Abstract
Conventional epithelioid hemangioendotheliomas (EHE) have a distinctive morphologic appearance and are characterized by a recurrent t(1;3) translocation, resulting in a WWTR1-CAMTA1 fusion gene. We have recently encountered a fusion-negative subset characterized by a somewhat different morphology, including focally well-formed vasoformative features, which was further investigated for recurrent genetic abnormalities. Based on a case showing strong TFE3 immunoreactivity, FISH analysis for TFE3 gene rearrangement was applied to the index case as well as to 9 additional cases, selected through negative WWTR1-CAMTA1 screening. A control group, including 18 epithelioid hemangiomas, 9 pseudomyogenic HE and 3 epithelioid angiosarcomas, was also tested. TFE3 gene rearrangement was identified in 10 patients, with equal gender distribution and a mean age of 30 years old. The lesions were located in somatic soft tissue in 6 cases, lung in 3 and one in bone. One case with available frozen tissue was tested by RNA sequencing and FusionSeq data analysis to detect novel fusions. A YAP1-TFE3 fusion was thus detected, which was further validated by FISH and RT-PCR. YAP1 gene rearrangements were then confirmed in 7 of the remaining 9 TFE3-rearranged EHEs by FISH. No TFE3 structural abnormalities were detected in any of the controls. The TFE3-rearranged EHEs showed similar morphologic features with at least focally, well-formed vascular channels, in addition to a variably solid architecture. All tumors expressed endothelial markers, as well as strong nuclear TFE3. In summary we are reporting a novel subset of EHE occurring in young adults, showing a distinct phenotype and YAP1-TFE3 fusions.
Keywords: TFE3, YAP1, epithelioid hemangioendothelioma, WWTR1
INTRODUCTION
Epithelioid vascular tumors encompass a wide histologic spectrum, including epithelioid hemangioma, a benign tumor; epithelioid hemangioendothelioma (EHE), a low grade malignant tumor; and epithelioid angiosarcoma, a high grade malignant tumor (Wenger and Wold, 2000; O’Connell et al., 2001; Fletcher et al., 2013). A recurrent t(1;3)(p36.23;q25.1) has recently been identified in most EHEs of different anatomic locations and grades of malignancy (Errani et al., 2011; Tanas et al., 2011). The translocation results in the fusion of CAMTA1 on 1p36.23 to WWTR1 on 3q25.1. This recurrent translocation has not been detected in any of the morphologic mimics of EHE, such as epithelioid hemangioma, epithelioid angiosarcoma or pseudomyogenic (epithelioid sarcoma-like) HE, and thus can serve as a useful molecular diagnostic tool in challenging cases.
In the course of WWTR1-CAMTA1 screening in a large series of EHE, we identified a fusion-negative subset that shows distinctive morphologic features, such as well-formed vaso-formative features with mature lumina lined by epithelioid cells with abundant eosinophilic cytoplasm. This detailed investigation was triggered by an index case showing strong TFE3 immunoreactivity, which prompted screening for TFE3 gene rearrangement in the index case as well as in other WWTR1-CAMTA1 fusion-negative epithelioid vascular tumors.
MATERIAL AND METHODS
Patient Selection and Tumor Characteristics
The files of the corresponding authors were searched for the diagnosis of epithelioid hemangioendothelioma (EHE), which showed unusual morphologic features such as abundant eosinophilic cytoplasm, mature vascular channel formation and were negative for WWTR1 and CAMTA1 rearrangements by FISH. Screening for these specific morphologic features was triggered by an index case that had these exact histologic characteristics, and due to its pseudo-alveolar pattern was tested and found to be diffusely positive for TFE3 by immunohistochemistry. Hematoxylin and eosin (H&E) stained slides from all cases were reviewed by two sarcoma pathologists (CRA and CDMF). Immunostains for endothelial markers (CD31 and/or ERG) and TFE3 were performed (pre-diluted from Ventana Medical Systems, Inc., Tucson, AZ) or available for review in all cases. Clinical information was obtained from review of patient’s clinical charts or from referring pathologists (see Acknowledgements) in all cases.
A control group, including 18 epithelioid hemangiomas, 9 pseudomyogenic (epithelioid sarcoma-like) HEs and 3 high grade epithelioid angiosarcomas, was also tested for TFE3 gene rearrangements.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking TFE3 and YAP1. BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes (Green 496 dUTP for telomeric probes and Orange 552 dUTP for centromeric probes, Enzo, Plymouth Meeting, PA) in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Watertown, MA, USA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
RNA Sequencing
Total RNA was prepared for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (10 μg) extracted from case. The mRNA was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer (Illumina). To reduce the inclusion of artifact chimeric transcripts into the sequencing library, an additional gel size-selection step was introduced prior to the adapter ligation step (Quail et al., 2008). Size-ranges captured were 300-350 bp during the first size-selection step and then 400-450 bp for the second size-selection step after the ligation of the adapters. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2000 (Illumina). A total of about 268 million paired-end reads were generated, corresponding to about 27 billion bases.
