Abstract
Cytokinesis and ciliogenesis are fundamental cellular processes that require strict coordination of microtubule organization and directed membrane trafficking. These processes have been intensely studied, but there has been little indication that regulatory machinery might be extensively shared between them. Here, we show that several central spindle/midbody proteins (PRC1, MKLP-1, INCENP, centriolin) also localize in specific patterns at the basal body complex in vertebrate ciliated epithelial cells. Moreover, bioinformatic comparisons of midbody and cilia proteomes reveal a highly significant degree of overlap. Finally, we used temperature-sensitive alleles of PRC1/spd-1 and MKLP-1/zen-4 in C. elegans to assess ciliary functions while bypassing these proteins' early role in cell division. These mutants displayed defects in both cilia function and cilia morphology. Together, these data suggest the conserved re-use of a surprisingly large number of proteins in the cytokinetic apparatus and in cilia.
Keywords: Ciliogenesis, cytokinesis, PRC1, INCENP, MKLP-1, bioinformatics, cilia midbody
Introduction
Cilia are microtubule-based organelles that project from cells, and mutations of genes associated with ciliogenesis underlie a broad spectrum of disorders in humans (Marshall, 2008; Sharma et al., 2008). As such, understanding the full complement of proteins required for establishing and maintaining cilia structure and function will be important. Genomic and proteomic approaches have defined the basal body and cilia proteomes, but many proteins essential for ciliogenesis are not present in these datasets (Gherman et al., 2006; Gray et al., 2009; Hayes et al., 2007; Inglis et al., 2006; Park et al., 2006). Functional screens might be expected to reveal these other important ciliogenesis factors (e.g. (Kim et al., 2010)), but many proteins have pleiotropic functions that may render their role in cilia invisible to such screens. Indeed, recent reports demonstrate exocytic roles for some proteins required for ciliogenesis (Finetti et al., 2009; Gray et al., 2009).
Cilia assembly and function require strictly coordinated microtubule organization and membrane delivery (Gray et al., 2009; Nachury et al., 2007; Park et al., 2008; Sorokin, 1968). Cells face a similar challenge during cytokinesis, as proteins in the central spindle must both organize the microtubule bundles and direct membrane traffic into the nascent cleavage furrow (Albertson et al., 2008; Glotzer, 2009; Gromley et al., 2005). In fact, a small number of regulatory proteins have been implicated in both processes (e.g. CP110, the exocyst, BBS6, RhoA; (Gromley et al., 2005; Kim et al., 2005; Pan et al., 2007; Park et al., 2008; Shah et al., 2008; Spektor et al., 2007; Zuo et al., 2009)).
The protein network that includes PRC1, INCENP, MKLP-1, and centriolin acts at the central spindle and midbody and has been studied extensively for its role in cytokinesis (Glotzer, 2009; Gromley et al., 2005; Jiang et al., 1998; Kieserman et al., 2008; Kurasawa et al., 2004). In this paper, we report novel roles for these proteins, showing that they localize to basal bodies in ciliated epithelial cells and control cilia function and morphology. Our data suggest the possibility that a surprising amount of cytokinetic regulatory machinery may also serve important functions related to cilia.
Results and Discussion
We have developed a method to identify novel ciliogenesis factors based upon differential gene expression in the multi-ciliated epithelial cells of the Xenopus embryo epidermis (Fig. 1A; see also (Hayes et al., 2007)). Extending this system to protein localization, we identified the microtubule-bundling protein PRC1 as localizing prominently to the base of cilia in multi-ciliated cells (Fig. 1B, b′ arrow). PRC1 is one member of a well-defined protein network acting at the central spindle and the midbody during cytokinesis (Glotzer, 2005; Glotzer, 2009; Jiang et al., 1998) (Fig. 1B, arrowhead). These proteins organize microtubule bundles, position the cleavage furrow, and also trigger vesicle recruitment for cell abscission (Glotzer, 2005; Glotzer, 2009; Gromley et al., 2005; Jiang et al., 1998).
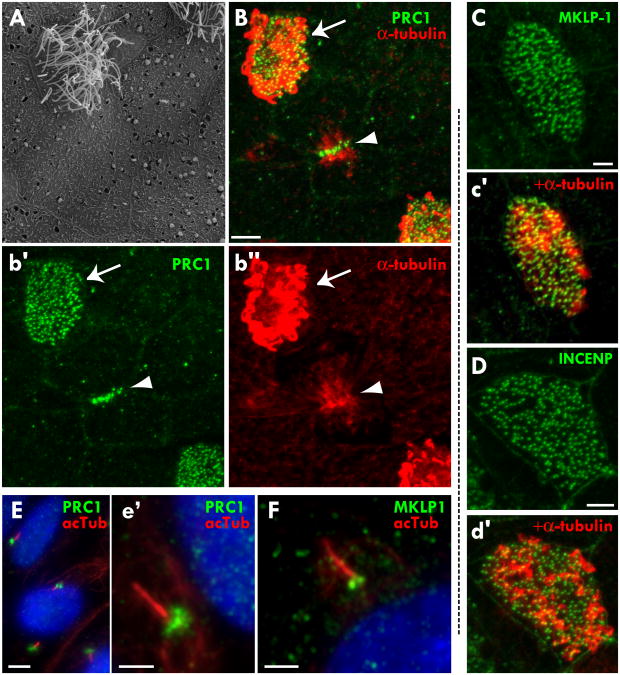
Figure 1. Central spindle/midbody proteins localize to basal bodies in multi-ciliated epithelial cells.
