Summary
The Drosophila Yorkie (Yki) protein and its mammalian homolog Yes-associated protein (YAP) are potent growth promoters and YAP overexpression is associated with multiple types of cancer [1,2]. Yki and YAP are transcriptional co-activators and function as downstream effectors of the Hippo tumor suppressor pathway [1–4]. The regulation of Yki and YAP by the Hippo signaling pathway has been extensively investigated, however, how they regulate gene expression is poorly understood. To identify additional regulators of Yki activity we performed a genome-wide RNAi screen in Drosophila S2 cells. In this screen, we identified the conserved protein Mask (Multiple ankyrin repeats single KH domain) as a novel promoter of Yki activity in vitro and validated this function in vivo in Drosophila. We found that Mask is required downstream of the Hippo pathway for Yki to induce target gene expression and that Mask forms complexes with Yki. The human Mask homolog MASK1 complexes with YAP and it is required for full activity of YAP and elevated MASK1 expression is associated with worsened outcomes for breast cancer patients. We conclude that Mask is a novel cofactor for Yki/YAP required for optimal Yki/YAP activity during development and oncogenesis.
Results and Discussion
Mask is a positive regulator of Yki activity in vitro
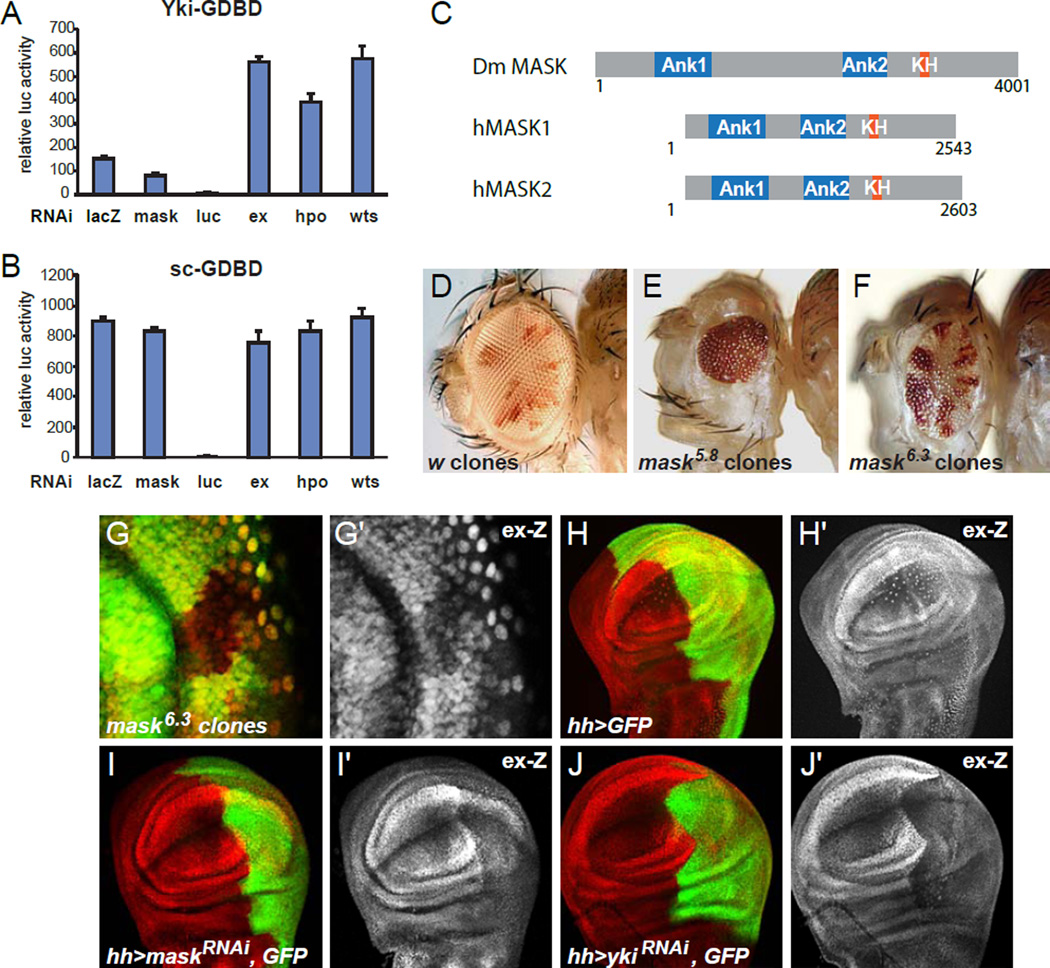
To identify new components of the Hippo pathway and potential regulators of Yki activity, we performed a genome-wide RNAi screen in S2 cells using a Yki-dependent luciferase assay [5, 6]. In this assay a fusion protein between Yki and the GAL4 DNA Binding domain (Yki-GDBD) drives expression of a UAS-luciferase construct. Co-expression of Expanded (Ex) was used to activate the Hippo pathway and diminish baseline Yki activity [6]. In a secondary assay, we eliminated false positives that have general effects on transcription by testing for effects on a fusion protein between Scute (Sc) and the GDBD. Sc-GDBD is not regulated by Hippo signaling but induces luciferase expression levels comparable to the Yki-GDBD construct [6]. In this screen, we identified the known Hippo pathway components Hpo, Ex, Wts, Kibra, Sav, Mats as well as actin regulators [6] as negative regulators of Yki activity, which validated the screening strategy and supported its utility for identifying novel components of the pathway (Fig. 1A). In addition to these negative Yki regulators we identified Multiple ankyrin repeats single KH domain (Mask) as a protein that promotes Yki activity (Fig. 1A). Knockdown of mask resulted in a 50% reduction of Yki-GDBD