Analysis of RNA Sequencing Results with FusionSeq
All reads were independently aligned with the CASAVA 1.8 software provided by Illumina against the human genome sequence (hg19) and a splice junction library, simultaneously. The splice junction library was generated by considering all possible junctions between exons of each transcript. We considered the University of California, Santa Cruz (UCSC) Known Genes annotation set (Hsu et al., 2006) to generate this library via RSEQtools, a computational method for processing RNA-seq data (Habegger et al., 2011). The mapped reads were converted into Mapped Read Format (Habegger et al., 2011) and analyzed with FusionSeq (Sboner et al., 2010) to identify potential fusion transcripts. FusionSeq is a computational method successfully applied to paired-end RNA-seq experiments for the identification of chimeric transcripts (Pflueger et al., 2011; Tanas et al., 2011; Pierron et al., 2012; Mosquera et al., 2013). Briefly, paired-end reads mapped to different genes are first selected to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, is then applied to remove spurious fusion transcript candidates. Once a confident list of fusion candidates is generated, they are ranked with several statistics to prioritize the experimental validation. In this case, we used the DASPER score (Difference between the observed and Analytically calculated expected SPER): a higher DASPER score indicates a greater likelihood that the fusion candidate is authentic and did not occur randomly. See Sboner A, et al. (Sboner et al., 2010) for further details about FusionSeq.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An aliquot of the RNA extracted above from frozen tissue (Trizol Reagent; Invitrogen, Carlsbad, CA) from the TFE3-rearranged EHE1 that was investigated for RNAseq, was used to confirm the novel fusion transcript identified by FusionSeq. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). RT-PCR was performed using the advantage 2 PCR kit (Clonetech, Mountain View, CA) for 32 cycles at a 66.6°C annealing temperature, using the following primers: YAP1 Ex1.4 fwd 5’-CCTGGAGGCGCTCTTCAACG-3’; TFE3 exon 4 (5’-GAGTGTGGTGGACAGGTACTG-3’); TFE3 exon 6 rev 5’GTTGCTGACAGTGATGGCTGG3’; TFE3 exon 8 rev 5’-CGGGTCACTGGACTTAGGGATGAGA-3’; TFE3 exon 10 rev 5’-CCTGCCCTCCTCCTCAATGTCC-3’. The PCR product was confirmed by agarose gel electrophoresis with ethidium bromide staining, and then sequenced using the Sanger method. In one additional case (EHE2) with tissue available, RNA was extracted from paraffin tissue and subjected to RT-PCR using YAP1 exon 1 forward and TFE3 exon 4 reverse, listed above.
DNA Polymerase Chain Reaction (PCR) to Determine the Genomic Breakpoint of YAP1-TFE3
Genomic DNA was extracted from frozen tissue by Phenol/Chloroform assay and quality was confirmed by electrophoresis. 0.5 microgram of DNA was amplified using the Advantage 2 PCR Kit (Clonetech) and the following primers: YAP1 forward primer at intron 1 (5’-CGGTCCACTTCAGTCTCCT -3’) and TFE3 reverse primers at exon 4 (5’-GAGTGTGGTGGACAGGTACTG -3’) at 64.5°C annealing temperature. The PCR product was sequenced as previously described.
RESULTS
Pathologic Characteristics and Clinical Follow-up
The 10 patients included in the study had an equal gender distribution and ranged from 14-50 years old at initial diagnosis (mean 30 years). The most common presentation was in somatic soft tissue in 6 cases (limbs, 3; head and neck 2; and trunk, 1), followed by lung in 3 cases (two of them being multifocal, Fig 3A) and one case in bone (T2 vertebral body, Fig 3 B) (Table 1).
Fig. 3.