(A) SEM of the ciliated epidermis of the Xenopus laevis tadpole showing a multi-ciliated cell and surrounding mucus secreting goblet cells. (B) Merged image of immunostaining of PRC1 (green) and α-tubulin (red) in the epidermis of a X. laevis tadpole. Arrow points to a ciliated cell with cilia protruding from the surface and a punctate pattern of PRC1 under the cilia. Arrowhead points to a dividing cell in mid-telophase with PRC1 localization at the central spindle. (b′) Same image as B showing only PRC1 localization. (b″) Same image as B showing only α-tubulin localization. (C) Immunostaining of MKLP-1 in a ciliated cell. (c′) Merged image of immunostaining of MKLP-1 and α-tubulin to reveal cilia. (D) Immunostaining of INCENP in a ciliated cell. (d′) Merged image of immunostaining of INCENP and α-tubulin to reveal cilia. (E,e′) Immunostaining of PRC1 (green) and acetylated tubulin (red) in human RPE serum starved cells shows PRC1 localizing to the base of primary cilia. (F) Immunostaining of MKLP-1 (green) and acetylated tubulin (red) in RPE cells also shows MKLP-1 localizing to the base of primary cilia. Nuclei stained with Hoechst. Scale bars B = 10μm, C,D = 3μm, E = 5μm, e′,F = 2μm.
The localization of PRC1 near basal bodies was surprising, since central spindle proteins have not previously been implicated in ciliogenesis. We therefore asked if other proteins in this network also localized to basal bodies. We found that MKLP-1, INCENP, centriolin, CENP-E, and Survivin also localized strongly to the base of cilia in Xenopus multi-ciliated cells (Fig. 1C, c′, D, d′; Fig. 3C, c″; data not shown).
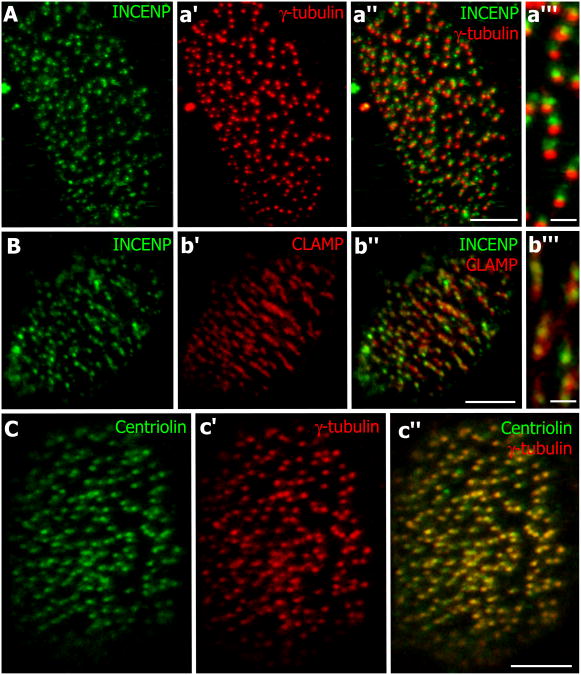
Figure 3. INCENP co-localizes with the rootlet and Centriolin with the basal body.
(A) Immunostaining of INCENP (green). (a′) Same image as A showing immunostaining of γ-tubulin (red). (a″) Merged image INCENP and γ-tubulin. (a‴) Immunostaining shows INCENP localizing proximal to, but not with, γ-tubulin. (B) Immunostaining of INCENP (green). (b′) Same image as B showing CLAMP-RFP localization. (b″) Merged image of INCENP and CLAMP-RFP. (b‴) Immunostaining shows INCENP co-localizing with CLAMP-RFP. (C) Centriolin (green) localization in a multi-ciliated cell. (c′) Same image as C showing γ-tubulin (red) localization. (c″) Merged image of Centriolin and γ-tubulin shows co-localization. Scale bars a′,b′,c′ = 5μm, a″,b″ = 1μm.
The PRC1 and MKLP1 antibodies used for immunostaining in Xenopus were ineffective in western blots with Xenopus embryo lysates, and some antibodies made in rabbits can non-specifically label centrosomes. Therefore, to verify the observed localization patterns, we obtained additional antibodies against these proteins (see Materials and Methods) and tested them in a different cell type, human RPE cells. Immunostaining for PRC1, MKLP-1, INCENP and Survivin revealed that each of these proteins prominently localized to the base of primary cilia in serum-starved human RPE cells in culture (Fig. 1E, e′, F; Supp. Fig. 1, and data not shown). These data therefore suggest that central spindle proteins localize to the base of both specialized motile cilia in Xenopus multiciliated cells and to the base of primary cilia in human RPE cells.
We further defined the localization of these central spindle proteins in multi-ciliated cells by co-immunostaining with γ-tubulin, a marker of the basal body. Interestingly, PRC1 did not localize to the basal body itself, but rather lay immediately adjacent to it in a planar polarized fashion (Fig. 2B, b′, b″). This pattern was intriguing because planar polarization of basal bodies in multi-ciliated cells is central to directed ciliary beating and fluid flow (Fig. 2A; (Frisch and Farbman, 1968; Mitchell et al., 2007; Park et al., 2008)).
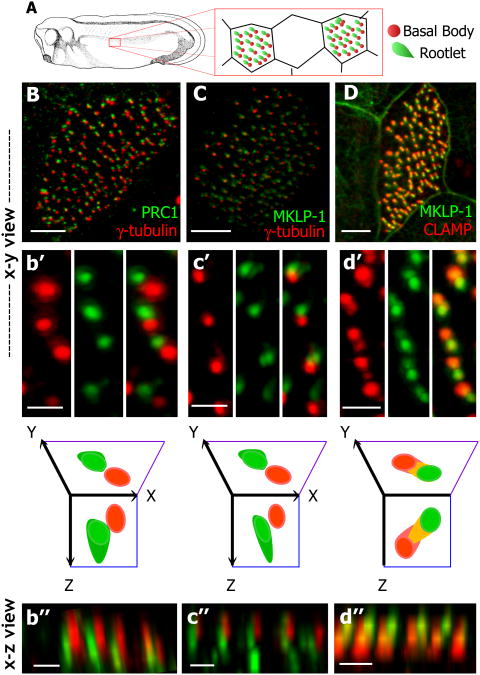
Figure 2. Spatially distinct localization of central spindle/midbody proteins at the basal body.