induced luciferase expression, while it did not affect Sc-GDBD induced reporter expression (Fig. 1B). These results indicate that the reduction in Yki activity caused by mask knockdown is specific and is not the consequence of general effects on transcription. We focused our further studies on Mask because knockdown of mask had some of the strongest effects on Yki-GDBD activity observed in the screen (apart from knockdowns that resulted in cell death) and because Mask also had robust and specific effects on Yki activity in vivo.
Figure 1. Mask is a positive regulator of Yki activity.
(A) Luciferase (luc) activity assay to measure the activity of a Yki-GAL4-DNA-binding-domain (Yki-GDBD) fusion protein on a UAS-luc reporter plasmid normalized with Renilla Luc expressed from a constitutive promoter. S2 cells were transfected with UAS-luc, Yki-GDBD and HA-Ex expressing plasmids in the presence of dsRNAs as indicated. (B) Similar as (A) but using a Scute (Sc)-GDBD construct instead of Yki-GDBD. (C) Schematic of Drosophila and human Mask proteins indicating conserved domains. (D–F) Adult flies with (D) white control clones, (E) mask5.8 null clones, and (F) mask6.3 hypomorphic clones. (G) Confocal image of a third instar antennal imaginal disc showing mask6.3 hypomorphic clones marked by the lack of GFP and stained for ex-lacZ expression (red, gray in G’). (H–J) Confocal images of third instar wing imaginal discs expressing GFP alone or together with maskRNAi or ykiRNAi in the posterior compartment driven by hh-GAL4 and stained for ex-lacZ (red or grey in the ’ panels).
Mask is required for normal tissue growth and Yki target gene expression in Drosophila
Mask is a large protein of 4001 amino acids that contains two clusters of ankyrin repeats (Ank1, and Ank2) and a single KH domain in its C-terminus (Fig. 1C) [7]. Ankyrin repeats are known to mediate protein-protein interactions, while KH domains can mediate protein-nucleic acid interactions [8, 9]. Previous studies identified mutations in mask as modifiers of Epidermal Growth Factor (EGF) signaling and a role for Mask in photoreceptor differentiation, cell survival and proliferation in Drosophila [7], although the molecular mechanism of Mask action was not known. Mask is conserved in humans, where there are two homologs, MASK1 (also known as ANKHD1) and MASK2 (also known as GTAR or ANKRD17) (Fig. 1C) [7, 10]. MASK1 is overexpressed in acute leukemia patient cells and cell lines [11] and regulates cell cycle progression and proliferation in multiple myeloma [12]. MASK2 is essential for vascular integrity during embryogenesis [13].