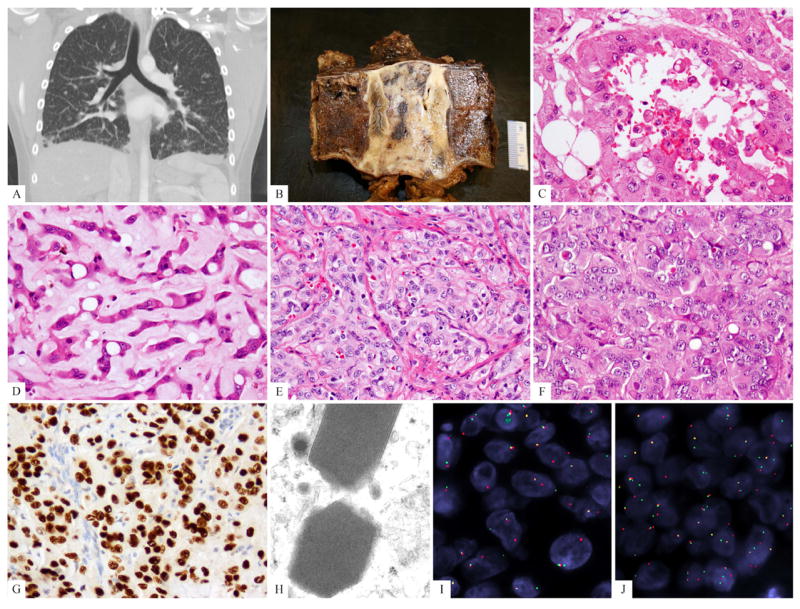
Clinical and pathologic spectrum of TFE3-rearranged EHE. (A) CT scan showing bilateral ground-glass opacities suggestive of interstitial lung disease, most predominantly in the lower lobes but diffusely present (EHE9); (B) Gross appearance of the T2 vertebral en-bloc resection showing an ill-defined white-gray lesion (EHE3). (C, D). Biphasic morphologic appearance showing a pseudo-alveolar component with abundant, densely eosinophilic cytoplasm and the other resembling classis EHE, with cord-like arrangement and myxoid stroma (EHE3); (E) foamy cytoplasm (histiocytoid), mild nuclear pleomorphism (EHE7); (F) predominantly solid and nested growth pattern, showing densely eosinophilic cytoplasm, rare vacuoles, and lack of significant intervening stroma (EHE4); (G) strong TFE3 immunostaining; (H) ultrastructural study showing distinctive rhomboid crystals with a periodicity ranging from 9.05-11.63, mean of 10.34 nm, (46,000 magnification, EHE10). (I) TFE3 and (J) YAP1 break-apart signals by FISH (EHE 7) (green, telomeric; red, centromeric).
Table 1.
Clinicopathologic Features of TFE3-rearranged EHE
| EHE# | Age/Sex | Location | Morphologic features | LR/Mets | Follow-up |
|---|---|---|---|---|---|
| EHE1 | 35/M | Soft tissue/cervical | Lumen formation, vacuoles, moderate nuclear pleomorphism, nuclear pseudo-inclusions, stromal inflammation; Late recurrence showed a sarcomatous/spindle cell component | Locoregional recurrence/ mets to LN, soft tissue, bone | NED, 22 years |
| EHE2 | 30/F | Soft tissue/popliteal | Lumen formation (pseudo-alveolar), vacuoles, mild nuclear pleomorphism, nuclear pseudo-inclusions, stromal inflammation | Mets to liver | AWD, 6 mo |
| EHE3 | 50/M | Bone/ T2 vertebral body | Biphasic appearance with one component showing dilated, mature lumen formation (pseudo-alveolar) and other with cords and single files growth | No | NED, 12 mo |
| EHE4 | 33/F | Soft tissue/inframammary fold | Lumen formation and nested growth, densely eosinophilic cytoplasm, nuclear pseudoinclusions, vacuoles, mild nuclear atypia | LR and axillary LN met at autopsy | DWD¥, 3mo |
| EHE5 | 27/M | Soft tissue/ supra-clavicular | Mostly solid, with only focal lumen formation, foamy cytoplasm (histiocytoid), rare vacuoles, mild nuclear pleomorphism | No | NED, 9 mo |
| EHE6 | 14/M | Inguinal LN | Extensive lumen formation and focal solid growth, moderate nuclear pleomorphism, rare vacuoles | No | NED, 3 mo |
| EHE7 | 25/F | Soft tissue/arm | Mostly solid, with only focal lumen formation, foamy cytoplasm (histiocytoid), rare vacuoles, mild nuclear pleomorphism | No | Recent case; N/A |
| EHE8 | 29/F | Lung* | Lumen formation, moderate nuclear pleomorphism, necrosis | Mets to bone, ST | DOD, 17 years |
| EHE9 | 22/F | Lung | Mostly solid, with only focal lumen formation, foamy cytoplasm (histiocytoid), rare vacuoles, mild nuclear pleomorphism | No | DWD, 6 mo |
| EHE10 | 36/M | Lung | Lumen formation and nested growth, densely eosinophilic cytoplasm | Mets to bone | AWD, 23 years |
LR, local recurrence; Mets, metastasis; LN, lymph node;
per report diagnosed as ‘intravascular bronchioloalveolar tumor’ in 1987 (slides not available for review); ST, soft tissue, NED, no evidence of disease, DOD, dead of disease, DWD, dead with disease;
chemotherapy toxicity developed post-partum; N/A, not available; mo, months.
