-FLAG antibody (Sigma), or anti-His antibody (Upstate), as described previously (15). The anti-Btk pY551 antibody was also used to detect the phosphorylation of Tyr511 of Itk. Kinetic parameters for full-length wild-type and mutant Itk proteins were derived from radioactive assays using 32P-ATP and a biotinylated Peptide B substrate that were described previously(15). The substrate Peptide B (aminohexanoyl biotin-EQEDEPEGIYGVLF-NH2) is an artificial tyrosine kinase substrate that was generated by combining sequences that are derived from hematopoetic-lineage cell–specific protein and a peptide library of optimized Lck peptide substrates (38). The tyrosine residue targeted by Itk and Btk is in bold.
Jurkat cell experiments
Jurkat E6-1 cells were maintained in RPMI-1640 medium supplemented with 10 % fetal bovine serum (FBS) and penicillin and streptomycin (each at 100 U/ml, Invitrogen). The cells were electroporated at 975 mF and 260 V with a Biorad Gene Pulser II with 10 μg of the appropriate DNA. Twenty-four hours after transfection, the cells were rinsed once with RPMI-1640 medium. Then, 1 × 106 cells were incubated at 37°C for 10 min and stimulated with an Anti-T-cell receptor (clone C305) antibody (6.5 mg/ml, Millipore) for 0.5, 1, and 2 min. The cells were lysed on ice in RIPA buffer [25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) with 1× EDTA-free Halt Protease and Phosphatase Inhibitor cocktail (ThermoScientific). For the unstimulated controls, C305 antibody (6.5 mg/ml) was added after cell lysis. The cell lysate was centrifuged and the supernatant was incubated with 20 μl of anti-FLAG resin (Sigma) for 2 hours at 4°C. The resin was rinsed and boiled in SDS-PAGE sample buffer. The extent of phosphorylation of Itk Tyr511 was monitored by Western blotting analysis with anti-Btk pY511 antibody, as described earlier. The in vitro kinase assay was performed by incubating the immunoprecipitated Itk enzyme with the purified C-terminal SH2-linker fragment of PLC-γ1 at a final concentration of 10 μM in the kinase assay buffer for 1 hour at room temperature. The reaction mixture was boiled in SDS-PAGE sample buffer. Kinase activity was monitored by Western blotting analysis with an anti- PLC-γ1 pY783 antibody, as described previously (37).
Mice
Rag1−/− OT-II transgenic C57BL/6 mice (model 4234-F/M) were purchased from Taconic and were crossed with Itk−/− C57BL/6 mice (39). Animals were housed at the University of Massachusetts Medical School animal facility in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines.
Retrovirus production
Complementary DNAs (cDNAs) encoding wild-type, kinase-deficient (K390R) (19), and hyperactive Itk were cloned into the pMiT vector (40). Human embryonic kidney (HEK) 293T cells were co-transfected with the X-tremeGENE 9 DNA transfection reagent (Roche), 1 μg of the pCL-Eco packaging vector, 0.1 μg of pmaxGFP reporter (Amaxa), and 2 μg of each of the the pMiT2 Itk-derivatives or 2 μg of empty pMiT-v5 vector. Cell supernatants were harvested two to four times during the 48 to 72 hours after infection and were stored at −80°C before being used for infections.
T cell purification and retroviral infection
Spleen and lymph node cells were isolated from wild-type and Itk−/− OT-II transgenic mice. CD4+ T cells were purified and stimulated on plates coated with anti-CD3 antibody (αCD3, clone 145-2C11, 0.8 μg/ml) and anti-CD28 antibody (αCD28, clone 37.51, 2 μg/ml). After two days, cells were mixed with 1 ml of retrovirus-containing supernatant with polybrene (8 μg/ml) and were centrifuged at 1600g for 2.5 hours at room temperature. Cells were then cultured in medium containing IL-2, IL-4, and IL-7 (each at 10 ng/ml) for 2 to 5 days. For the pERK1/2 assay, infected Thy1.1-expressing cells were enriched with αCD90.1(Thy1.1) microbeads before being cultured in cytokine-containing medium.