To investigate a potential role for Mask in Hippo signaling in vivo, we first tested whether there is a general requirement for mask in tissue growth. We found that larvae with mosaic eye discs in which most cells were homozygous mutant for the null allele mask5.8 [7], produced flies with small eyes that lacked mutant cells and had only a few heterozygous cells (Fig. 1D,E). Eye discs mosaic for the hypomorphic mask6.3 allele [7] also produced small eyes but contained homozygous cells, although their amounts were still far below control eyes (Fig. 1D,F). In imaginal discs, mask null clones grew poorly in all areas of developing eye and wing discs, and were eventually eliminated during late stages of development, while hypomorphic clones grew at reduced rates (Fig. S1A–C) [7]. Broad knockdown of mask during eye development by ey-GAL4 driven expression of genome-encoded shRNAs (ey-GAL4>UAS-maskRNAi) resulted in adult flies that had severely reduced eyes (Fig. S1E–G). Similarly, knock down of mask expression in the developing wing pouch using the nub-GAL4 driver resulted in smaller wings with some wing margin defects (Fig. S1H–K). Notably, four different non-overlapping mask-RNAi constructs showed similar effects although with varying strengths (Fig. S1) and mask mRNA levels were reduced even by a moderate RNAi line as measured by RT-qPCR (Fig. S1M). On the other hand, overexpression of Mask using nub-GAL4 driving different EY insertions in mask did not affect wing size (data not shown). These data indicate that Mask is generally required for normal tissue growth, similar to the requirement for Yki [5].
Based on our in vitro data and the requirement for Mask in tissue growth, we hypothesized that Mask acts in the Hippo pathway. To test this hypothesis, we examined the expression of Yki target genes in cells with reduced mask expression. Clones of cells mutant for the null allele mask5.8 were generally composed of only very few cells and thus difficult to analyze (Fig. S1B). However, clones mutant for the hypomorphic mask6.3 allele were larger and showed reduced expression of the expanded-lacZ (ex-Z) reporter, a commonly used and sensitive readout for Yki activity (Fig. 1G) [14]. Likewise, when mask expression was knocked down in the posterior compartment of developing wing discs by RNAi, ex-lacZ was strongly reduced when compared to control discs and to its expression in the anterior compartment (Fig. 1H,I). In addition, compartment size was reduced (Fig. 1H,I). This result is strikingly similar to the reduced expression of ex-lacZ and small compartment size observed when yki expression was knocked down in the posterior compartment using RNAi (Fig. 1J) [15]. Knockdown of mask also caused reduction of other Yki target genes such as diap1-lacZ (Fig. S2A,B). To support the specificity of the RNAi induced phenotypes, we repeated these experiments using different UAS-RNAi lines targeting non-overlapping regions of mask and each showed similar phenotypes (Fig. S2C,D). However, levels of Cubitus interruptus (Ci) (Fig. S2E,F) and a phosphorylated form of Mothers against Dpp (pMad) (Fig. S2G–H) used as Hedgehog and Dpp pathway readouts respectively, were not notably affected indicating that the effects of Mask knockdown are specific. Thus, Mask is required for the activation of Yki target genes.
Mask forms complexes with Yki and is required for Yki activity in vivo
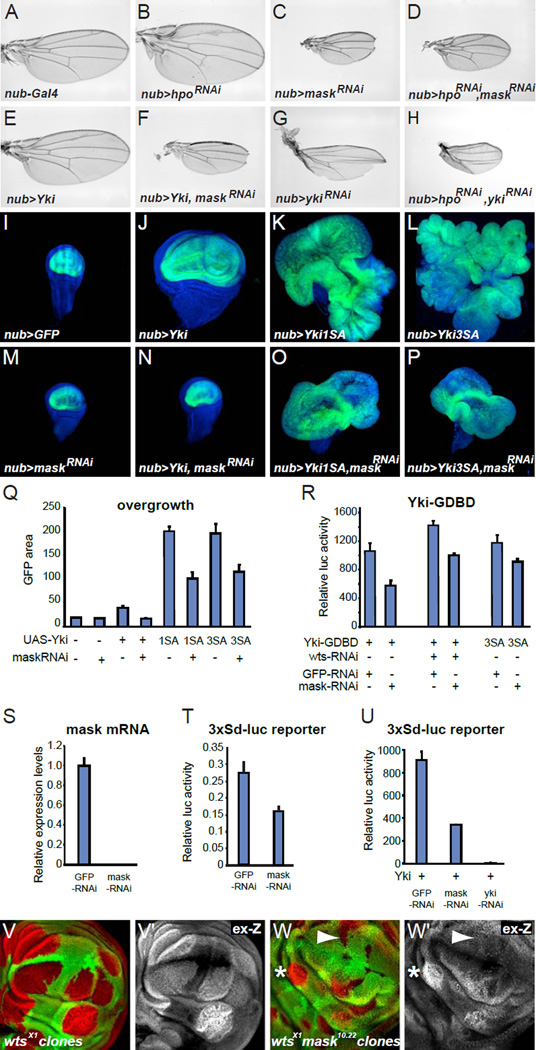
If Mask is necessary for normal Yki activity in vivo, then mask knockdown should modify Hippo pathway phenotypes in a manner similar to yki knockdown. To determine if that is the case and to place the action of Mask within the Hippo pathway, we decreased Hippo pathway activity using RNAi against fat, ex, and hpo alone or in combination with knockdown of mask or yki. Knockdown of fat, ex, or hpo in developing wing discs by the nub-Gal4 driver causes hyperactivation of Yki and resulted in overgrown adult wings (Fig. 2A,B and data not shown) [16, 17]. Conversely, knockdown of yki or mask resulted in wings that were much smaller than normal (Fig. 2C,G) [15]. Simultaneous knockdown of hpo, ex, or fat together with yki produced the yki knockdown phenotype of small wings showing that Yki activity is required for the overgrowth caused by Hippo pathway inactivation as expected (Fig. 2H). Similarly, when mask was knocked down together with fat, ex, or hpo, wing overgrowth was suppressed and the adult wings were small (Fig. 2D and data not shown). These results indicate that Mask functions within the Hippo pathway downstream of Fat, Ex, and Hpo.