All tumors showed evidence of mature vessel lumen formation, in addition to intra-cytoplasmic vacuoles (blister cells). The extent of vasoformative features was quite variable, from prominent and readily discernible open lumens (Figs.1A, 2A), to a focal and subtle finding in two cases (see Table 1 for summary of morphologic findings). In addition to lumen formation the tumors often showed a solid growth pattern, with back-to-back tumor cells with minimal intervening stroma (Figs. 3E,F). This contrasts with the often abundant myxochondroid or hyaline type stroma separating the cells of conventional EHE in cords and single files. One case showed a distinctive nested appearance. One other tumor showed a biphasic growth, with one component showing dilated and well-formed blood vessels lined by epithelioid cells with abundant eosinophilic cytoplasm, reminiscent of a pseudo-alveolar architecture, while the other component was composed of cords and single cells separated by a myxoid stroma, resembling a classic EHE (Figs. 3C,D). No other cases showed significant areas that resembled conventional EHE morphology.
Fig. 1.
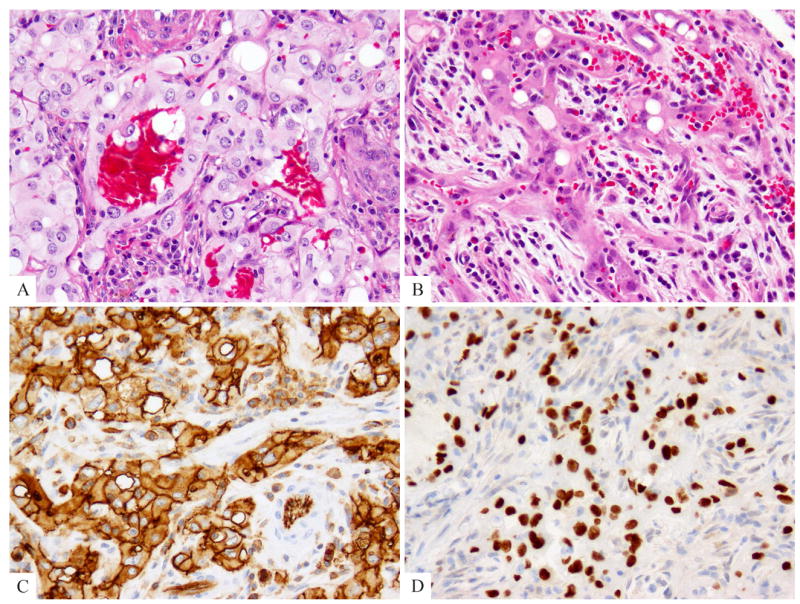
Index case displaying a distinctive pseudo-alveolar pattern was tested for TFE3 immunohistochemistry and gene rearrangements (EHE2). (A, B) Morphologic appearance showing mature vessel lumen formation lined by epithelioid endothelial cells with moderate amount of densely to lightly eosinophilic cytoplasm, focal intra-cytoplasmic vacuoles and stromal inflammation including eosinophils. (C) CD31 and (D) TFE3 strong reactivity.
Fig. 2.
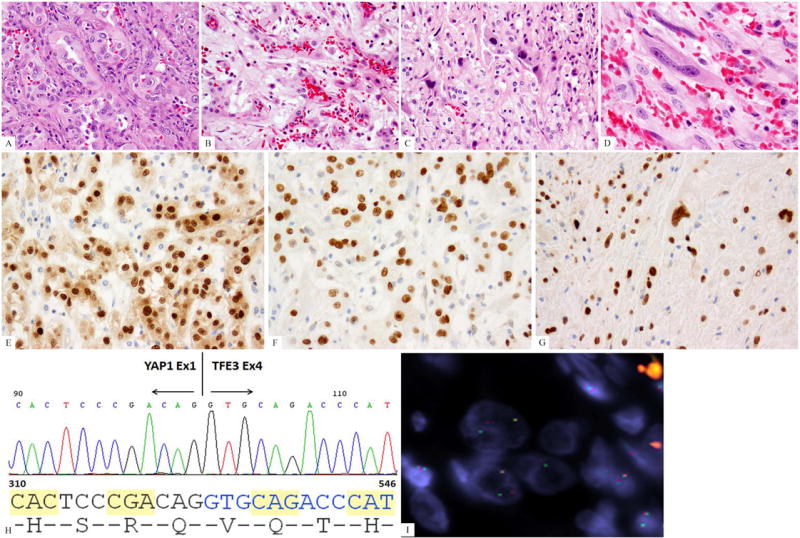
Morphologic progression of EHE after multiple local recurrences (EHE1). (A) Histologic appearance of the primary tumor showing well-formed vasoformative features, with epithelioid cells with moderate nuclear pleomorphism, nuclear indentations and pseudo-inclusions; (B-D). Histologic appearance of the latest recurrence 20 years after the initial diagnosis showing a spectrum of well-differentiated to solid to frankly malignant and spindle cell areas; (E) ERG immunostaining showing strong nuclear reactivity; (F, G) TFE3 strong reactivity in both primary as well as latest sarcomatous recurrence; (H) ABI sequence from the RT-PCR product showing YAP1 exon1 being fused to TFE3 exon 4. (I) YAP1 break-apart by FISH (green, telomeric; red signals, centromeric).