T cell stimulation assays
For IL-2 stimulation assays, cells were stimulated for 5 to 6 hours on plates coated with αCD3 and αCD28 antibodies in the presence of GolgiStop (Life Technologies) and GolgiPlug (Life Technologies). Cells were incubated with fluorescently tagged antibodies specific for Thy1.1, CD4, and TCRβ, fixed and permeabilized with the BD Cytofix/Cytoperm kit (BD Biosciences), and then were incubated with anti-IL-2 antibody acccording to the manufacturer’s protocol. For pERK analysis, Thy1.1+ cells were incubated with biotinylated anti-CD3 antibody for 5 min, and then were stimulated for the indicated times with streptavidin at 37°C. Reactions were stopped by the addition of 4% paraformaldehyde warmed to 37°C. After fixation, cells were permeabilized with 90% methanol at −20°C, and cells were stained with anti-pERK1/2 antibody (BD Biosciences), as previously described (41). For positive controls, cells were stimulated for 4 min with PMA (75 ng/ml, Sigma-Aldrich) and ionomycin (600 ng/ml, Calbiochem). Samples were analyzed with a BD LSRII flow cytometer (BD Biosciences), and data were analyzed with FlowJo (TreeStar Inc.) software. Statistical analysis was performed with the student’s t test in Graphpad Prism 5 software.
Western blotting analysis
Cells were lysed in RIPA buffer, and proteins were resolved by SDS-PAGE and then transferred to membranes. Membranes were incubated with anti-Itk antibody (clone 6k237; US Biological) or an anti-ERK antibody (Cell Signaling Technology) as a loading control. Band intensities were analyzed with a Versadoc Imager and Quantity One software (Bio-Rad).
Deuterium exchange reactions
Kinase-deficient (K430R) forms of Btk and Btk_Itk loop-6 mutant (L542M, S543T, V555T, R562K, S564A, and P565S) proteins carrying a Y617P mutation (23) were purified from bacteria. Deuterium labeling was initiated with an 18-fold dilution of an aliquot (80 pmoles) of Btk and Btk_Itk loop-6 protein into a buffer containing 99.9 % D2O, 20 mM Tris, 150mM NaCl, 10% glycerol, pD 8.01 (pD = pH + 0.4). At specific time points, the labeling reaction was quenched with the addition of an equal volume of quench buffer [150 mM potassium phosphate (pH 2.47),]. Quenched samples were immediately frozen on dry ice until required for liquid chromatography-mass spectrometry (LC-MS) analysis. Quenched samples were rapidly thawed and injected immediately into a Waters nanoACQUITY with HDX technology for online pepsin digestion and ultra performance liquid chromatography (UPLC) separation of the resulting peptic peptides, and were analyzed as reported previously (42). All mass spectra were acquired with a WATERS SYNAPT HDMSE mass spectrometer. The data were analyzed with DynamX software. Relative deuterium amounts for each peptide were calculated by subtracting the average mass of the undeuterated control sample from that of the deuterium-labeled sample for isotopic distributions corresponding to the +1, +2, or +3 charge state of each peptide. The data were not corrected for back exchange and are therefore reported as relative (43, 44). Calculations of the percentage deuterium incorporation by each peptide also took into account the lack of an exchangeable amide in proline residues.
NMR analysis
Uniformly 15N-labeled isolated kinase domains of Btk or of Btk containing the Itk activation segment (Btk_Itk loop-6) were produced in E. coli BL21(DE3) cells, as described previously (23). Kinase domain samples for NMR analysis contained a Y617P mutation that increases protein production in bacteria (23). The Btk_Itk loop-6 construct contains six mutations in the activation segment (L542M, S543T, V555T, R562K, S564A, and P565S). 15N-Phenylalanine and 15N-tyrosine selectively labeled samples were produced by growing the E. coli BL21(DE3) cells in modified minimal media as described earlier (23). The purified proteins were concentrated and dialyzed into NMR buffer, which consisted of 50 mM Bicine (pH 8.0), 75 mM NaCl, 2 mM DTT, 5% glycerol and 0.02% NaN3. All NMR spectra were collected at 298 K on a Bruker AVII 700 spectrometer equipped with a 5-mm HCN z-gradient cryoprobe operating at a 1H frequency of 700.13 MHz. Selective amino acid labeling was pursued because of difficulties in preparing a uniformly labeled sample of this protein required for conventional assignment methods. Chemical shift assignments for F540, F559, Y545, and Y551 residues in Btk and Btk_Itk loop-6 were determined by comparing the tyrosine or phenylalanine selectively labeled 1H-15N TROSY-HSQC spectra of the wild-type protein to that of the respective alanine mutant in both the Btk and Btk_Itk loop-6 backgrounds (figs. S4 and S5). All data were analyzed with NMRView J software (45). {1H}-15N NOEs were obtained from the ratio of the volumes of TROSY-based experiments recorded with and without 1H saturation. NOE = Vsat/Vnosat, where Vsat and Vnosat are peak volumes measured with and without proton saturation, respectively (46). The uncertainties in the NOEs were obtained from the standard deviation between the intensities of duplicate experiments.