Figure 2. Mask is required for full Yki activity independent of Wts phosphorylation.
(A–H) Pictures of wings from flies expressing the indicated constructs driven by the wing specific nubbin-GAL4 driver. “maskRNAi" refers to the VDRC v33396 maskRNAi line. (I–P) Confocal images of third instar wing discs with nubbin-GAL4 driving GFP plus the indicated constructs. GFP is in green and nuclei are stained with DAPI displayed in blue. (Q) Quantification of the GFP expressing area of imaginal discs with the genotypes shown in (I–P) (median area of GFP pixels ± S.E.M., n=5). (R) Luciferase assay on S2 cells transfected with plasmids expressing UAS-luc and wild-type Yki-GDBD or Yki3SA-GDBD fusion proteins in the presence of the indicated dsRNAs. (S) Quantification of mask mRNA levels by RT-qPCR from S2 cells transfected with dsRNA targeting GFP or mask. (T) Luciferase assay on S2 cells transfected with the Yki/Sd responsive 3xSd-luc reporter, which has multimerized Sd binding sites, in the presence of the indicated dsRNAs and normalized to beta-galactosidase activity expressed from a constitutive promoter. (U) Luciferase assay on S2 cells transfected with the 3X-Sd-luc reporter and Yki expressing plasmids in the presence of the indicated dsRNAs. (V) wtsx1 null mutant clones in a wing disc marked by lack of GFP and stained for ex-lacZ expression (red, gray in V’). (W) mask10.22 wtsx1 double mutant clones in a wing disc stained for ex-lacZ expression (red, gray in W’). The arrowhead indicates a clone with low levels of ex-lacZ expression and the asterisk indicates a clone with upregulated ex-lacZ expression.
We then tested whether Yki activity itself was dependent upon Mask. To investigate this, we first overexpressed Yki with and without knockdown of mask. Overexpression of wild-type Yki is sufficient to induce wing overgrowth (Fig. 2E) [5], but in combination with knockdown of mask, the adult wings exhibited the maskRNAi phenotype (Fig. 2F), namely small wings with distal scalloping (Fig. 2C). Thus, overexpressed Yki requires Mask to cause overgrowth. However, wild-type Yki can still be regulated by Wts phosphorylation and this experiment thus does not discriminate between direct effects on Yki and indirect effects through modulating Wts. We therefore repeated this experiment with mutant versions of Yki, namely Yki1SA and Yki3SA, in which the main or all three Wts phosphorylation sites were mutated to alanine [18]. Overexpression of Yki1SA or Yki3SA by nub-GAL4 caused severe overgrowth phenotypes in discs (Fig 2.I–L), which were not able to produce adult wings. Co-expression of mask-RNAi partially rescued these overgrowth phenotypes indicating that even non-phosphorylatable Yki requires Mask for its growth inducing activity (Fig 2.M–P, quantified in Q). To further quantify the effects of Mask on the activity of Yki independent of Wts phosphorylation, we used the Yki-GDBD luc assay in S2 cells. We found that Mask was again required for the full activity of Yki with wtsRNAi and of unphosphorylatable Yki3SA (Fig. 2R). However, mask knockdown did not completely inactivate wild-type or mutant Yki even though mask mRNA levels were nearly undetectable in mask RNAi cells (Fig. 2S). Therefore, Mask is not absolutely required for Yki activity. This conclusion was further confirmed in S2 cells, where knock down of mask reduced the activity of a reporter construct containing binding sites for Scalloped (Sd), a transcription factor that interacts with Yki, by about 50% (Fig 2.T,U). This was true when the reporter was driven by endogenous levels of Yki and Sd (Fig. 2T), or when Yki was overexpressed (Fig. 2U). We next analyzed the clonal growth and levels of ex-lacZ expression in wts single mutant and wts, mask double mutant clones. wts null mutant clones grew large and consistently had high levels of ex-lacZ expression (Fig. 2V). In contrast, wts, mask double null mutant clones were generally smaller and about half of these clones had no or weak upregulation of ex-lacZ expression (Fig. 2W arrowhead), although the other half had elevated levels (Fig. 2W asterisk). These in vivo data thus show that Mask is required for the full wts mutant phenotype but that loss of Wts can still enhance the activity of Yki even in the absence of Mask. Altogether the in vivo and in vitro experiments show that Mask is required for full activity of Yki and that at least part of this function is independent (in parallel or downstream) of the activity of Wts.