(A) Early tailbud stage Xenopus laevis embryo and diagram showing the planar polarization of cilium structures in the multi-ciliated cells of the epithelium, with the rootlet (green) positioned on the anterodorsal face of the basal body (red). (B) PRC1 (green) and γ-tubulin (red) immunostaining shows that PRC1 is proximal to the basal body on the anterior dorsal side, (b′) x-y view of PRC1 and γ-tubulin with diagram below showing section views. (b″) x-z plane view. (C) Co-immunostaining against MKLP-1 (green) and γ-tubulin (red) shows MKLP-1 proximal to but not co-localizing with the basal body on the anterior dorsal side. (c′) x-y plane view of MKLP-1 and γ-tubulin localization with diagram below showing section views. (c″) x-z plane view. (D) MKLP-1 co-localizes with rootlet marker CLAMP in a polarized manner. (d′) x-y view showing co-localization between MKLP-1 and CLAMP, with diagram below showing x-z and x-y slices and overlap. (d″) x-z plane view. Scale bars B,C,D = 5μm, b′-d″= 1μm.
Basal body planar polarity is manifested in the stereotyped positioning of accessory structures, such as the ciliary rootlet and basal foot (Frisch and Farbman, 1968; Mitchell et al., 2007). In Xenopus multi-ciliated cells, rootlets are consistently positioned anterodorsally to the basal body, opposite from the direction of fluid flow (Fig. 2A; (Mitchell et al., 2007; Park et al., 2008)). In X/Y projections, PRC1, MKLP-1 and INCENP were consistently present on the anterodorsal side of the γ-tubulin-positive basal body (Fig. 2B, b′, C, c′; Fig. 3A-a″), suggesting that these proteins may localize in the vicinity of the ciliary rootlet.
Rootlets in Xenopus multi-ciliated cells project basolaterally into the cytoplasm (Steinman, 1968). Likewise, X/Z projections from 3D datasets revealed that PRC1 and MKLP1 signals extended basolaterally into the cytoplasm from the anterodorsal face of the γ-tubulin-positive basal body (Fig. 2b″, c″). Moreover, the microtubule-binding protein, CLAMP marks the vicinity of the ciliary rootlet (Park et al., 2008), and co-immunostaining demonstrated that MKLP-1 and INCENP each partially co-localized with CLAMP-RFP (Fig. 2D-d″; Fig. 3B-b″). Together, these data suggest that central spindle proteins localize in a planar polarized fashion at the base of cilia in the vicinity of the rootlet.
The rootlet is implicated as a key site where vesicular traffic is coordinated with microtubule-based structures during ciliogenesis (Fariss et al., 1997; Gray et al., 2009; Park et al., 2008; Yang and Li, 2005). The localization of central spindle proteins near this site suggested a potential parallel between ciliogenesis and cytokinesis, as the central spindle also governs vesicle delivery during cytokinesis (Albertson et al., 2008; Gromley et al., 2005). To further explore this parallel, we examined centriolin, which localizes to the midbody ring, immediately adjacent to the central spindle proteins, and which directs the recruitment of vesicles (Gromley et al., 2005). We found that centriolin was also localized to the base of cilia in multi-ciliated cells (Fig. 3C-c″). Interestingly, centriolin did not localize near the rootlet, but rather co-localized with γ-tubulin at the basal body itself (Fig. 3c″). In summary, localization experiments in Xenopus multi-ciliated cells revealed that several components of the protein network governing cytokinesis localize to spatially distinct domains at the basal body that reflect to some degree their function in the midbody.
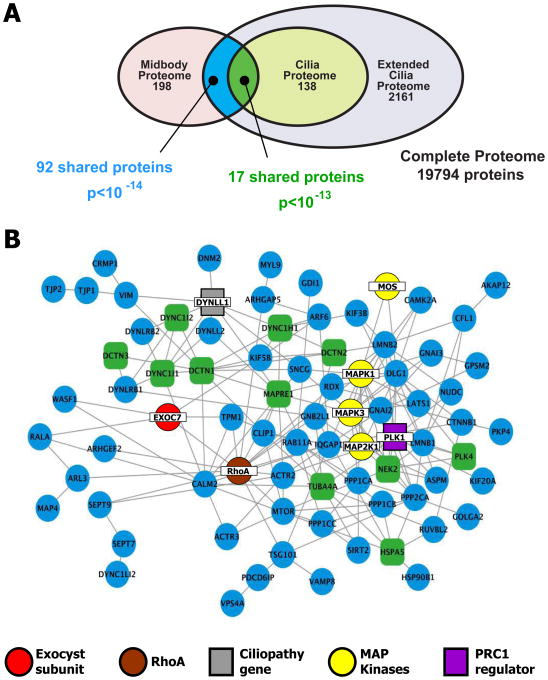
To more comprehensively explore the potential link between cytokinesis and ciliogenesis, we used bioinformatic approaches to compare the midbody proteome, as defined by proteins known to localize to the midbody (Ren et al., 2009; Skop et al., 2004) with the cilia/basal body proteome, as defined by mass spectrometry and comparative genomics (Gherman et al., 2006; Inglis et al., 2006). We found that the two proteomes overlap significantly (Fig. 4A, blue overlap; p ≤ 10-14, hypergeometric probability). The cilia/basal body proteome includes proteins identified directly, as well as many predicted by comparative genomics and other means (Gherman et al., 2006; Inglis et al., 2006). To more stringently examine the overlap of these proteomes, we re-examined the overlap using only centrosome or basal body proteins as determined directly by mass-spectroscopy. Again, we observed far greater overlap than could be expected by chance (Fig. 4A, green overlap, p ≤ 10-13, hypergeometric probability).
