SUMMARY
Polar bears are uniquely adapted to life in the High Arctic and have undergone drastic physiological changes in response to Arctic climates and a hyperlipid diet of primarily marine mammal prey. We analyzed 89 complete genomes of polar bear and brown bear using population genomic modeling and show that the species diverged only 479–343 thousand years BP. We find that genes on the polar bear lineage have been under stronger positive selection than in brown bears; nine of the top 16 genes under strong positive selection are associated with cardiomyopathy and vascular disease, implying important reorganization of the cardio-vascular system. One of the genes showing the strongest evidence of selection, APOB, encodes the primary lipoprotein component of low-density lipoprotein (LDL); functional mutations in APOB may explain how polar bears are able to cope with life-long elevated LDL levels that are associated with high risk of heart disease in humans.
INTRODUCTION
The polar bear (Ursus maritimus) is uniquely adapted to the extreme conditions of life in the High Arctic and spends most of its life out on the sea ice. In cold Arctic climates, energy is in high demand. Lipids are the predominant energy source and the polar bear has a lipid-rich diet throughout life. Young nurse on milk containing c. 27% fat (Hedberg et al., 2011) and adults feed on a marine mammal diet, primarily consisting of seals and their blubber (Thiemann et al., 2008). Polar bears have substantial adipose deposits under the skin and around organs, which can comprise up to 50% of the body weight of an individual, depending on its nutritional state (Atkinson and Ramsay, 1995; Atkinson et al., 1996).
The polar bear is most closely related to the brown bear (Ursus arctos), a widely distributed omnivore found in a variety of habitats across the Holarctic (Fig. 1). The two species differ fundamentally in their ecology, behavior and morphology, reflecting adaptations to different ecological niches. Despite ample data there is still no consensus regarding when the two species diverged. Inferences based on the fossil record suggest polar bears diverged from brown bears some 800–150 thousand years ago (kya) (Kurtén, 1964). Estimates based on genomic data span an order of magnitude from 5–4 million years ago (Mya) (Miller et al., 2012) to c. 600 kya (Hailer et al., 2012), depending on assumptions about effective population size and migration in the period when the two populations were drifting apart. Establishing a reliable time frame for when the polar bear emerged as a species is essential for our understanding of what evolutionary processes drove speciation, and how fast novel adaptations to extreme environments can arise in a large mammal.
Fig. 1. Sampling localities.
Polar and brown bear distributions are shown in blue and brown shading, respectively. See also Table S2.
Here, we apply a population genomic framework to analyze complete nuclear genomes of polar bear and brown bear populations (Fig. 1, Tables S1, S2) to (i) estimate when polar bears and brown bears diverged; (ii) infer the joint demographic history of the two species to elucidate what happened after they diverged; and (iii) detect genes under positive selection in polar bears to gain insight into polar bear evolution and the genetic background of its unique adaptations to life in the High Arctic.
To address these issues, we deep-sequenced and de novo assembled a polar bear reference genome at a depth of 101X (Extended Experimental Procedures Section Polar Bear Reference Genome and de novo Assembly) and re-sequenced at 3.5X to 22X coverage 79 Greenlandic polar bears and ten brown bears from Fennoscandia, mainland US and the Admiralty, Baranof, and Chichagof (ABC) Islands off the coast of Alaska (Fig. 1) (Extended Experimental Procedures Section Samples, Tables S1, S2, Fig. S1).
RESULTS AND DISCUSSION
Joint demographic history of polar bears and brown bears
To infer the joint demographic history of polar bears and brown bears, we used a novel method based on identity by state (IBS) tracts of DNA shared within and between populations (Harris and Nielsen, 2013) and ∂a∂i (diffusion approximation for demographic inference (Gutenkunst et al., 2009)), which infers demographic parameters based on a diffusion approximation to the site frequency spectrum. The two models differed in their individual parameter estimates (Table S3); in part reflecting the fact that the IBS tract method uses both recombination rate and mutation rate, and ∂a∂i uses only the latter. However, despite the inherent uncertainty in the genome-wide mutation rate estimate, which we calibrated using deep fossil divergence dates (Fig. S2A), the estimates from the two models are in fact quite similar with regards to divergence time, relative effective population sizes and direction of gene flow.
We find evidence of smaller long-term effective population sizes in polar bears than in brown bears (Fig. 2A). Genetic diversity is a function of effective population size, and the number of private SNPs in polar bears (2.6 million, Fig. S1B) is about one third of that in brown bears (7.7 million, Fig. S1C). Similarly, patterns of linkage disequilibrium (LD) can be informative about demographic history (Reich et al., 2001) and we find a slower rate of LD decay in polar bears (Fig. S3A).
Fig. 2. Demographic inference.
Joint demographic model for polar bear and North American brown bear populations inferred using the identity by state (IBS) tract method (A). Joint past population is in grey, polar bear in blue and brown bear in brown. Estimated effective population sizes are indicated and the migration rate is in genetic replacements per generation. The recent brown bear population size has been downscaled by a factor of 20, the recent polar bear population size is to scale. (B, C) Distribution of IBS tract length from our observed data (solid line) and from model prediction (dotted line) inferring gene flow from polar bear into brown bear (B) or using a simple isolation-with-migration (IM) model (C), which does not account for past population size changes. There are only two black dotted curves in (C) because the IM model constrains the within-polar bear and within-brown bear tract lengths to be the same. See also Fig. S4B and Table S3.