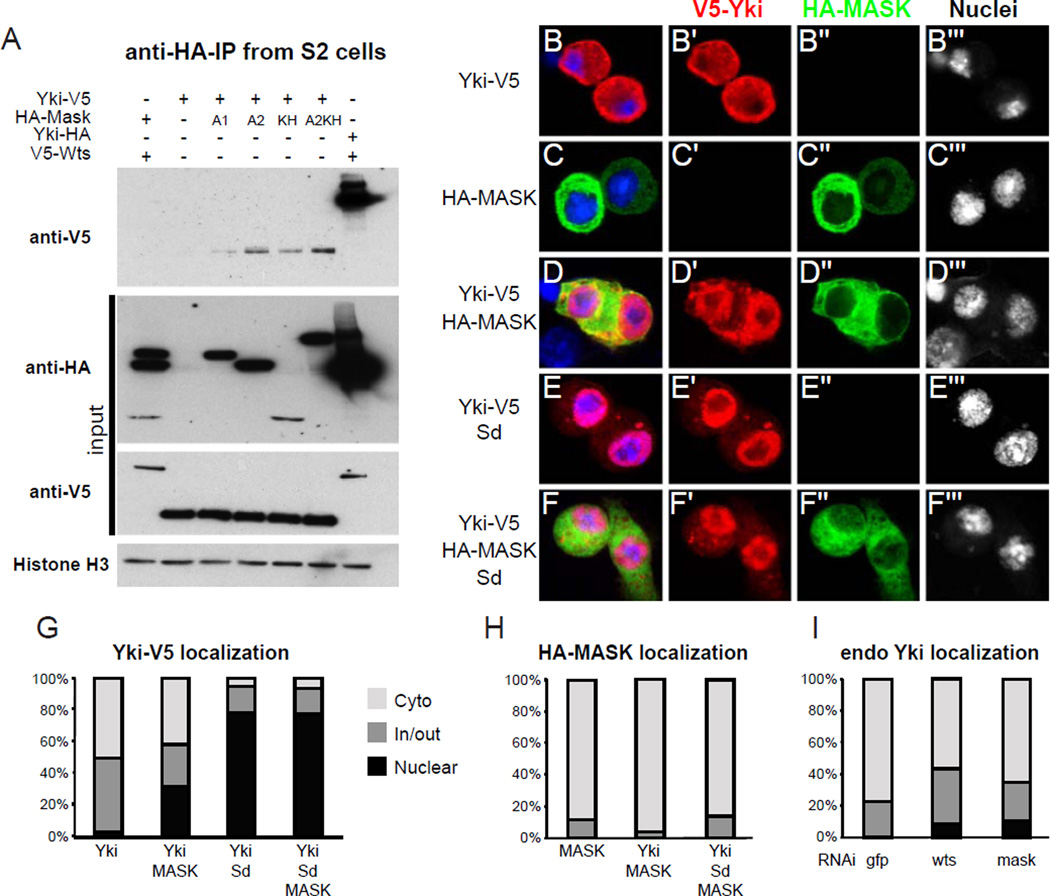
The above data indicate that the activities of Mask and Yki may be closely linked. To determine whether Mask can physically interact with Yki, we co-immunoprecipitated V5-tagged Yki with HA-tagged Mask constructs. We found that the Ank2 and KH domains, and a construct that contained the Ank2-KH domains together, formed complexes with Yki, whereas Ank1 complexed weakly (Fig. 3A). We also tested for interaction with Wts, but found that none of the Mask domains bound to Wts (Fig. 3A). This experiment also demonstrates the specificity of the interaction between Mask and Yki. Together with our in vivo and S2 cell data, we conclude that Mask represents a novel component of the Hippo pathway that positively regulates Yki activity, likely through a direct interaction with Yki.