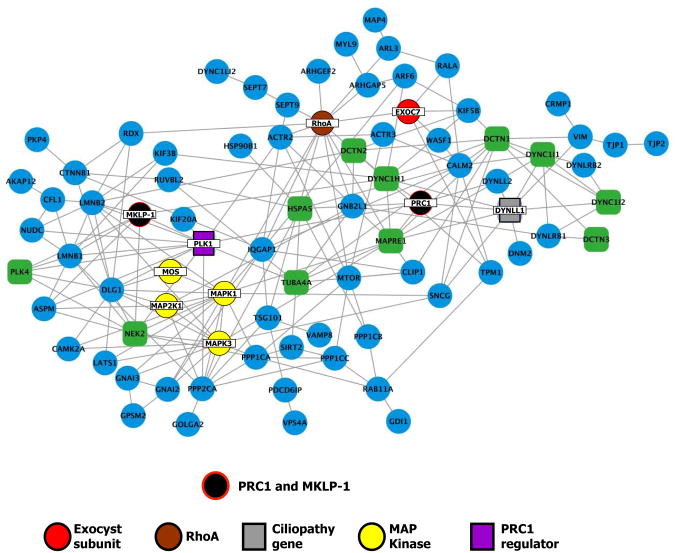
Figure 4. Bioinformatic analyses reveal a large set of common proteins involved in cytokinesis and ciliogenesis.
(A) Venn diagram showing the overlap between the midbody and basal body/ciliary proteomes. The overlap between midbody proteome and either core and extended basal body/cilia proteomes (see Supplemental Methods) is significantly greater than expected by chance (hypergeometric test). (B) A network diagram of gene-gene associations is drawn for proteins present in both the midbody proteome and core (green squares) or extended (blue circles) basal body/cilia proteome. Lines connect proteins likely to function together on the basis of co-expression, co-regulation, and/or protein-protein interactions, as described in the Supplemental Methods. The core cilia proteome is preferentially connected by network associations to midbody proteins (p ≤ 10-8, see Supplemental Methods). This cluster contains several notable proteins, including exocyst subunits (red), RhoA (brown), several MAP kinases (yellow), PLK1 (purple), and the human ciliopathy gene DynLL1 (gray).
To ask if this overlap between midbody and basal body proteomes is biologically relevant, we examined the positioning of these proteins in a functional gene network. This network is composed of functional interactions among human genes (determined as a probabilistic combination of evidence for co-regulation, physical and/or genetic interactions, co-evolution, and interactions between orthologs in other species) and serves as a useful indicator of proteins operating in the same biological processes and pathways (Gray et al., 2009; Lee et al., 2008b; Li et al., 2009). Proteins found in the midbody and cilia/basal body proteomes were much more strongly interconnected within the probabilistic network than would be expected by chance (Fig. 4B; p ≤ 10-8, Z-score test). In fact, 80% of the overlapping proteins clustered together tightly within the network (Fig. 4B).
This cluster of proteins included not only the few already known to play roles in both cytokinesis and ciliogenesis (e.g. exocyst, RhoA (Glotzer, 2005; Gromley et al., 2005; Pan et al., 2007; Park et al., 2008; Zuo et al., 2009) (Fig. 4B, red/brown)), but also several additional proteins of interest. For example, Plk1 was present in this cluster within the network (Fig. 4B, purple); Plk1 interacts with PRC1 at the central spindle (Neef et al., 2007) but has not been previously implicated in cilia. The cluster also contains several MAP kinases, whose connection in this network may shed light on the mechanisms by which FGF/MAP kinase signaling, which is well-known to control cell division, also controls cilia length (Berman et al., 2003; Burghoorn et al., 2007; Neugebauer et al., 2009) (Fig. 4B, yellow). Finally, the cluster contains a known human ciliopathy gene, Dynll1 (Horvath et al., 2005).
PRC1 and other central spindle proteins are not present in this cluster because they have not previously been associated with cilia or basal bodies. In order to link our bioinformatic analyses to our immunostaining results, we added PRC1 and MKLP-1 to the overlapping proteome set and re-examined the connectivity in our network. Both PRC1 and MKLP-1 were tightly embedded in this cluster of connected genes (Fig. 5).
Figure 5. PRC1 and MKLP-1 are tightly linked in the cluster of cytokinesis and ciliogenesis genes in a probabilistic gene network.
A network diagram of gene-gene associations is drawn for PRC1, MKLP-1 and the proteins present in both the midbody proteome and core (green squares) or extended (blue circles) basal body/cilia proteome. Lines connect proteins likely to function together on the basis of co-expression, co-regulation, and/or protein-protein interactions, as described in the Supplemental Methods.
Together with our immunostaining data, these bioinformatic analyses suggest the possibility that a functionally analogous protein network acts at both the central spindle/midbody to control cytokinesis and at the basal body to control cilia structure or function. However, the essential role for these proteins in cell division would render their role in cilia invisible to standard loss-of-function approaches in vivo. To circumvent this problem, we obtained a temperature-sensitive mutant of PRC1 in the worm, C. elegans. This PRC1 allele, spd-1, has normal function at 15°C but is non-functional at 25°C (O'Connell et al., 1998; Verbrugghe and White, 2004). Worms carrying this mutation were reared at the permissive temperature through early development and then shifted to the restrictive temperature at the time when cilia are thought to begin assembling on sensory neurons (Perkins et al., 1986; Swoboda et al., 2000). We then assessed the function and morphology of neuronal cilia in these animals.