The tumor cells showed moderate to voluminous cytoplasm, which ranged from having a foamy or feathery quality, with a distinctive histiocytoid appearance in four cases (Fig. 3E), to densely eosinophilic cytoplasm in two cases (Fig. 3F), reminiscent of an oncocytic phenotype. Two cases showed abundant stromal chronic inflammatory cells and scattered eosinophils (Fig. 1B). The latter finding may simulate the diagnosis of epithelioid hemangioma, however, the nuclear cytomorphology was overall more atypical and in keeping with a malignant neoplasm. All tumors showed at least mild nuclear atypia and in three cases it showed focal moderate nuclear pleomorphism, with hyperchromatic and markedly indented and irregular nuclear contours. Densely eosinophilic nuclear pseudo-inclusions were noted in two cases (Fig. 2 A). Mitotic activity was not increased in most cases, and did not exceed 3MF/10HPFs. Necrosis was present only in the two cases that progressed to a spindle, sarcomatoid phenotype after more than 15 years of follow-up and indolent clinical behavior (Figs. 2B-D). All tumors were diffusely and strongly positive for CD31 and/or ERG (Figs. 1C, 2E), and all showed nuclear reactivity for TFE3 (Figs. 1D, 2F,G).
In one case, tissue for ultrastructural examination was available (EHE10, bone metastasis, Fig. 3H) showing distinctive membrane-bound, cytoplasmic rhomboid crystals. Rare tumor cells showed both Weibel-Palade bodies and crystal structures within the same cell. The periodicity of 10nm in average and the overall appearance were indistinguishable from the crystal structures seen in other TFE3-rearranged tumors, such as alveolar soft tissue sarcoma or pediatric renal cell carcinomas.
Of the 6 patients with follow-up information available for more than one year, 5 had evidence of metastatic disease: two loco-regionally to the lymph nodes or to adjacent bone and soft tissue and three distantly (to liver, soft tissue and bone). To date only one patient succumbed of disease, who presented with pulmonary multifocal disease and progressed after a 17 year-course with widespread metastases to bone and soft tissue. An additional patient who presented with multifocal lung lesions died with disease 6 months after diagnosis, the course being complicated by pneumonia. One additional patient died with disease due to chemotherapy toxicity.
RNA-seq and Fusion Seq identifies a novel YAP1-TFE3 fusion
The sample with frozen material was RNA-sequenced to identify potential fusion candidates. YAP1-TFE3 fusion transcript was selected by FusionSeq as the top candidate. Alignment of the reads suggested a fusion of YAP1 exon 1 with exon 4 of TFE3 (Fig. 4). The RT-PCR confirmed the presence of a fusion transcript of YAP1 exon 1 to exon 4 of TFE3 (Figs. 2H, 4 B and Supplem Fig 1A). In one additional case paraffin material was available for RNA extraction and RT-PCR assay, which revealed an identical YAP1-TFE3 fusion transcript, using YAP1 exon 1 forward primer and TFE3 exon 4 reverse primer (results not shown).
Fig. 4.
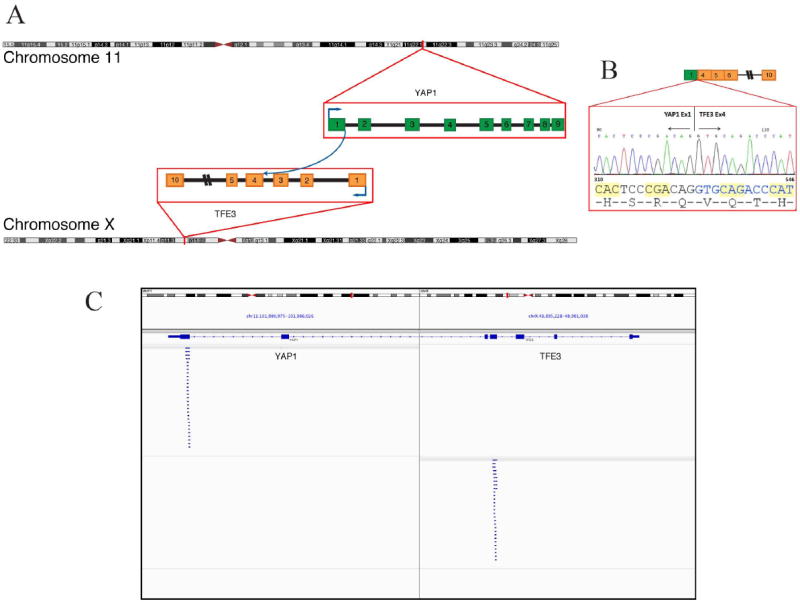
YAP1-TFE3 gene fusion. (A) Schematic representation of the YAP1-TFE3 fusion indicating the loci that are fused together. (B) Experimental validation of the fusion shows the sequence of the junction between exon 1 of YAP1 and exon 4 or TFE3. (C) Integrative Genome Viewer (IGV) snapshot of the reads supporting the fusion candidate as determined by RNA-seq.