Figure 3. Mask complexes with Yki but localizes independently of Yki.
(A) Immunoprecipitation of lysates from S2 cells transfected with Yki-V5 and different HA-Mask or V5-Wts and HA-Yki constructs. Top shows an anti-V5 western blot from an anti-HA immunoprecipitation. Panels below that show anti-V5 and anti-HA western blots to detect expression of the input proteins, and an anti-Histone H3 western blot as a loading control. Lane 1: V5-Wts co-expressed with HA-tagged Ank1, Ank2 and KH domains of Mask. Lane 2: Yki-V5 alone. Lane 3–6: Yki-V5 co-expressed with the Ank1, Ank2, KH, or Ank2-KH constructs respectively. Lane 7: Yki-HA co-expressed with V5-Wts as a positive control. (B–F) Confocal images of S2 cells transfected with the constructs indicated on the left and stained for the antigens indicated on the top. Nuclei were labeled with DAPI shown in blue. (G) Quantification of the localization of Yki-V5 in (B–F). (H) Quantification of the localization of Mask in (C,D) and (F). (I) Quantification of endogenous Yki localization in S2 cells transfected with dsRNAs targeting GFP, wts or mask.
We then assayed whether Mask and Yki influence each other’s subcellular localization. Mask and Yki are localized mainly in the cytoplasm in imaginal disc cells and in S2 cells [5, 7] and Yki translocates to the nucleus when the Hippo pathway is inactivated [5]. However, we did not observe significant effects on Mask localization in wts mutant clones (Fig. S3A,B). We then tested whether in S2 cells nuclear translocation of Yki is sufficient to recruit Mask into the nucleus. For this experiment we analyzed the localization of tagged Yki and tagged Ank2-KH Mask proteins. We found that co-expression of Sd efficiently recruited Yki into the nucleus but not Mask (Fig. 3F, quantified in G and H). Thus, Mask and Yki do not form obligatory complexes and the regulation of Mask localization may be independent of the Hippo pathway. Conversely, knockdown of mask caused a slight increase in nuclear localization of Yki in S2 cells, similar to the effect of wts knockdown (Fig. 3I). Knockdown of mask in imaginal discs did not noticeably affect the localization pattern of Yki, but it resulted in an increase of Yki protein levels (Fig. S3C,D) However, even though mask RNAi cells have elevated levels of Yki protein, they have less Yki activity, further supporting that Mask is required for full activity of Yki.
MASK1 is required for full YAP activity and MASK1 expression levels correlate with breast cancer patient outcomes
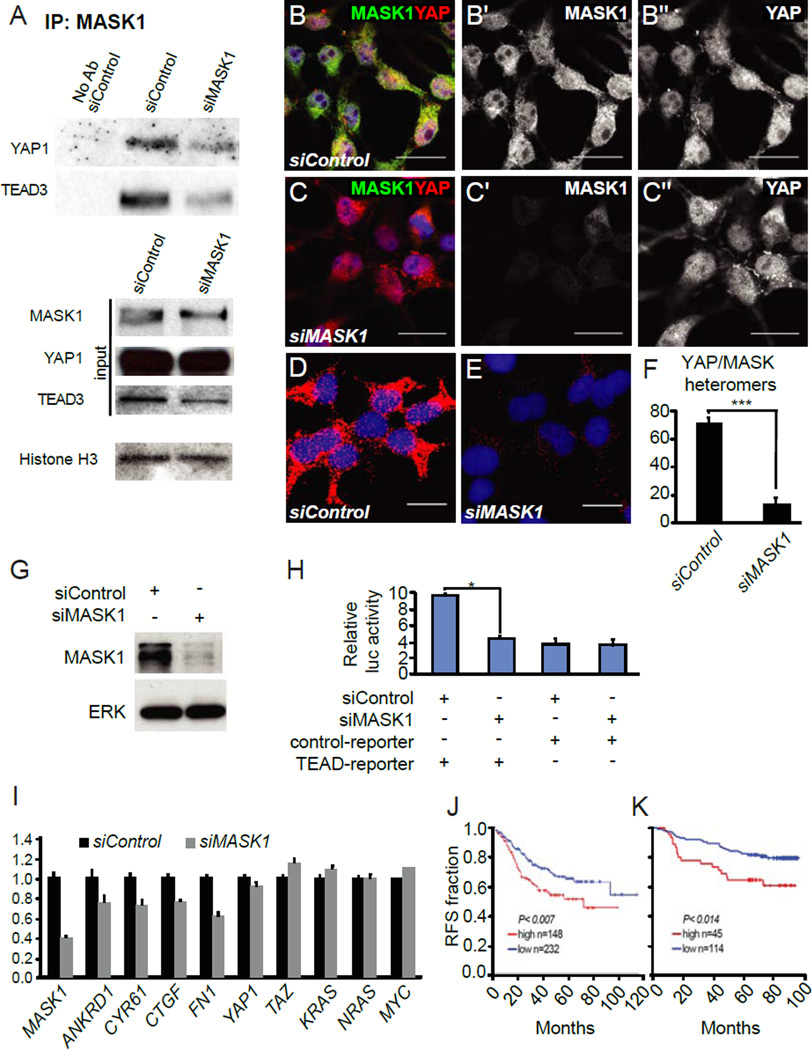
In mammals, the Hippo pathway controls organ size and cell proliferation by regulating the activity of the Yki homologs YAP and TAZ, which can complex with the Sd-homologous TEAD transcription factors to activate target gene expression [1, 3–5]. We found that immunoprecipitation of endogenous MASK1 co-purified YAP indicating that MASK1 and YAP form complexes (Fig. 4A). In addition, we found that immunoprecipitation of MASK1 also co-purified endogenous TEAD3 (Fig. 4A). Knockdown of MASK1 significantly reduced the