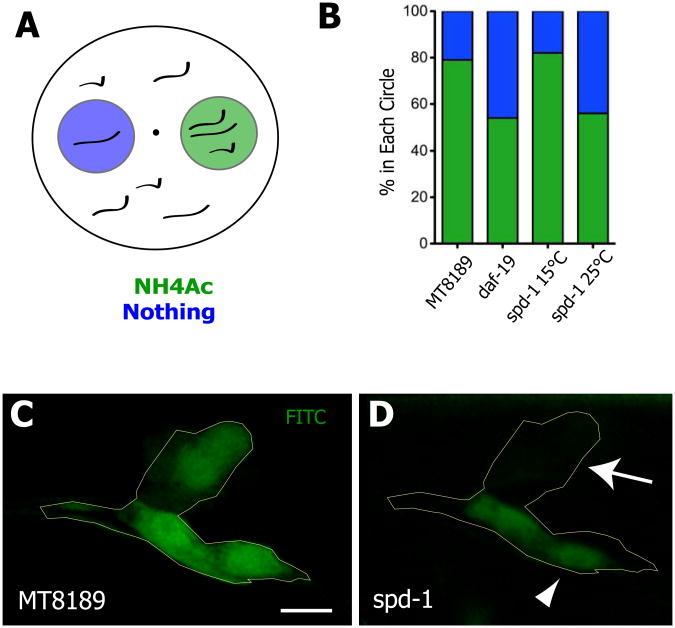
The proper functioning of neuronal sensory cilia is essential for worms to detect chemoattractants (Perkins et al., 1986), so we initially assayed the role of PRC1 in cilia function by scoring chemotaxis of spd-1 worms at permissive and restrictive temperatures. Because spd-1 mutant worms display a mild uncoordinated phenotype (O'Connell et al., 1998), we designed a quantification method that uncouples effects on movement and chemotaxis (Fig. 6A). Worms were placed equidistant from circles containing the attractant or containing no attractant (Fig. 6A), and chemotaxis was quantified as the number of worms successfully moving into the chemoattractant circle as compared to the number successfully moving into the empty circle. In this assay, spd-1 worms at the permissive temperature chemotaxed normally (Fig. 6B). Importantly however, when shifted to the restrictive temperature, spd-1 worms failed to chemotax (Fig. 6B). The chemotaxis defect observed with spd-1 mutant worms at the restrictive temperature was comparable to that of daf-19 depleted worms, which are known to lack cilia (Fig. 6B; (Perkins et al., 1986; Swoboda et al., 2000)).
Figure 6. PRC1 is essential for cilia function in C. elegans.
(A) Diagram on left shows the experimental set-up: worms were placed in the middle of the plate and then the number present in a circle with chemoattractant (green) or without (blue) was scored. (B) Graph shows that spd-1 mutants at 25° chemotax ineffectively compared to spd-1 mutants at 15°C (p<0.0001, Fisher's exact test) and MT8189 controls (p<0.0001, Fisher's exact test), and this defect is comparable to worms fed RNAi against daf-19 (p-value n.s.), which lack cilia (Perkins et al., 1986). For the chemotaxis assay n>200 for all strains. (C) FITC dye filling in neurons of a control worm. (D) Defective neuronal filling with FITC dye in spd-1 worms at 25°C. Outlines in C and D represent approximate position of neuronal cell bodies. Some cells fail to fill (arrow), while others display reduced filling (arrowhead). Overall, fluorescence intensity in spd-1 mutant worms at 25°C was reduced to 57% of control levels (see Supp. Fig. 4).
To confirm a defect in cilia function, we next exploited the finding that functional cilia mediate the uptake of dye by sensory neurons (Fig. 6C; (Hedgecock et al., 1985)). We observed that many neuronal cell bodies in spd-1 worms at restrictive temperatures failed to fill with dye (Fig. 6D, arrow). Moreover, those neurons in spd-1 mutants that did fill were consistently less brightly labeled than were control neurons (Fig. 6D, arrowhead). We quantified dye intensity in several neurons from several animals and observed that spd-1 mutant worms at restrictive temperature displayed a 43% reduction in fluorescence intensity as compared to controls (Supp. Fig. 2B). Similar phenotypes - both loss of dye filling and reduction in intensity of dye filling - are observed in bona fide cilia mutants in C. elegans (Perkins et al., 1986). Thus, two distinct assays indicate that cilia function was impaired in the absence of wild-type PRC1.
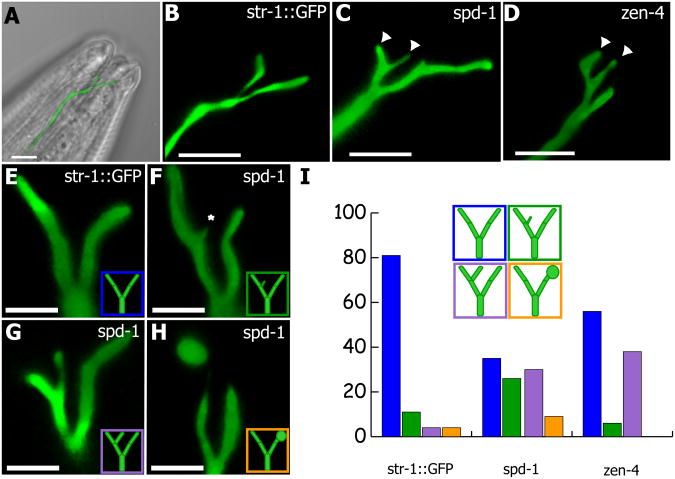
To examine the role of PRC1 in governing cilia morphology, we crossed the spd-1 worms into the str-1∷GFP strain, with which the cilia on AWB neurons can be visualized (Mukhopadhyay et al., 2007; Mukhopadhyay et al., 2008). The AWB cilia develop a complex architecture that is perturbed by mutation of genes involved in ciliogenesis, thus providing a sensitive assay for candidate genes (Mukhopadhyay et al., 2007; Mukhopadhyay et al., 2008; Murayama et al., 2005). AWB cilia typically display a two-pronged structure, though there is some variability in control worms, and cilia with ectopic branches are occasionally observed (Fig. 7A, B, E, I; and see (Mukhopadhyay et al., 2007; Mukhopadhyay et al., 2008)). In spd-1(str-1∷GFP) mutant worms at restrictive temperature, we observed a dramatic increase in the number of cilia with ectopic branches (Fig. 7C, F, G, I). Moreover, we observed an increased frequency of large bulges on sensory cilia of mutant worms (Fig. 7H, I). These phenotypes in AWB cilia, both bulges and ectopic projections, have been previously observed following mutation of genes involved in ciliogenesis (Mukhopadhyay et al., 2007; Mukhopadhyay et al., 2008).
Figure 7. PRC1 and MKLP1 control morphology of sensory cilia inC. elegans.
(A,B) Wild-type worms expressing GFP behind the str-1 promoter in the AWB neurons. The majority (81%) of wild-type str-1∷GFP worms have no ectopic projections (blue, E) on their AWB cilia. In spd-1 worms at 25°C (C), however, over half (56%) have small (F) to large (G) ectopic projections (green, purple, asterisk), while a minority have an additional phenotype of GFP accumulation at the distal tip (H, orange). In zen-4 worms (D) at 25°C there is also an increase in ectopic projections (42%). (I) Percentages of worms with cilia morphology. For cilia morphology str-1∷GFP, n = 27, spd-1(str-1∷GFP), n = 43, zen-4(str-1∷GFP), n = 38. For A-C scale bars = 5μm, D-G scale bars = 2μm.