Fluorescence In Situ Hybridization (FISH)
All ten cases investigated showed TFE3 break-apart signal (Fig. 3G). Eight of the 10 also showed YAP1 rearrangement. The two cases that did not show structural abnormality of YAP1 were tested for WWTR1 fusion but were negative (Errani et al., 2011). None of the cases included in the control group showed structural or copy number abnormalities of TFE3.
DNA PCR for the YAP1-TFE3 Genomic Breakpoint
A 1233bp amplified product was obtained by DNA PCR using the YAP1 intron 1 forward primer and TFE3 exon 4 reverse primer (Supplem Fig. 1B). By direct sequencing the first portion of YAP1 intron 1 (1- 1331bp) was linked to 24 bp of anti-paralleled segment (1354-1377 bp) of YAP1 intron1, which was subsequently fused to 3’portion of TFE3 intron 3 (336-66bp) (Supplem Fig. 1C).
DISCUSSION
Conventional epithelioid hemangioendothelioma (EHE) has a distinctive morphologic appearance, characterized by epithelioid cells arranged in cords and single cells in a myxochondroid or sclerotic stroma, typically lacking well-formed vasoformative properties. The common t(1;3) chromosomal translocation in these lesions results in a fusion protein which includes the 14-3-3 binding protein and WW domains of WWTR1 and the transcription factor immunoglobulin (TIG)-like DNA-binding domain, ankyrin (ANK) repeats and IQ domains of CAMTA1 (Errani et al., 2011; Tanas et al., 2011). Sharing amino acid sequence homology with YAP (Yes-associated protein), WWTR1 contains a conserved WW domain able to interact with the PDZ domain (Kanai et al., 2000). The WW domain of WWTR1 is capable of interacting with PPXY motifs (Pro-Pro-X-Tyr) and a coiled-coil C-terminal domain that recruits core components of the transcriptional machinery (Hong et al., 2005).
In contrast, the morphologic hallmark of TFE3-rearranged EHE includes voluminous eosinophilic cytoplasm with mild to moderate cytologic atypia and more overt vasoformative features. These histologic findings are distinct from either classic EHE, typically lacking mature vessel formation (Errani et al., 2011), or epithelioid hemangioma, which show similar mature lumen formation but has relatively bland cytomorphology (Errani et al., 2012). The presence of a YAP1 gene rearrangement in this EHE subset, which shares significant functional and sequence homology with WWTR1, is noteworthy. The transcriptional co-activator YAP is a major downstream effector of the Hippo pathway (Dong et al., 2007). Lats1/2 inhibit YAP by direct phosphorylation at S127, which results in YAP binding to 14-3-3 and cytoplasmic sequestration (Dong et al., 2007; Zhao et al., 2007; Hao et al., 2008). Similar to WWTR1, YAP acts mainly through TEAD family transcription factors to stimulate expression of genes that promote proliferation and inhibit apoptosis (Zhao et al., 2008). Phosphorylation of YAP S381 by Lats1/2 kinases can also promote its ubiquitination-dependent degradation (Zhao et al., 2010). Sustained YAP expression results in hyperplasia and eventual tumor development (Dong et al., 2007). Although abnormal activation of YAP and WWTR1 (TAZ) has been associated with human cancers (Overholtzer et al., 2006; Zender et al., 2006; Zhao et al., 2007; Steinhardt et al., 2008), suggesting an important role for the Hippo pathway in tumorigenesis, the mechanism of YAP1 dysregulation in the tumorigenesis of this EHE subset appears distinct. The fusion transcript retains the very proximal portion of the YAP1 amino-terminal (encoded by exon 1, proline-rich domain, see Fig. 5), while losing the S127 14-3-3 binding site, WW domain and its C-terminal transactivation domain. Based on these findings, the most plausible explanation is that YAP1 provides a stronger promoter to the oncogenic TFE3 function.
Fig 5.