amount of co-immunoprecipitated YAP and TEAD3, while the total amount of YAP in the cell lysates was not affected and TEAD3 was slightly lower (Fig. 4A). We next wanted to determine the cellular localization of these proteins. MASK1 mainly localized to the cytoplasm, while YAP was evenly distributed in nuclei and cytoplasm in our assay conditions (Fig. 4B,C). We then used the proximity ligation assay (PLA) to visualize the localization of the heteromers of MASK with YAP or TAZ. Such heteromers were readily detected in the cytoplasm but also observed in the nuclei of HEK293T cells treated with non-targeting siRNAs (Fig. 4D,F and Fig.S4), while the number of complexes was significantly reduced upon MASK knock down (Fig 4.E,F and Fig.S4). Together, these data indicate that endogenous MASK1 and YAP form complexes.
Figure 4. MASK1 interacts with YAP and TEAD3, regulates YAP activity and its expression levels correlate with breast cancer patient outcomes.
(A) Lysates of HEK293T cells treated with non-targeting or MASK1 siRNAs were immunoprecipitated for endogenous MASK1 and immunoblotted for YAP1 or TEAD3. (B,C) Endogenous MASK1 (green, grey in B′ and C′) and YAP (red, grey in B′′ and C′′) localization in HEK293T cells treated with non-targeting (B) or MASK1 siRNAs (C). (D–E) MASK1/YAP heteromers (red) detected by in situ proximity ligation assay (PLA) using MASK1 and YAP antibodies in cells treated with non-targeting (D) or MASK1 siRNA (E). (F) Quantification of the PLA signals in (D) and (E). (G) Western blot demonstrating the level of siRNA knock down of MASK1 protein in HEK293T cells. (H) Relative luciferase activity of a luciferase reporter plasmid with TEAD binding sites and the control vector in HEK293T cells co-transfected with MASK1 siRNA and a non-targeting siRNA. The error bars represent mean ± SE (n=6) (* p > 0.005). (I) RT-qPCR analysis of mRNA expression of YAP target genes in HEK293T cells treated with non-targeting or MASK1 siRNAs. (J) Kaplan-Meier graphs showing a significant association of MASK1 expression levels with Relapse Free Survival (RFS) in independent datasets of breast cancer patients (p>0.007 and p>0.014, log-rank test). Scale bars are 20 µm.
To examine whether MASK1 is required for the transcriptional activity of YAP/TEAD complexes, we assayed the ability of MASK1 to modulate the activity of a commonly used TEAD luciferase reporter [19]. We transfected HEK293T cells with si-MASK1, which resulted in a significant reduction of MASK1 expression (Fig. 4G) and significantly reduced TEAD-dependent luciferase reporter activity when compared with a non-targeting siRNA control (p>0.005) (Fig. 4H). We also assayed for YAP/TEAD target gene expression by RT-qPCR in HEK293T cells transfected with si-MASK1 or non-targeting siRNA. We found that the expression of the YAP target genes CTGF, ANKRD1, FN1, CYR61 were significantly reduced while non-target genes such as KRAS, NRAS and MYC, as well as YAP and TAZ themselves, were not affected (Fig. 4I). Together, these data indicate that MASK1 is required for full activity of YAP/TEAD.