Finally, because PRC1 acts at the central spindle together with MKLP-1 (Kurasawa et al., 2004) and because both proteins localize to the base of cilia (Fig. 1B, C), we assessed the role of MKLP-1 in AWB cilia. Using the temperature-sensitive MKLP-1 mutant zen-4, we found that loss of MKLP-1 resulted in a significantly increased number of ectopically branched AWB cilia (Fig. 7D, I), consistent with our results for PRC1/spd-1.
Conclusions
Our data demonstrate that PRC1 and MKLP-1 are required for normal AWB cilia morphology in C. elegans. Coupled to the spatially-distinct patterns of localization of PRC1, MKLP-1, INCENP, centriolin, and other central spindle/midbody proteins at basal bodies in Xenopus motile cilia and in human primary cilia, our data suggest that these proteins play a conserved role in regulating cilia structure and function.
The function of the central spindle proteins in relation to cilia remains unknown. One role for these proteins in the central spindle is microtubule bundling, and so it is possible that they are required for centrosome or basal body structure; indeed, MKLP-1 and PRC1 can localize to centrosomes. However, in post-mitotic multi-ciliated cells in Xenopus, we found these proteins to localize adjacent to, but not in, the basal body, suggesting that they may serve a different function. Another possibility is that these proteins act in ciliary disassembly, as the Fa2p kinase regulates ciliary disassembly in Chlamydomonas but localizes to midbodies when expressed in mammalian cells (Mahjoub et al., 2004).
Another possible role for the central spindle proteins is in recruitment of exocyst-mediated vesicle traffic to the cytokinetic furrow (Gromley et al., 2005). Given the localization of exocyst components to the base of cilia and their implication in ciliogenesis (Park et al., 2008; Zuo et al., 2009), it is tempting to suggest that the central spindle proteins may influence cilia-related vesicle traffic. This model would be consistent with our localization data, as the rootlet is implicated as a site of vesicle trafficking (Fariss et al., 1997; Gray et al., 2009; Park et al., 2008; Yang and Li, 2005). While the mechanisms of action must be explored further, the identification of this new connection between the cytokinetic apparatus and cilia should accelerate our understanding of both, as further study in one context may now provide important new insights for understanding the other (Supp. Fig. 3).
Finally, our data may shed light on the etiology of human ciliopathies such as Bardet-Biedel Syndrome, as the BBS6 protein localizes prominently to the midbody during cytokinesis (Kim et al., 2005). Indeed, while mutation of this gene in humans leads to a disease that is classified as a ciliopathy, experimental manipulation of BBS6 has been found to have relatively modest effects on cilia morphology and function (Shah et al., 2008). By contrast, knockdown of BBS6 prevents cell abscission during cytokinesis (Kim et al., 2005). Likewise, knockdown of the intraflagellar transport protein IFT27 leads to both flagellar defects and cell division defects (Qin et al., 2007), and this protein has been reported to localize to vesicles associated with the cytokinetic furrow (Baldari and Rosenbaum, 2010). Together with these findings, our data raise the possibility of a connection between cytokinesis defects and ciliopathy phenotypes. Thus, while the pleiotropic roles of central spindle proteins will make the study of their roles in cilia challenging, further exploration of the functional connection between the cytokinetic machinery and cilia should be exciting.
Material and Methods
Xenopus embryo isolation and injection
Female Xenopus laevis were injected with 700 ml of human chorionic gonadotropin (HCG) hormone and stored at 18°C overnight. The next day, oocytes were isolated from mothers and fertilized (Sive et al., 2000). Embryos were dejellied with a 3% cysteine solution in 1/3× MMR. To image CLAMP localization embryos were injected on the ventral side of the embryo at the 4-cell stage with 300pg/per injection of CLAMP-RFP mRNA made using the mMessage kit (Ambion). Embryos were grown to stage 13, early neurula, and stage 30, tailbud, before fixation.
Immunohistochemistry on Xenopus embryos
Immunostaining for central spindle and passenger proteins was performed essential as previously described (Lee et al., 2008a).
Embryos were grown to appropriate stages and fixed in Dent's Fix (80% methanol/20% DMSO)(Becker and Gard, 2006) overnight at 4°C. Embryos were fully dehydrated in methanol and stored at -20°C overnight. Embryos were slowly rehydrated by 5 min washes in 75% methanol in PTW (PBS + 1% tween 20), 50% methanol in PTW, 25% methanol in PTW, and finally PTW. Embryos were bleached in a light box in a solution of 10% H2O2, 0.5% formamide, and 2.5% 2× SCC. Embryos were then washed 3× 5 min in TBST. Embryos were blocked in 300 ml of TBS + 10% FBS + 5% DMSO. Primary antibody was added; rabbit-anti-PRC1 (1:20) (BioLegend), rabbit-anti-MKLP1 (1:200)(Potapova et al., 2006), rabbit-anti-INCENP (1:800)(Potapova et al., 2006), Aurora-B (1:50)(Potapova et al., 2006), rabbit-anti-centriolin serum (1:100)(Gromley et al., 2005) or mouse-anti-γ-tubulin (1:250) (Abcam ab27076). Embryos were incubated overnight at 4°C. Embryos were washed 5× 1 hr in TBST at RT the next day. Embryos were then blocked as before for 1 hr and anti-rabbit Alexa fluor 488 (1:250) and anti-mouse Alexa fluor 555 (1:250) were added. Embryos were incubated again over night at 4°C and washed 5× 1 hr in TBST at RT the next day. For embryos that were being imaged for 〈-tubulin localization TBST was removed and 300 ml of mouse-anti-〈-tubulin antibody + blocking solution was added (1:250). Embryos were incubated for 1 hr at room temperature. Embryos were washed again for 3× 1 hr washes in TBST. Secondary antibodies were added as above. In some cases during the last wash a dilution of (1:2000) of DAPI was added to label nuclei.