Supplementary Material
Fig. S1. Mutational analysis of the kinase activation segment of Btk.
Fig. S2. Analysis of pERK1/2 in primary T cells transduced with retrovirus encoding Itk.
Fig. S3. HDX-MS analysis of Btk and Btk_Itk loop-6.
Fig. S4. Chemical shift assignment of Btk_Itk loop-6 kinase residues.
Fig. S5. Chemical shift assignment of the kinase residues of Btk.
Acknowledgments
We thank E. Huseby, UMMS for the pMiT vector.
Funding: This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases, AI043957 and AI075150 to A.H.A., and AI084987 and AI083505 to L.J.B.; National Institute of General Medical Sciences, GM086507 and GM101135 to J.R.E., a grant from the Sigrid Juselius Foundation (to I.K.), and a research collaboration with the Waters Corporation (to J.R.E.).
Footnotes
Author contributions: R.E.J., I.K., T.E.W., Q.X., and D.B.F. performed experiments; R.E.J., T.E.W., J.R.E., L.J.B., and A.H.A. designed experiments, analyzed data and wrote the paper.
Competing interests: none.
References and Notes
- 1.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 2.Izarzugaza JM, Krallinger M, Valencia A. Interpretation of the consequences of mutations in protein kinases: combined use of bioinformatics and text mining. Front Physiol. 2012;3:323. doi: 10.3389/fphys.2012.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chico LK, Van Eldik LJ, Watterson DM. Targeting protein kinases in central nervous system disorders. Nat Rev Drug Discov. 2009;8:892–909. doi: 10.1038/nrd2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 5.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 6.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, Nakagawa A, Nada S, Okada M, Tsukihara T. Structure of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2002;277:14351–14354. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 8.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Chu DH, Spits H, Peyron JF, Rowley RB, Bolen JB, Weiss A. The Syk protein tyrosine kinase can function independently of CD45 or Lck in T cell antigen receptor signaling. EMBO J. 1996;15:6251–6261. [PMC free article] [PubMed] [Google Scholar]
- 10.Latour S, Chow LM, Veillette A. Differential intrinsic enzymatic activity of Syk and Zap-70 protein-tyrosine kinases. J Biol Chem. 1996;271:22782–22790. doi: 10.1074/jbc.271.37.22782. [DOI] [PubMed] [Google Scholar]
- 11.Au-Yeung BB, Deindl S, Hsu LY, Palacios EH, Levin SE, Kuriyan J, Weiss A. The structure, regulation, and function of ZAP-70. Immunol Rev. 2009;228:41–57. doi: 10.1111/j.1600-065X.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 12.Boucheron N, Ellmeier W. The role of Tec family kinases in the regulation of T-helper-cell differentiation. Int Rev Immunol. 2012;31:133–154. doi: 10.3109/08830185.2012.664798. [DOI] [PubMed] [Google Scholar]
- 13.Horwood NJ, Urbaniak AM, Danks L. Tec family kinases in inflammation and disease. Int Rev Immunol. 2012;31:87–103. doi: 10.3109/08830185.2012.670334. [DOI] [PubMed] [Google Scholar]
- 14.Joseph RE, Xie Q, Andreotti AH. Identification of an allosteric signaling network within Tec family kinases. J Mol Biol. 2010;403:231–242. doi: 10.1016/j.jmb.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph RE, Min L, Andreotti AH. The Linker between SH2 and Kinase Domains Positively Regulates Catalysis of the Tec Family Kinases. Biochemistry. 2007;46:5455–5462. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 16.Mao C, Zhou M, Uckun FM. Crystal structure of Bruton’s tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked agammaglobulinemia. J Biol Chem. 2001;276:41435–41443. doi: 10.1074/jbc.M104828200. [DOI] [PubMed] [Google Scholar]
- 17.Marcotte DJ, Liu YT, Arduini RM, Hession CA, Miatkowski K, Wildes CP, Cullen PF, Hong V, Hopkins BT, Mertsching E, Jenkins TJ, Romanowski MJ, Baker DP, Silvian LF. Structures of human Bruton’s tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases. Protein Sci. 2010;19:429–439. doi: 10.1002/pro.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins J, Marcy A. Characterization of Itk tyrosine kinase: contribution of noncatalytic domains to enzymatic activity. Protein Expr Purif. 2001;22:211–219. doi: 10.1006/prep.2001.1447. [DOI] [PubMed] [Google Scholar]