Deregulation of the Hippo pathway is frequently detected in human cancers and increased YAP activity correlates with poor prognosis and patient survival [1,2]. Additionally, MASK1 is over-expressed in acute leukemias [11] and regulates cell cycle progression and proliferation in multiple myeloma cells [12] suggesting that it may have oncogenic activity. To examine whether there is a relationship between clinical prognosis and expression of MASK1 in human cancers, we analyzed gene expression profiles of different breast cancer data sets using probes that identify the first ankyrin domain of MASK1 mRNA. We found that low expression of MASK1 is associated with significantly better relapse-free survival in two independent data sets (p> 0.007, p>0.014) UNC (Fig. 4J) and Pawitan (Fig. 4K). These findings suggest that MASK1 levels are an important factor in the aggressiveness of cancer progression and patient outcomes.
In summary, we identified Mask as a novel component of the Hippo pathway. We found that Mask and its mammalian homolog MASK1 are required for full activity of the Hippo pathway effector Yki/YAP, and that MASK1 expression levels correlate with breast cancer patient outcomes. We found that Mask/MASK1 form complexes with Yki/YAP, however, the mechanism by which Mask/MASK1 potentiates Yki/YAP activity is currently not known. Our data suggest a model in which Mask/MASK1 acts in the nucleus to promote Yki/YAP’s transcriptional co-activator function, but Mask/MASK1 may also function in the cytoplasm to promote Yki/YAP activity. In any case, our data indicate that Mask/MASK1 acts in parallel to the Wts mediated phosphorylation and regulated localization and Mask/MASK1 may thus identify a novel regulatory branch of the Hippo pathway.
Supplementary Material
Highlights.
-
▪
Mask is required for tissue growth and activation of Yki target genes in Drosophila
-
▪
Mask forms complexes with the Hippo pathway effector Yki
-
▪
Human MASK complexes with YAP is required for full YAP activity in mammalian cells
-
▪
Mask expression levels correlate with worsened outcomes for breast cancer patients
Acknowledgements
We thank Michael Simon, Barry Thompson, Konrad Basler, and Jin Jiang for fly stocks, Mask antibodies and plasmids. We thank Molly Schroeder for images of adult eyflp; cl/+ control females, and Marlese Pisegna and Chao-Lin Chen for plating the dsRNA library into 384-well plates.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes four figures and Supplemental Experimental Procedures and can be found with this article on line.
References
- 1.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes & development. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO journal. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith RK, Carroll PM, Allard JD, Simon MA. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development. 2002;129:71–82. doi: 10.1242/dev.129.1.71. [DOI] [PubMed] [Google Scholar]
- 8.Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends in biochemical sciences. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 9.Valverde R, Edwards L, Regan L. Structure and function of KH domains. The FEBS journal. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 10.Poulin F, Brueschke A, Sonenberg N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. The Journal of biological chemistry. 2003;278:52290–52297. doi: 10.1074/jbc.M310761200. [DOI] [PubMed] [Google Scholar]
- 11.Traina F, Favaro PM, Medina Sde S, Duarte Ada S, Winnischofer SM, Costa FF, Saad ST. ANKHD1, ankyrin repeat and KH domain containing 1, is overexpressed in acute leukemias and is associated with SHP2 in K562 cells. Biochimica et biophysica acta. 2006;1762:828–834. doi: 10.1016/j.bbadis.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Dhyani A, Duarte AS, Machado-Neto JA, Favaro P, Ortega MM, Olalla Saad ST. ANKHDI regulates cell cycle progression and proliferation in multiple myeloma cells. FEBS letters. 2012 doi: 10.1016/j.febslet.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 13.Hou SC, Chan LW, Chou YC, Su CY, Chen X, Shih YL, Tsai PC, Shen CK, Yan YT. Ankrd17, an ubiquitously expressed ankyrin factor, is essential for the vascular integrity during embryogenesis. FEBS letters. 2009;583:2765–2771. doi: 10.1016/j.febslet.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature cell biology. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Developmental cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Developmental cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS biology. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 20.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome biology. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, Sartor CI, Rieger-House T, Bernard PS, Carey LA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast cancer research : BCR. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.