Prior to divergence, we find a 10-fold decline in the global joint ancestral population (Table S3). Polar bears declined in population size after the split from brown bears, although we were unable to confidently estimate the timing of the bottleneck. However, both the IBS tract method and ∂a∂i indicate that the population size decrease in polar bears was either of a greater magnitude or of a longer duration than in brown bears, in agreement with our other indicators of relative population sizes.
The age of the polar bear as a species
To reliably estimate when polar bears and brown bears diverged, we used the IBS tract method (Harris and Nielsen, 2013) and ∂a∂i (Gutenkunst et al., 2009), which both take past population size changes into account. Both approaches indicated that the two species diverged only c. 479–343 kya (Fig. 2A, Table S3). Because genotyping errors appear as singletons and given that in both methods singletons lead to increasing divergence times estimates, we can conclude that the polar bear likely emerged closer to the lower bound of our estimate. Our date greatly decreases the age of polar bear origin, and agrees with fossil evidence (Kurtén, 1964; Miller et al., 2012).
We assessed the effect of using our more complex demographic models versus simpler models by analyzing our data using a simple isolation-with-migration model similar to that used by (Hailer et al., 2012; Miller et al., 2012). The procedure yielded an older divergence date in the range of 1.6–0.8 Mya (Table S3). However, we found that our complex model with a more recent divergence time estimate was a better fit to our empirical data (Fig. 2B, C). Discrepancies between our divergence date and previous genomic estimates highlight the impact of accounting for past population size changes on divergence time estimates, suggesting that models that do not account for past population size changes have the potential to overestimate divergence times (e.g. Hailer et al., 2012; Miller et al., 2012).
The timing of polar bear origin coincides with Marine Isotope Stage (MIS) 11. MIS 11 was a warm period, which spanned c. 424–374 kya. It was the longest interglacial in half a million years (Dickson et al., 2009) and lasted almost 50 kyr (de Vernal and Hillaire-Marcel, 2008). The period was associated with a substantial decrease in Greenland ice-sheet volume; DNA from the basal part of the Dye 3 ice core from southern central Greenland (Willerslev et al., 2007) and abundant spruce pollen from the shore off southwest Greenland (de Vernal and Hillaire-Marcel, 2008) both suggest that boreal coniferous forest developed at least over southern Greenland. Such a prolonged interglacial could have enabled brown bears to colonize northern latitudes that were previously uninhabitable for the species, setting the stage for future allopatric speciation, as subsequent climatic and environmental change caused population isolation (Stewart et al., 2010).
Gene flow between polar bears and brown bears after divergence
Based on morphology and a phylogenetic analysis of their nuclear genomes, polar bears and brown bears are monophyletic sister species (Fig. S2B; Pagès et al., 2008). Nevertheless, the mitochondrial genomes of brown bear are paraphyletic and extant polar bear sequences are recovered as a monophyletic sister clade to the brown bear population from Alaska’s ABC Islands, within the diversity of brown bear (Fig. S2C). The consensus has been that this pattern reflects female-mediated gene flow from brown bears into polar bears c. 150 kya, with subsequent fixation of the brown bear mitochondrial lineage in polar bears (Lindqvist et al., 2010). However, this was not supported by a recent genomic study, which presented evidence that gene flow historically took place from polar bears into ABC brown bears, and not the other way around (Cahill et al., 2013).
Based on the IBS tract method, we find strong evidence of continuous gene flow from polar bears into North American brown bears after the species diverged (Fig. 2A, Table S3). We used the IBS tract method to compare likelihoods of two scenarios with parsimonious one-way gene flow, finding that gene flow from polar bears to North American brown bears explained the data better than the reverse scenario (Figs 2B, S4B, Table S3). In the former scenario, we estimate a migration rate of 0.0018% genetic replacement per generation. As a complementary approach, we used ∂a∂i to infer the parameters of a model with asymmetric two-way gene flow between polar bears and North American brown bears. With this approach, we observe non-zero migration in both directions, but infer a substantially higher migration rate in the polar-to-brown bear direction (Table S3). These results suggest that the major direction of introgression has historically been from polar bears into North American brown bears, in agreement with (Cahill et al., 2013).
We were unable to confidently infer when admixture took place. With the IBS tract method, we estimate gene flow from the timing of the polar bear bottleneck 319 kya to 148 kya (Fig. 2A), but the method has limited power to detect migration that occurred very close to the initial divergence time. With ∂a∂i, we infer continuous gene flow until the present. We see no admixture using classical structure analyses (Tang et al., 2005) (Fig. S4A), suggesting admixture is not a recent or current phenomenon. However, we note that the number of analyzed brown bear samples is limited, and none of our brown bear samples originate from regions where polar and brown bears are currently sympatric (i.e. where recently admixed individuals are most likely to be found).
To further investigate the question of admixture, we split the genomic data into 100 kbp regions (Table S4) and calculated the length distribution of regions that were introgressed between species, we find that the longest blocks were a maximum of 1.1 Mbp length. If admixture between species had taken place within the last hundreds of generations, we would expect longer tracts of shared DNA (Gravel, 2012; Pool and Nielsen, 2009). Hence the limited length of admixture blocks supports the hypothesis that admixture was an old event, and that enough time has passed for recombination to break up the long stretches of introgressed DNA.