- 19.Heyeck SD, Wilcox HM, Bunnell SC, Berg LJ. Lck phosphorylates the activation loop tyrosine of the Itk kinase domain and activates Itk kinase activity. J Biol Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 20.Wilcox HM, Berg LJ. Itk phosphorylation sites are required for functional activity in primary T cells. J Biol Chem. 2003;278:37112–37121. doi: 10.1074/jbc.M304811200. [DOI] [PubMed] [Google Scholar]
- 21.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinh M, Grunberger D, Ho H, Tsing SY, Shaw D, Lee S, Barnett J, Hill RJ, Swinney DC, Bradshaw JM. Activation mechanism and steady state kinetics of Bruton’s tyrosine kinase. J Biol Chem. 2007;282:8768–8776. doi: 10.1074/jbc.M609920200. [DOI] [PubMed] [Google Scholar]
- 23.Xie Q, Joseph RE, Fulton DB, Andreotti AH. Substrate recognition of PLCgamma1 via a specific docking surface on Itk. J Mol Biol. 2013;425:683–696. doi: 10.1016/j.jmb.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 25.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 26.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 27.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 28.Dixit A, Yi L, Gowthaman R, Torkamani A, Schork NJ, Verkhivker GM. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One. 2009;4:e7485. doi: 10.1371/journal.pone.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang HM, Yu X, Greig MJ, Gajiwala KS, Wu JC, Diehl W, Lunney EA, Emmett MR, Marshall AG. Drug binding and resistance mechanism of KIT tyrosine kinase revealed by hydrogen/deuterium exchange FTICR mass spectrometry. Protein Sci. 2010;19:703–715. doi: 10.1002/pro.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valiaho J, Smith CI, Vihinen M. BTKbase: the mutation database for X-linked agammaglobulinemia. Hum Mutat. 2006;27:1209–1217. doi: 10.1002/humu.20410. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min L, Joseph RE, Fulton DB, Andreotti AH. Itk tyrosine kinase substrate docking is mediated by a nonclassical SH2 domain surface of PLCgamma1. Proc Natl Acad Sci USA. 2009;106:21143–21148. doi: 10.1073/pnas.0911309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol. 2010;2:a002287. doi: 10.1101/cshperspect.a002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douhan J, 3rd, Miyashiro JS, Zhou X, Cole DC, Wu PW, Collins M, Dunussi-Joannopoulos K. A FLIPR-based assay to assess potency and selectivity of inhibitors of the TEC family kinases Btk and Itk. Assay Drug Dev Technol. 2007;5:751–758. doi: 10.1089/adt.2007.9982. [DOI] [PubMed] [Google Scholar]
- 36.Joseph RE, Andreotti AH. Bacterial expression and purification of Interleukin-2 Tyrosine kinase: Single step separation of the chaperonin impurity. Protein Expr Purif. 2008;60:194–197. doi: 10.1016/j.pep.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph RE, Min L, Xu R, Musselman ED, Andreotti AH. A remote substrate docking mechanism for the tec family tyrosine kinases. Biochemistry. 2007;46:5595–603. doi: 10.1021/bi700127c. [DOI] [PubMed] [Google Scholar]
- 38.Park YW, Cummings RT, Wu L, Zheng S, Cameron PM, Woods A, Zaller DM, Marcy AI, Hermes JD. Homogeneous proximity tyrosine kinase assays: scintillation proximity assay versus homogeneous time-resolved fluorescence. Anal Biochem. 1999;269:94–104. doi: 10.1006/abio.1999.4029. [DOI] [PubMed] [Google Scholar]
- 39.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell TC, Hildeman D, Kedl RM, Teague TK, Schaefer BC, White J, Zhu Y, Kappler J, Marrack P. Immunological adjuvants promote activated T cell survival via induction of Bcl-3. Nat Immunol. 2001;2:397–402. doi: 10.1038/87692. [DOI] [PubMed] [Google Scholar]
- 41.Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- 42.Choi SH, Wales TE, Nam Y, O’Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–349. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 46.Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Mutational analysis of the kinase activation segment of Btk.
Fig. S2. Analysis of pERK1/2 in primary T cells transduced with retrovirus encoding Itk.
Fig. S3. HDX-MS analysis of Btk and Btk_Itk loop-6.
Fig. S4. Chemical shift assignment of Btk_Itk loop-6 kinase residues.
Fig. S5. Chemical shift assignment of the kinase residues of Btk.