Image acquisition of Xenopus embryos
All imaging was performed using a Zeiss LSM5 PASCAL microscope with either 40× planNeofluar (1.3na), 63× planApochromat (1.4na) oil immersion objectives or 100× Fluar (1.3na). Confocal optical slices were collected and maximum intensity projections made with Zeiss LSM5 software. For presentation purposes, some images have been processed with Abode Photoshop.
Cell Culture and Imaging
RPE-hTERT cells were cultured in DMEM/F12 (10% FBS/PS/Glut). Confluent cells were ciliated by serum starvation in (0.2% FCS) for 48 hours. Cells were fixed in methanol (ice cold) for 5 minutes on ice or 4% PFA for 15 minutes at room temperature. Following fixation cells were blocked for 1 hr in 3% BSA/0.1% Triton-X100 and incubated with primary antibodies; rabbit-anti-PRC1 (1:50) (Abcam, ab21437), rabbit-anti-MKLP-1 (1:100) (Santa Cruz, sc-22793; H-110), and anti-acetylated-tubulin (1:10,000) (Sigma-Aldrich T6793; clone 6-11B-1) 2-4 hrs at RT in 3% BSA. Secondary antibody incubation was performed for 1 hr at room temperature using Jackson Immunoresearch secondary antibodies (1:500) and Hoechst (1:100,000) (33342, Invitrogen). Samples were embedded in Fluoromount-G (Beckman Coultier). Imaging was performed using an Everest deconvolution workstation (Intelligent Imaging Innovations) equipped with a Zeiss AxioImager.Z1 microscope and a CoolSnapHQ-cooled CCD camera (Roper Scientific) and a 63× PlanApochromat, NA 1.4 objective and processed using SlideBook 5 (Intelligent Imaging Innovations) and ImageJ.
Proteome sets
Cilia proteins were downloaded from the ciliary proteome database at http://v3.ciliaproteome.org/cgi-bin/index.php (Gherman et al., 2006). The core basal body proteome set was defined as centrosome and centriole proteins identified by mass spectrometry (MS) (Andersen et al., 2003; Keller et al., 2005). For the extended set, basal body, flagellum, and cilium proteins identified by either MS or comparative genomics were also included (Avidor-Reiss et al., 2004; Li et al., 2004; Liu et al., 2007). Human midbody proteins were downloaded from MiCroKit 3.0, a manually curated database of protein subcellular localizations, experimentally verified by fluorescence microscopy (http://microkit.biocuckoo.org, (Ren et al., 2009)). The set of all human Entrez proteins were retrieved from the Ensembl database at www.ensembl.org (Hubbard et al., 2009).
Proteome Overlap Analysis
The hypergeometric distribution was used to assess if the midbody and cilia proteomes share a significant number of proteins. The cumulative p-value was calculated using
| (1) |
where m and n are the number of proteins in the midbody and cilia proteomes, k is the number of overlapping proteins, and N is the size of the human proteome.
Network Analysis
The functional network of human genes was prepared as described previously (Gray et al., 2009) and is searchable at:
http://www.functionalnet.org/humannet/
As a measure of connectivity, we asked whether the proteins most strongly connected to midbody proteins in the gene network tended to be ciliary proteins using a method of Gaussian field label propagation (Mostafavi et al., 2008). First, a bias score was assigned to each gene in the network: +1 to known midbody genes and k to all other genes. The value k is defined as:
| (2) |
where n is the number of midbody genes and N is the total number of genes in the network. The connectivity score was then computed as:
| (3) |
where W is an association matrix where entry wij is the network edge weight between genes i and j, yi is the bias node of i, and f is the vector of final scores where entry fi indicatives how strongly connected gene i is to the set of midbody genes. Each gene was rank-ordered by its final score as described previously (Lee et al., 2008b), and true positive and false positive rates were calculated as a function of rank. From the resultant ROC curve, the area under the ROC curve (AUC) was calculated. To obtain the p-value, the mean and standard deviation of AUCs obtained from 1000 random sets were calculated and a one-tailed Z-test was performed.
C. elegans strains and assays
C. elegans strains were cultured in standard growth conditions on nematode growth medium with Escherichia coli strain OP50. Except where indicated, all worms were reared at 25°C. All assays were performed at 25°C. Temperature sensitive strains were maintained at 15C with the exception of assays. Strains used were lin-15B(n765), spd-1(oj5), zen-4(or153) and kyls104(str-1p∷gfp). spd-1(oj5) and zen-4(or153) strains expressing GFP behind the str-1 promoter were made using standard mating techniques. All strains were provided by the Caenorhabditis Genetics Center.
Temperature sensitive spd-1 and zen-4 mutant strains were grown at 15°C. Embryos were shifted to the restrictive temperature of 25°C when they were between the 2-fold stage and L1. This timing allowed PRC1/MKLP-1 function to be disrupted shortly after the commencement of daf-19 expression – seen to be from the 2-fold stage to early L2 (Swoboda et al., 2000) – while avoiding the early embryonic cell divisions. spd-1 adult worms that had been raised at either the permissive or restrictive temperatures were assayed using chemotaxis plates set up in a variation on previous protocols (Troemel et al., 1997). Briefly, adult worms were washed from OP50 fed plates using M9 buffer, and washed again with M9 until all bacteria were removed. Worms were then placed on the center of the agar plate, equidistant from two circles – one soaked with 50μl of 100mM ammonium acetate attractant, the other empty – and allowed to migrate for 1 hour, at which time the worms within each circle were counted. FITC filling was performed using standard protocols (Hedgecock et al., 1985), and the fluorescent intensity of confocal projections was measured using Image Pro Plus.
Imaging of C. elegans cilia and neurons
Worms for cilia and neuron imaging were mounted on agar pads and anesthetized with sodium azide. Confocal images of the AWB neuron cilia and dye-filled cell bodies were obtained using a Zeiss LSM5 PASCAL microscope with 63× plan Apochromat (1.4na) or 100× Fluar (1.3na) oil-immersion objectives. Confocal optical slices were collected using Zeiss LSM5 software and 3-dimensional reconstructions of the cilia were made using Imaris x64 4.5.2.