In order to determine whether gene flow happened before or after the divergence of brown bear populations from different parts of the Holarctic, we used the D statistic (Durand et al., 2011; Green et al., 2010). We find evidence of gene flow between polar bears and all brown bear populations, suggesting that some gene flow took place prior to the divergence of the brown bear populations (Table S5). The strongest evidence is found with brown bears from the ABC Islands and the weakest with brown bear populations from North America and Fennoscandia, suggesting gene flow continued between polar bears and ABC brown bears also after the brown bear populations diverged. In addition, we find evidence of recent migration between brown bear populations. Our data included six brown bear samples from the ABC Islands (Fig. 1, Table S2). One of these individuals (ABC06) was from Admiralty, the island located closest to the US mainland. The mitochondrial genome of ABC06 clustered with the other five ABC individuals from Baranof and Chichagof Islands, as a sister group to the polar bear (Fig. S2C). We observe substantial levels of gene flow between polar bears and the Baranof and Chichagof individuals using the D statistic, as expected (Table S5). However, we find no signal of polar bear admixture in ABC06, which clustered with the Glacier National Park individual from Montana in the principal component analysis (Fig. S3D). We do not find evidence of polar bear admixture in the Glacier NP individual either, the mitochondrial genome of which clustered with European brown bears (Fig. S2C). The patterns in ABC06 reflect migration between the Admiralty Island and mainland US, in agreement with previous inferences based on nuclear microsatellites (Paetkau et al., 1999).
Genes under positive selection in polar bears
Despite being closely related species, the polar bear differs from the brown bear in ecology, behavior and morphology, and is a prime example of what happens when a species evolves through selection and adaptation to a novel environment/lifestyle. Our remarkably recent divergence time estimate of only c. 479–343 kya, coupled with stable isotope analysis of an ancient jawbone from Svalbard that indicates that polar bears were adapted to a marine diet and life in the high Arctic by at least 110 kya (Lindqvist et al., 2010), provides us with an unprecedented timeframe for rapid evolution. Assuming an average generation time of 11.35 years (Cronin et al., 2009; De Barba et al., 2010), the distinct adaptations of polar bears may have evolved in less than 20,500 generations; this is truly exceptional for a large mammal. In this limited amount of time, polar bears became uniquely adapted to the extremities of life out on the Arctic sea ice, enabling them to inhabit some of the world’s harshest climates and most inhospitable conditions.
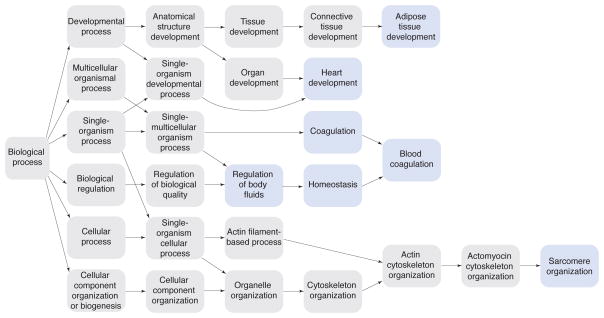
The observation of rapid evolutionary changes in the polar bear genome raises the question of what signatures of selection are to be found in the extant genomes. We find that the enrichment categories of the top candidate genes under positive selection (i.e. the genes that showed greater values for all of our three test statistics homogeneity test, Hudson-Aguade-Kreitman test and Fst estimation in the polar bear, Table S6) are associated with sarcomere organization, blood coagulation, heart development and adipose tissue development (Fig. 3, Extended Experimental Procedures Section Positive Selection). In brown bears, we do not find significant enrichment categories for the top 20 candidate genes under selection.
Fig. 3. Enrichment analysis.
Gene Ontology enrichment analysis for putative genes under positive selection in the polar bear lineage. We ranked genes based on their homogeneity test score by first considering genes where the ratio between polymorphisms and divergence was lower in the polar bear than in the brown bear samples. We used the web application GOrilla (http://cbl-gorilla.cs.technion.ac.il) to detect biological process terms enriched with top genes in the ranked list. Blue shading indicates biological categories significantly enriched with genes under positive selection in the polar bear lineage, after correction for multiple tests.
In general, we also find evidence of more positive selection acting on the polar bear lineage than on the brown bear lineage. Polar bears had markedly higher values in the distribution of our primary test score, the homogeneity test, compared to brown bears (Fig. 4A). These patterns may only in part be explained by variation in sample size and effective population size (Fig. 4B). Overall, our data support a scenario of polar bears evolving rapidly and being under strong positive selection following the divergence from brown bears.
Fig. 4. Positive selection analysis.
(A, B) Distribution of the homogeneity test scores for the top-50 genes in polar bear (blue) and brown bear (brown). We compared the observed distribution versus the expected distribution under neutrality, using the demographic model presented in Table S3; (C, D) Predicted functional impact of polar bear-specific protein substitutions. We reported the functional classification and probability of being damaging for polar bear-specific missense mutations located in the top 20 genes under positive selection, according to the two metrics HumanDiv and HumanVar computed by PolyPhen-2. See also Table S7.
Genes associated with adipose tissue development and fatty acid metabolism
The enrichment of genes associated with adipose tissue development (Fig. 3) reflects the crucial role lipids play in the ecology and life history of polar bears; the species is adapted to cope with a diet rich in fatty acids (e.g. (Smith, 1980; Stirling and Archibald, 1977)) and has substantial adipose deposits (Atkinson and Ramsay, 1995; Atkinson et al., 1996). Cholesterol levels in blood plasma of polar bears are extreme (e.g. Ormbostad, 2012); in humans, elevated cholesterol levels are a major risk factor for the development of cardiovascular disease (Cannon et al., 2010). It remains an enigma how polar bears are able to deal with such lifelong elevated levels of cholesterol.