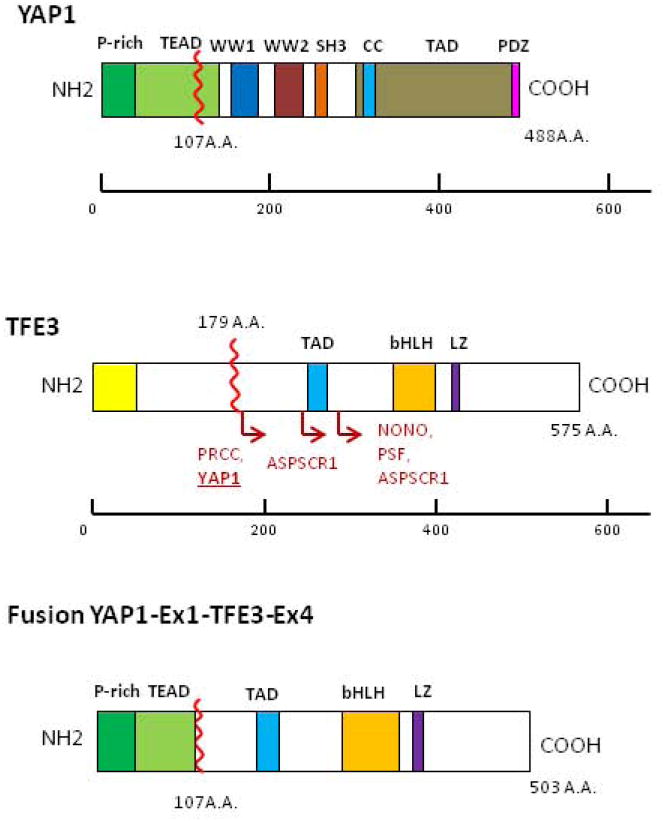
Protein domains of YAP1, TFE3 and projected YAP1-TFE3 fusion protein. Schematic representations of YAP1 showing prolin-rich domain (P-rich), TEAD binding domain (TEAD), WW1/WW2 domains, SRC homology 3 domain (SH3), coiled-coil domain (CC), transactivation domain (TAD), PSD-95/DLG1/ZO-1 (PDZ) domain and for TFE3 showing common domains: glutamine-rich domain (Gln-rich); activation domain (AD); basic, a positively charged domain; helix-loop-helix (HLH) and domain; leucine zipper (LZ) domain. For TFE3, locations of fusions in some cancer types are shown. ASPSCR1, alveolar soft part sarcoma chromosome region, candidate 1; NONO, non-POU domain containing, octamer-binding; PRCC, papillary renal cell carcinoma; PSF, splicing factor proline/glutamine-rich.
The transcription factor E3 (TFE3) belongs to the MiT family of transcription factors, together with MITF, TFEB, and TFEC sharing in common a helix-loop-helix leucine zipper dimerization motif, a transactivation domain and basic region involved in DNA contact and binding. Because of their sequence homology, all MiT family members bind to identical DNA recognition sequences (CA[T/C]GTG) termed E-boxes. In the Xp11 translocation-associated renal carcinomas the amino-terminal portion of transcription factorE3 (TFE3) fuses to any of several gene partners, including PRCC, NONO, SFPQ, and CLTC (Sidhar et al., 1996; Clark et al., 1997; Argani et al., 2003). In alveolar soft part sarcoma, TFE3 is fused to the ASPL gene (Ladanyi et al., 2001). The common feature of all TFE3 fusion proteins is preservation of the bHLH-LZ and transcriptional activation domains of TFE3, which is also the case with the YAP1-TFE3 fusion (Fig. 5). The various TFE3 fusion partners are typically expressed at a consistently high-level in the given tumor type, suggesting that mis-expression of TFE3 is sufficient to promote tumorigenesis. Because dysregulation of the MiT family in cancer uniformly preserves the DNA-binding domain, it is likely that these factors promote oncogenesis by altering target gene expression.
TFE3 immunoexpression was uniformly present in all cases with a strong and diffuse nuclear pattern of staining, suggesting that this can be applied as a useful marker and as a method of screening epithelioid vascular tumors for the presence of TFE3-rearrangements. The presence of diffuse expression of CD31 and/or ERG endothelial markers helps in the distinction from other TFE3-positive neoplasms, such as alveolar soft part sarcoma, PEComa, and Xp11-translocation positive renal cell carcinomas.
The clinical follow-up available in this small series of patients suggests that the TFE3-rearranged EHE is a clinically indolent tumor with a substantial long term risk of distant metastasis. One patient succumbed of disease with widespread bone and soft tissue metastases, 17 years after the initial diagnosis of ‘intravascular bronchioloalveolar tumor’. Only the metastatic lesions were available for review in this latter case, showing a rather aggressive histology with significant cytologic atypia and necrosis. A different patient, who developed local recurrences and loco-regional metastases, remains alive without evidence of disease 22 years after the initial diagnosis, although the morphologic appearance of his latest recurrence appears focally more frankly malignant when compared to the primary tumor. These phenotypic changes to a high grade component and/or aggressive clinical behavior in these two cases after a prolonged time interval suggests the acquisition of additional secondary genetic events.