Supplementary Material
Acknowledgments
We thank Phil Abitua for technical assistance. All strains were provided by the Caenorhabditis Genetics Center which is funded by the NIH/NCRR. This work was supported by grants to E.M.M. from the NSF, NIH, Welch Foundation (F-1515), Texas Institute for Drug and Diagnostic Development, and a Packard Fellowship; to R.H.G and S.G.B. from a VIDI/Aspasia grant from Dutch Organisation for Scientific Research (VIDI 91766354) and EU FP7 consortium SYSCILIA; and to J.B.W. from the NIH/NIGMS, The March of Dimes, The Burroughs Wellcome Fund, the American Asthma Foundation, and the Texas Advanced Research Program. J.B.W. is an Early Career Scientist of the Howard Hughes Medical Institute.
References
- Albertson R, Cao J, Hsieh TS, Sullivan W. Vesicles and actin are targeted to the cleavage furrow via furrow microtubules and the central spindle. J Cell Biol. 2008;181:777–90. doi: 10.1083/jcb.200803096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–39. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Baldari CT, Rosenbaum J. Intraflagellar transport: it's not just for cilia anymore. Curr Opin Cell Biol. 2010;22:75–80. doi: 10.1016/j.ceb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker BE, Gard DL. Visualization of the cytoskeleton in Xenopus oocytes and eggs by confocal immunofluorescence microscopy. Methods Mol Biol. 2006;322:69–86. doi: 10.1007/978-1-59745-000-3_6. [DOI] [PubMed] [Google Scholar]
- Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol. 2003;13:1145–9. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, Jansen G. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:7157–62. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariss RN, Molday RS, Fisher SK, Matsumoto B. Evidence from normal and degenerating photoreceptors that two outer segment integral membrane proteins have separate transport pathways. J Comp Neurol. 1997;387:148–56. doi: 10.1002/(sici)1096-9861(19971013)387:1<148::aid-cne12>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–9. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch D, Farbman AI. Development of order during ciliogenesis. Anat Rec. 1968;162:221–32. doi: 10.1002/ar.1091620209. [DOI] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–2. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–9. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RS, Abitua PB, Wlodarczyk BJ, Szabo-Rogers HL, Blanchard O, Lee I, Weiss GS, Liu KJ, Marcotte EM, Wallingford JB, Finnell RH. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nature Cell Biology. 2009;11:1225–32. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Hayes JM, Kim SK, Abitua PB, Park TJ, Herrington ER, Kitayama A, Grow MW, Ueno N, Wallingford JB. Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev Biol. 2007;312:115–30. doi: 10.1016/j.ydbio.2007.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–70. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Horvath J, Fliegauf M, Olbrich H, Kispert A, King SM, Mitchison H, Zariwala MA, Knowles MR, Sudbrak R, Fekete G, Neesen J, Reinhardt R, Omran H. Identification and analysis of axonemal dynein light chain 1 in primary ciliary dyskinesia patients. Am J Respir Cell Mol Biol. 2005;33:41–7. doi: 10.1165/rcmb.2004-0335OC. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–85. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–8. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kieserman EK, Glotzer M, Wallingford JB. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol. 2008;18:116–23. doi: 10.1016/j.cub.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–51. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Ou YY, Badano JL, Esmail MA, Leitch CC, Fiedrich E, Beales PL, Archibald JM, Katsanis N, Rattner JB, Leroux MR. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci. 2005;118:1007–20. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. Embo J. 2004;23:3237–48. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, EK K, Gray RS, Park TJ, Wallingford JB. Whole-Mount Fluorescence Immunocytochemistry on Xenopus Embryos. Cold Spring Harbor Protocols. 2008a doi: 10.1101/pdb.prot4957. [DOI] [PubMed] [Google Scholar]
- Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008b;40:181–8. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–52. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Qasim Rasi M, Quarmby LM. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol Biol Cell. 2004;15:5172–86. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. The cell biological basis of ciliary disease. J Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO Journal. 2007;26:2966–80. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Lu Y, Shaham S, Sengupta P. Sensory signaling-dependent remodeling of olfactory cilia architecture in C. elegans. Dev Cell. 2008;14:762–74. doi: 10.1016/j.devcel.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama T, Toh Y, Ohshima Y, Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J Mol Biol. 2005;346:677–87. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–13. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–44. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–4. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell KF, Leys CM, White JG. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 1998;149:1303–21. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci. 2007;120:1868–76. doi: 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–11. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–9. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–87. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Daum JR, Pittman BD, Hudson JR, Jones TN, Satinover DL, Stukenberg PT, Gorbsky GJ. The reversibility of mitotic exit in vertebrate cells. Nature. 2006;440:954–8. doi: 10.1038/nature04652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Liu Z, Gao X, Jin C, Ye M, Zou H, Wen L, Zhang Z, Xue Y, Yao X. MiCroKit 3.0: an integrated database of midbody, centrosome and kinetochore. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AS, Farmen SL, Moninger TO, Businga TR, Andrews MP, Bugge K, Searby CC, Nishimura D, Brogden KA, Kline JN, Sheffield VC, Welsh MJ. Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc Natl Acad Sci U S A. 2008;105:3380–5. doi: 10.1073/pnas.0712327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Current Topics in Developmental Biology. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, N.Y: 2000. [Google Scholar]
- Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–6. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–30. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–90. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Steinman RM. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am J Anat. 1968;122:19–55. doi: 10.1002/aja.1001220103. [DOI] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–21. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–9. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Verbrugghe KJ, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14:1755–60. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Yang J, Li T. The ciliary rootlet interacts with kinesin light chains and may provide a scaffold for kinesin-1 vesicular cargos. Exp Cell Res. 2005;309:379–89. doi: 10.1016/j.yexcr.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Zuo X, Guo W, Lipschutz JH. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–9. doi: 10.1091/mbc.E08-07-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.