The enriched categories may highlight the genes that have been important in polar bear adaptation to a lipid-rich diet. A top gene in our selection scan was APOB (Table 1), which produces apolipoprotein B (apoB), the primary lipid-binding protein of chylomicrons and low-density lipoproteins (LDL) (Whitfield et al., 2004). LDL cholesterol is a major risk factor for heart disease and is also known as “bad cholesterol”. ApoB enables the transport of fat molecules in blood plasma and lymph and acts as a ligand for LDL receptors, facilitating the movement of molecules such as cholesterol into cells (Benn, 2009). The extreme signal of APOB selection implies an important role for this protein in the physiological adaptations of the polar bear. The gene is ranked second using our homogeneity test score, has an Fst ranking in the top 3% of the empirical distribution, and the ratio of fixed-to-polymorphic mutations in the polar bear lineage is 1:2, compared with 1:162 in the brown bear lineage—an 80-fold reduction.
Table 1. Top-20 genes under positive selection in polar bears.
We used several statistics to analyse the coding regions of 19,822 genes annotated across the polar and brown bear population samples, using the giant panda (Ailuropoda melanoleuca) genome sequence as an outgroup. Genes were ordered based on their homogeneity test score; we only considered genes with (i) a significant nominal p-value for the HKA test for selection in the polar bear lineage only, and (ii) a ranked Fst over the 90th percentile. See also Tables S6, S7.
| Gene | Length (bp) | Homogeneity test score | HKA test p-value | Fst |
|---|---|---|---|---|
| TTN | 99,416 | 16.76 | 2.47E-03 | 0.93 |
| APOB | 13,264 | 13.16 | 1.54E-05 | 0.89 |
| OR5D13 | 871 | 8.08 | 4.93E-10 | 0.82 |
| FCGBP | 5,216 | 6.36 | 8.20E-04 | 0.85 |
| XIRP1 | 3,848 | 6.05 | 1.50E-05 | 0.88 |
| COL5A3 | 4,402 | 5.89 | 1.38E-02 | 0.81 |
| LYST | 11,172 | 5.58 | 1.08E-03 | 0.89 |
| ALPK3 | 3,007 | 5.34 | 1.32E-05 | 0.91 |
| VCL | 3,106 | 4.87 | 7.51E-03 | 0.82 |
| SH3PXD2B | 2,458 | 4.34 | 2.81E-05 | 0.88 |
| EHD3 | 1,230 | 4.28 | 1.62E-03 | 0.90 |
| IPO4 | 1,260 | 4.18 | 1.81E-04 | 0.86 |
| ARID5B | 3,109 | 4.14 | 1.31E-02 | 0.84 |
| ABCC6 | 3,346 | 4.02 | 9.26E-03 | 0.85 |
| LAMC3 | 1,885 | 3.93 | 9.25E-03 | 0.85 |
| CUL7 | 2,701 | 3.86 | 4.43E-03 | 0.83 |
| C15orf55 | 3,001 | 3.86 | 1.71E-02 | 0.89 |
| POLR1A | 4,499 | 3.85 | 1.89E-02 | 0.82 |
| AIM1 | 4,344 | 3.8 | 2.03E-02 | 0.92 |
| OR8B8 | 965 | 3.71 | 7.37E-06 | 0.87 |
Due to a lack of appropriate functional studies of polar bears, we were unable to directly identify causal variants. Nevertheless, we assessed the impact of polar bear–specific substitutions on human proteins for top-20 genes under positive selection by computational predictions: a large proportion (c. 50%) of mutations were predicted to be functionally damaging (Fig. 4C, D, Table S7). Substantial work has been done on the functional significance of APOB mutations in other mammals. In humans and mice, genetic APOB variants associated with increased levels of apoB are also associated with unusually high plasma concentrations of cholesterol and LDL, which in turn contribute to hypercholesterolemia and heart disease in humans (Benn, 2009; Hegele, 2009). In contrast with brown bear, which has no fixed APOB mutations compared to the giant panda genome, we find nine fixed missense mutations in the polar bear (Fig. 5A). Five of the nine cluster within the N-terminal βα1 domain of the APOB gene, although the region comprises only 22% of the protein (binomial test p-value = 0.029). This domain encodes the surface region and contains the majority of functional domains for lipid transport. We suggest that the shift to a diet consisting predominantly of fatty acids in polar bears induced adaptive changes in APOB, which enabled the species to cope with high fatty acid intake by contributing to the effective clearance of cholesterol from the blood.
Fig. 5. The apoB and LYST protein sequences.
The distribution of fixed non-synonymous polar bear mutations (blue arrows) compared to the brown bear, using the giant panda sequence as an outgroup. (A) Mutations predicted to affect protein structure based on apoB alignments across 20 vertebrate species, using the SIFT algorithm (Sim et al., 2012), are indicated with hollow circles on arrows. The grey curve shows the cubic smoothing spline of the amino acid conservation scores; higher scores indicate higher conservation across 20 vertebrate species. The x-axis shows the amino acid position from the N-terminal, the five domains are based on the human apoB sequence (Prassl and Laggner, 2009). (B) The same representation as in panel A, but for the LYST protein sequence. The domains are based on [http://www.ebi.ac.uk/interpro/protein/LYST_HUMAN]. (C) Mapping of polar bear-specific substitutions and Chediak-Higashi syndrome causing variants on the protein structure of LYST N-terminal domain.