The differential diagnosis includes primarily other epithelioid vascular tumors. Epithelioid hemangioma shares the well-formed vasoformative features, but the degree of cytologic and nuclear atypia in the TFE3-rearranged EHE is clearly in keeping with a malignant neoplasm, as are the more solidly cellular areas. Conventional EHE, showing WWTR1-CAMTA1 fusion, has cytoplasm which is more glassy/hyaline and typically lacks mature vessel formation, its vasoformative features being limited to the intra-cytoplasmic lumina (the so-called ‘blister cells’). TFE3-overexpressing EHE may also be confused with epithelioid angiosarcoma, due to its solid growth and abundant eosinophilic cytoplasm; however, the high mitotic activity, frequently more amphophilic cytoplasm and areas of necrosis seen commonly in angiosarcoma are not present in this entity. Additionally, as noted in the index case, the pseudo-alveolar pattern and strong reactivity for TFE3 may raise the possibility of an alveolar soft part sarcoma; however, these tumors are diffusely positive for most vascular markers applied, including CD31 and ERG, which can help with this distinction.
In summary, we are reporting recurrent TFE3 oncogenic activation secondary to gene rearrangements and common fusion with YAP1 in what appears to be a distinctive subset of EHE. The 10 cases illustrated here show significant morphologic similarity and clinically follow an indolent course, despite a high propensity for metastasis, further confirming that they almost certainly represent a distinct and reproducible entity. Whether these lesions are indeed a variant of EHE or a separate, distinct tumor type awaits clinicopathologic and molecular assessment of a larger number of cases.
Supplementary Material
(A) Gel electrophoresis of RT-PCR showing 3 different amplicons of YAP1-TFE3 fusion transcripts based on the three different TFE3 reverse primers (exons 6, 8 and 10); (B) gel electrophoresis of DNA PCR for confirming the genomic break of YAP1-TFE3 showing a 1233 bp size band; (C) Sanger sequencing showing YAP1 intron 1 (position 1331 bp) is fused to a 24 bp antiparallel fragment of intron 1, followed by TFE3 intron 4 (starting at position 336 bp).
Acknowledgments
We would like to thank the following pathologists for providing cases: Prof. Franco Bertoni, Bologna, Italy, Dr. Penelope Korkolopoulou, Athens, Greece, Dr. Thomas Mentzel, Friedrichshafen, Germany, Dr. Randall S. Smith, Jackson, MS and Prof. Nives Jonjic, Rijeka, Croatia. Also thanks to Ms Jackie Pittman for expertise with ultrastructural studies, Alyne Manzo for composite figure preparation, and Milagros Soto for editorial assistance.
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA).
Footnotes
Conflict of interest: none
References
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argani P, Lui MY, Couturier J, Bouvier R, Fournet JC, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Panicek DM, Healey JH, Antonescu CR. Epithelioid hemangioma of bone and soft tissue: a reappraisal of a controversial entity. Clin Orthop Relat Res. 2012;470:1498–1506. doi: 10.1007/s11999-011-2070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Zhang L, Sung YS, Hajdu M, Singer S, Maki RG, Healey JH, Antonescu CR. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CD, Bridge JA, Hogendoorn PC, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2013. [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, Argani P, Healey JH, Ueda T, Yoshikawa H, Meloni-Ehrig A, Sorensen PH, Mertens F, Mandahl N, van den Berghe H, Sciot R, Dal Cin P, Bridge J. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, Wexler LH, Laquaglia MP, Edelman M, Sreekantaiah C, Rubin MA, Antonescu CR. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013 doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JX, Nielsen GP, Rosenberg AE. Epithelioid vascular tumors of bone: a review and proposal of a classification scheme. Adv Anat Pathol. 2001;8:74–82. doi: 10.1097/00125480-200103000-00003. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, Cao X, Barrette T, Tewari AK, Chee MS, Chinnaiyan AM, Rickman DS, Demichelis F, Gerstein MB, Rubin MA. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhar SK, Clark J, Gill S, Hamoudi R, Crew AJ, Gwilliam R, Ross M, Linehan WM, Birdsall S, Shipley J, Cooper CS. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–1338. doi: 10.1093/hmg/5.9.1333. [DOI] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Wenger DE, Wold LE. Malignant vascular lesions of bone: radiologic and pathologic features. Skeletal Radiol. 2000;29:619–631. doi: 10.1007/s002560000261. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Gel electrophoresis of RT-PCR showing 3 different amplicons of YAP1-TFE3 fusion transcripts based on the three different TFE3 reverse primers (exons 6, 8 and 10); (B) gel electrophoresis of DNA PCR for confirming the genomic break of YAP1-TFE3 showing a 1233 bp size band; (C) Sanger sequencing showing YAP1 intron 1 (position 1331 bp) is fused to a 24 bp antiparallel fragment of intron 1, followed by TFE3 intron 4 (starting at position 336 bp).