Genes associated with cardiovascular function
We find that nine out of the top 16 genes showing the strongest evidence of positive selection in polar bears are directly related to heart function in humans (Table 1). Mutations in all nine genes, including APOB, are associated with either atherosclerosis or cardiomyopathy in humans and other mammalian model organisms. TTN encodes Titin, an abundant protein of striated muscle, which includes cardiac muscle tissue; mutations in TTN are associated with familial dilated cardiomyopathy (Herman et al., 2012). XIRP1, also known as Cardiomyopathy-associated protein 1, is associated with the development of cardiac muscle cells (van der Ven et al., 2006). ALPK3 encodes a kinase and plays a role in cardiomyocyte differentiation; knockout genes in mice show both hypertrophic and dilated forms of cardiomyopathy (Van Sligtenhorst et al., 2012). VCL encodes vinculin, a cytoskeletal protein associated with cell-cell and cell-matrix junctions, which is also the major talin-binding protein in platelets. Defects in VCL are associated with dilated cardiomyopathy in humans (Olson et al., 2002). EHD3 encodes a class of cardiac trafficking proteins and plays a role in endocytic transport (Galperin et al., 2002). Regulation of EHD3 plays a role in a molecular pathway related to heart failure (Gudmundsson et al., 2012). ARID5B is involved in pathogenesis of atherosclerosis and adipogenesis (Wang et al., 2012). ABCC6 is associated with transport of molecules across membranes and is associated with premature atherosclerosis (Trip et al., 2002), and CUL7 plays a role in vascular morphogenesis (Arai et al., 2003).
Based on this evidence, we argue that potentially important re-organization of the cardiovascular system has taken place in polar bears since their divergence from brown bears, which may be related to polar bear ecology. Chronically elevated serum cholesterol, particularly LDL, contribute to the degenerative accumulation of plaques in the arteries, which can lead to progressive narrowing or blocking of blood vessels (Klop et al., 2013). Alternatively, smaller plaques may rupture and cause a clot to form and obstruct blood flow, leading to reduced blood supply of the heart muscle and eventually heart attack. Changes in behavior, including long distance swimming (Pagano et al., 2012), may also have imposed selection on other aspects of the cardio-vascular system, including cardiac morphology.
Genes associated with white fur
A white phenotype is usually selected against in natural environments, but is common in the Arctic (e.g. beluga whale, arctic hare and arctic fox), where it likely confers a selective advantage. A key question in the evolution of polar bears is which gene(s) cause the white coat color phenotype. The white fur is one of the most distinctive features of the species and is caused by a lack of pigment in the hair. We find evidence of strong positive selection in two candidate genes associated with pigmentation, LYST and AIM1 (Table 1). LYST encodes the lysosomal trafficking regulator Lyst. Melanosomes, where melanin production occurs, are lysosome-related organelles and have been implicated in the progression of disease associated with Lyst mutation in mice (Trantow et al., 2010). The types and positions of mutations identified in LYST vary widely, but Lyst-mutant phenotypes in cattle, mice, rats and mink are characterized by hypopigmentation, a melanosome defect characterized by light coat colour (Kunieda et al., 1999; Runkel et al., 2006; Gutiérrez-Gil et al., 2007). LYST contains seven polar bear-specific missense substitutions, in contrast to only one in brown bear. One of these, a glutamine to histidine change within a conserved WD40-repeat containing domain, is predicted to significantly affect protein function (Fig. 5B, Table S7). Three polar bear changes in LYST are located in proximity to the N-terminal structural domain, and map close to human mutations associated with Chediak-Higashi syndrome, a hair and eyes depigmentation disease (Fig. 5C). We predict that all these protein-coding changes, possibly aided by regulatory mutations or interactions with other genes, dramatically suppress melanin production and transport, causing the lack of pigment in polar bear fur. Variation in expression of the other colour-associated gene, AIM1 (absent in melanoma 1), has been associated with tumor suppression in human melanoma (Trent et al., 1990), a malignant tumor of melanocytes that affects melanin pigment production.
Conclusions
Our study reveals the strength of using a population genomic approach to resolve the evolutionary history of a non-model organism in terms of divergence time, demographic history, selection and adaptation. We find it remarkable that a majority of the top genes under positive selection in polar bears have functions related to the cardiovascular system and most of them to cardiomyopathy, in particular when considering their divergence from brown bears no more than c. 479–343 kya. Such a drastic genetic response to chronically elevated levels of fat and cholesterol in the diet has not previously been reported. It certainly encourages a move beyond the standard model organisms in our search for the underlying genetic causes of human cardiovascular diseases.
EXTENDED EXPERIMENTAL PROCEDURES
Detailed Extended Experimental Procedures can be found in the Extended Extended Experimental Procedures in the Supplementary Information (SI).
Samples and data
We deep-sequenced and de novo assembled a polar bear reference genome at a depth of 101X using the Illumina HiSeq 2000 sequencing platform (Table S1; Extended Experimental Procedures Section Polar Bear Reference Genome and de novo Assembly). The scaffold N50 size of the genome was c. 16 Mb (http://dx.doi.org/10.5524/100008). In addition, we generated complete genomes of multiple polar bears from three management areas around Greenland (Kane Basin and Baffin Bay in Northwest Greenland, and Scoresbysound/Ittoqqortoormiit in Central East Greenland) and several brown bears from Fennoscandia, mainland US and the Admiralty, Baranof, and Chichagof (ABC) Islands off the coast of Alaska (Fig. 1, Table S2; Extended Experimental Procedures Section Samples). We resequenced 18 polar bear and 10 brown bear genomes at high coverage (an average sequencing depth of ~22X), and an additional 61 polar bear genomes at low coverage (an average sequencing depth of 3.5X) (Extended Experimental Procedures Section Data Generation and QC Measures). We filtered data with a dedicated pipeline and removed low quality reads as well as sites showing unusual coverage compared to the empirical distribution, base quality score bias (p < 1e-5), strand bias (p < 1e-5), and deviation from Hardy-Weinberg Equilibrium (p < 1e-3) (Extended Experimental Procedures Section Data Generation and QC Measures). We analyzed the data within a population genomic framework (SI).
Divergence time and joint demographic history of polar bears and brown bears
We applied two approaches to estimate reliably when polar bears and brown bears diverged. Importantly, both methods incorporate past population size changes. We used a novel method based on identity by state (IBS) tracts of DNA shared within and between populations (Harris and Nielsen, 2013) and ∂a∂i (diffusion approximation for demographic inference (Gutenkunst et al., 2009)), which infers demographic parameters based on a diffusion approximation to the site frequency spectrum (Extended Experimental Procedures Section Demographic History).
Gene flow between polar bears and brown bears after divergence
To fully elucidate patterns of gene flow between polar and brown bear populations since their divergence, we used several methods: (i) the IBS tract method; (ii) ∂a∂i; and (iii) D statistics, also known as the ABBA-BABA test (Durand et al., 2011; Green et al., 2010) (Extended Experimental Procedures Section Gene Flow and Introgression).
Genes under positive selection in polar bears
We used several complementary approaches to investigate evolutionary changes in protein sequences and analyzed the coding regions of 19,822 genes annotated across the polar bear and brown bear samples, using the giant panda genome sequence (Li et al., 2009) as an outgroup (Extended Experimental Procedures Section Positive Selection). We (i) computed a homogeneity test statistic to identify genes with a low polymorphism-to-divergence ratio in polar bears relative to brown bears; (ii) used the Hudson-Aguade-Kreitman (HKA) test to verify that selection had acted specifically on the polar bear lineage and not on the brown bear lineage; (iii) estimated Fst to identify genes that were highly differentiated between polar bears and brown bears; and (iv) used a novel approach to estimate nucleotide diversity within species and divergence between species from low- to medium-quality sequencing data by taking genotype call uncertainty into account (Nielsen et al., 2012; Fumagalli et al., 2014). Because we were interested primarily in identifying completed sweeps unique to the polar bear, we did not apply haplotype-based tests aimed at identifying ongoing selective sweeps. We did not assign simulation-based p-values based on specific demographic models to the test statistics. Rather, we used the computed statistics to generate ranked lists of candidate genes, and then further subjected them to statistical enrichment analyses, an approach often referred to as outlier analyses (e.g. Voight et al., 2006).
Supplementary Material
HIGHLIGHTS.
Polar bears and brown bears diverged < 500,000 years ago
Genes on the polar bear lineage have been under stronger selection than brown bears
Strong selection in polar bears restructured metabolic and cardiovascular function
Dietary changes to fatty acids shaped variation in the APOB gene in polar bears
Acknowledgments
We thank the subsistence Greenland hunters for their valuable participation in obtaining the polar bear samples used in this study, which have been collected since 1999 through a number of projects funded under the DANCEA (Danish Cooperation for Environment in the Arctic) programme, including the CORE programme and the large scale IPY programme “BearHealth”. We thank the University of Alaska Museum of the North for providing the ABC brown bear samples and Ilpo Kojola, Katherine Kendall and John Waller for providing additional brown bear samples. We thank Zhaolei Zhang, Yaping Zhang, Fang Li and Weilin Qiu for help with the analyses. We also thank Xiaoning Wang (South China University of Technology) and the National Gene Bank Project of China and Shenzhen Municipal Government of China (NO.JC201005260191A, CXB201108250096A). EDL was supported by grants from the Danish Council for Independent Research | Natural Sciences (09-069307) and a Marie Curie International Outgoing Fellowship within the European Community 7th Framework Programme (PIOF-GA-2009-253376). MF and MS were supported by EMBO Long-term Postdoctoral Fellowships (ALTF-229-2011 and ALTF-1475-2010) and grant PJ008116 from Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea. KH was supported by a National Science Foundation Graduate Research Fellowship and a UC Berkeley Chancellors Fellowship. RN was supported by NIG grant • NIH grant 2R14003229-07. GAW was supported by NIH (5P-50-GM-081883 to the Duke Center for Systems Biology). CCM and MJO’C would like to thank the Irish Research Council for Science, Engineering and Technology (Embark Initiative to CCM (RS2000172) and Science Foundation Ireland Research Frontiers Program to MJO’C (EOB2673). LD was supported by grants from the Swedish Research Council and the BiodivERsA ERA-NET project Climigrate.
Footnotes
The data reported in this paper are tabulated in the Supporting Online Material. The data are archived at the following databases: Short raw reads have been deposited into the Short Read Archive under accession number SRA092289. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AVOR00000000. The version described in this paper is version AVOR01000000. The remaining data, including the gene set and SNPs, have been uploaded onto the GigaDB DOI: 10.5524/100008 (http://gigadb.org/dataset/100008).
Author contributions: EW, JW, GZ and RN conceived and supervised the project. EWB, RD and CH provided the polar bear samples. LD and LO obtained the brown bear samples. EDL extracted the samples. SL, BL, WH, XX performed SNP calling, population and gene function analyses. TSK provided support during the bioinformatic analyses of the sequencing data. BL, ZX, LZ, JL, ZW, WF, AD, CCM, MJOC, JOM performed analyses in genome sequencing, assembly, annotation, evolution and alignment. SL, MF and RN designed and performed the population genomic analyses. MF and KH performed the demographic inference analyses. EDL performed the mitogenome analyses with input form MF and RN. SL performed the introgression and selection analyses with input from MF, EDL, and RN. MF, MS, CB and GW performed the functional analyses of genes under selection. EDL produced the figures with input from KH and MS. EDL wrote the manuscript, with critical input from RN, MF and the remaining authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci USA. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SN, Ramsay MA. The effects of prolonged fasting of the body composition and reproductive success of female polar bears (Ursus maritimus) Funct Ecol. 1995;9:559–567. [Google Scholar]
- Atkinson SN, Nelson RA, Ramsay MA. Changes in the body composition of fasting polar bears (Ursus maritimus): the effect of relative fatness on protein conservation. Physiol Zool. 1996;69:304–316. [Google Scholar]
- Benn M. Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis. 2009;206:17–30. doi: 10.1016/j.atherosclerosis.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Green RE, Fulton TL, Stiller M, Jay F, Ovsyanikov N, Salamzade R, St John J, Stirling I, Slatkin M, et al. Genomic Evidence for Island Population Conversion Resolves Conflicting Theories of Polar Bear Evolution. PLoS Genet. 2013;9:e1003345. doi: 10.1371/journal.pgen.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon CP, Shah S, Dansky HM, Davidson M, Brinton EA, Gotto AM, Jr, Stepanavage M, Liu SX, Gibbons P, Ashraf TB, et al. Safety of Anacetrapib in Patients with or at High Risk for Coronary Heart Disease. N Engl J Med. 2010;363:2406–2415. doi: 10.1056/NEJMoa1009744. [DOI] [PubMed] [Google Scholar]
- Cronin MA, Amstrup SC, Talbot SL, Sage GK, Amstrup KS. Genetic variation, relatedness, and effective population size of polar bears (Ursus maritimus) in the southern Beaufort Sea, Alaska. J Hered. 2009;100:681–690. doi: 10.1093/jhered/esp061. [DOI] [PubMed] [Google Scholar]
- De Barba M, Waits LP, Garton EO, Genovesi P, Randi E, Mustoni A, Groff C. The power of genetic monitoring for studying demography, ecology and genetics of a reintroduced brown bear population. Mol Ecol. 2010;19:3938–3951. doi: 10.1111/j.1365-294X.2010.04791.x. [DOI] [PubMed] [Google Scholar]
- de Vernal A, Hillaire-Marcel C. Natural Variability of Greenland Climate, Vegetation, and Ice Volume During the Past Million Years. Science. 2008;320:1622–1625. doi: 10.1126/science.1153929. [DOI] [PubMed] [Google Scholar]
- Dickson AJ, Beer CJ, Dempsey C, MASLIN MA, Bendle JA, McClymont EL, Pancost RD. Oceanic forcing of the Marine IsotopeStage 11 interglacial. Nat Geosci. 2009;2:428–433. [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T, Nielsen R. ngsTools: methods for population genetics analyses from Next-Generation Sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin E, Benjamin S, Rapaport D, Rotem-Yehudar R, Tolchinsky S, Horowitz M. EHD3: a protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic. 2002;3:575–589. doi: 10.1034/j.1600-0854.2002.30807.x. [DOI] [PubMed] [Google Scholar]
- Gravel S. Population genetics models of local ancestry. Genetics. 2012;191:607–619. doi: 10.1534/genetics.112.139808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MHY, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson H, Curran J, Kashef F, Snyder JS, Smith SA, Vargas-Pinto P, Bonilla IM, Weiss RM, Anderson ME, Binkley P, et al. Journal of Molecular and Cellular Cardiology. J Mol Cell Cardiol. 2012;52:1183–1190. doi: 10.1016/j.yjmcc.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Gil B, Wiener P, Williams JL. Genetic effects on coat colour in cattle: dilution of eumelanin and phaeomelanin pigments in an F2-Backcross Charolais × Holstein population. BMC Genet. 2007;8:56. doi: 10.1186/1471-2156-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer F, Kutschera VE, Hallström BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science. 2012;336:344–347. doi: 10.1126/science.1216424. [DOI] [PubMed] [Google Scholar]
- Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 2013;9:e1003521. doi: 10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg GE, Derocher AE, Andersen M, Rogers QR, DePeters EJ, Lönnerdal B, Mazzaro L, Chesney RW, Hollis B. Milk composition in free-ranging polar bears (Ursus maritimus) as a model for captive rearing milk formula. Zoo Biol. 2011;30:550–565. doi: 10.1002/zoo.20375. [DOI] [PubMed] [Google Scholar]
- Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klop B, Elte JWF, Cabezas MC. Dyslipidemia in obesity: mechanisms and potential targets. Nutrients. 2013;5:1218–1240. doi: 10.3390/nu5041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Nakagiri M, Takami M, Ide H, Ogawa H. Cloning of bovine LYST gene and identification of a missense mutation associated with Chediak-Higashi syndrome of cattle. Mamm Genome. 1999;10:1146–1149. doi: 10.1007/s003359901181. [DOI] [PubMed] [Google Scholar]
- Kurtén B. The Evolution of the Polar Bear. Ursus maritimus (Phipps) Acta Zool Fennica. 1964;108:1–26. [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2009;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist C, Schuster SC, Sun Y, Talbot SL, Qi J, Ratan A, Tomsho LP, Kasson L, Zeyl E, Aars J, et al. Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc Natl Acad Sci USA. 2010;107:5053–5057. doi: 10.1073/pnas.0914266107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci USA. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. SNP Calling, Genotype Calling, and Sample Allele Frequency Estimation from New-Generation Sequencing Data. PLoS ONE. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin Mutations Alter Actin Interaction in Dilated Cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- Ormbostad I. Relationships Between Persistent Organic Pollutants (POPs) and Plasma Clinical-Chemical Parameters in Polar Bears (Ursus maritimus) Svalbard, Norway: 2012. pp. 1–73. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Amstrup SC, Born EW, Calvert W, Derocher AE, Garner GW, Messier F, Stirling I, Taylor MK, Wiig O, et al. Genetic structure of the world’s polar bear populations. Mol Ecol. 1999;8:1571–1584. doi: 10.1046/j.1365-294x.1999.00733.x. [DOI] [PubMed] [Google Scholar]
- Pagano AM, Durner GM, Amstrup SC, Simac KS, York GS. Long-distance swimming by polar bears (Ursus maritimus) of the southern Beaufort Sea during years of extensive open water. Can J Zool. 2012;90:663–676. [Google Scholar]
- Pagès M, Calvignac S, Klein C, Paris M, Hughes S, Hänni C. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol Phy Evol. 2008;47:73–83. doi: 10.1016/j.ympev.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Inference of Historical Changes in Migration Rate From the Lengths of Migrant Tracts. Genetics. 2009;181:711–719. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassl R, Laggner P. Molecular structure of low density lipoprotein: current status and future challenges. European Biophysics Journal: EBJ. 2009;38:145–158. doi: 10.1007/s00249-008-0368-y. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Runkel F, Büssow H, Seburn KL, Cox GA, Ward DM, Kaplan J, Franz T. Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25. Mamm Genome. 2006;17:203–210. doi: 10.1007/s00335-005-0015-1. [DOI] [PubMed] [Google Scholar]
- Smith TG. Polar bear predation of ringed and bearded seals in the land-fast sea ice habitat. Can J Zool. 1980;58:2201–2209. [Google Scholar]
- Stewart JR, Lister AM, Barnes I, Dalen L. Refugia revisited: individualistic responses of species in space and time. Proc Soc Lond B Bio. 2010;277:661–671. doi: 10.1098/rspb.2009.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling I, Archibald WR. Aspects of Predation of Seals by Polar Bears. J Fish Res Bd Can. 1977;34:1126–1129. [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Thiemann GW, Iverson SJ, Stirling I. Polar bear diets and arctic marine food webs: insights from fatty acid analysis. Ecol Monogr. 2008;78:591–613. [Google Scholar]
- Trantow CM, Hedberg-Buenz A, Iwashita S, Moore SA, Anderson MG. Elevated Oxidative Membrane Damage Associated with Genetic Modifiers of Lyst-Mutant Phenotypes. PLoS Genet. 2010;6:e1001008. doi: 10.1371/journal.pgen.1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JM, Stanbridge EJ, McBride HL, Meese EU, Casey G, Araujo DE, Witkowski CM, Nagle RB. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science. 1990;247:568–571. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- Trip MD, Smulders YM, Wegman JJ, Hu X, Boer JM, Jacoline B, Zwinderman AH, Kastelein JJ, Feskens EJ, Bergen AA. Frequent Mutation in the ABCC6 Gene (R1141X) Is Associated With a Strong Increase in the Prevalence of Coronary Artery Disease. Circulation. 2002;106:773–775. doi: 10.1161/01.cir.0000028420.27813.c0. [DOI] [PubMed] [Google Scholar]
- van der Ven PFM, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, Micheel B, Fürst DO. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp Cell Res. 2006;312:2154–2167. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Van Sligtenhorst I, Ding ZM, Shi ZZ, Read RW, Hansen G, Vogel P. Cardiomyopathy in α-Kinase 3 (ALPK3)-Deficient Mice. Vet Pathol. 2012;49:131–141. doi: 10.1177/0300985811402841. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A Map of Recent Positive Selection in the Human Genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Watanabe M, Imai Y, Hara K, Manabe I, Maemura K, Horikoshi M, Ozeki A, Itoh C, Sugiyama T, et al. Associations of variations in the MRF2/ARID5B gene with susceptibility to type 2 diabetesin the Japanese population. J Hum Genet. 2012;57:727–733. doi: 10.1038/jhg.2012.101. [DOI] [PubMed] [Google Scholar]
- Whitfield AJ, Barrett PH, van Bockxmeer FM, Burnett JR. Lipid Disorders and Mutations in the APOB Gene. Clin Chem. 2004;50:1725–1732. doi: 10.1373/clinchem.2004.038026. [DOI] [PubMed] [Google Scholar]
- Willerslev E, Cappellini E, Boomsma W, Nielsen R, Hebsgaard MB, Brand TB, MMH, Bunce M, Poinar HN, Dahl Jensen D, et al. Ancient Biomolecules from Deep Ice Cores Reveal a Forested Southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.