Abstract
Targeted delivery of therapeutic genes to the tumor site is critical for successful and safe cancer gene therapy. The arginine grafted bio-reducible poly (cystamine bisacrylamide-diaminohexane, CBA-DAH) polymer (ABP) conjugated poly (amido amine) (PAMAM), PAM-ABP (PA) was designed previously as an efficient gene delivery carrier. To achieve high efficacy in cancer selective delivery, we developed the tumor targeting bio-reducible polymer, PA-PEG1k-RGD, by conjugating cyclic RGDfC (RGD) peptides, which bind αvβ3/5 integrins, to the PAM-ABP using polyethylene glycol (PEG,1kDa) as a spacer. Physical characterization showed nanocomplex formation with bio-reducible properties between PA-PEG1k-RGD and plasmid DNA (pDNA). In transfection assays, PA-PEG1k-RGD showed significantly higher transfection efficiency in comparison with PAM-ABP or PA-PEG1k-RGD in αvβ3/5 positive MCF7 breast cancer and PANC-1 pancreatic cancer cells. The targeting ability of PA-PEG1k-RGD was further established using a competition assay. To confirm the therapeutic effect, the VEGF siRNA expressing plasmid was constructed and then delivered into cancer cells using PA-PEG1k-RGD. PA-PEG1k-RGD showed 20-59% higher cellular uptake rate into MCF7 and PANC-1 than that of non-targeted polymers. In addition, MCF7 and PANC-1 cancer cells transfected with PA-PEG1k-RGD/pshVEGF complexes had significantly decreased VEGF gene expression (51-71%) and cancer cell viability (35-43%) compared with control. These results demonstrate that a tumor targeting bio-reducible polymer with an anti-angiogenic therapeutic gene could be used for efficient and safe cancer gene therapy.
Keywords: Targeted gene delivery, Cancer gene therapy, RGD peptides, VEGF siRNA, Bio-reducible polymer
Introduction
Various non-viral vectors have been studied as alternatives to viral vectors and have many advantages such as non-immunogenicity and flexibility of DNA loading [1, 2]. Unfortunately, many of these non-viral vectors show significant cellular toxcicity [3]. Representative cationic polymers with high transfection efficiency, such as high molecular weight poly (ethylenimine) (PEI) and poly (l-Lysine) (PLL) showed significant cytotoxicity due to excessive cellular accumulation of non-degradeable polymers with high charge density [4]. Although newly developed polymers have low toxicity, relatively limited efficiency necessitates further improvement for in vivo application [5]. Therefore, improving the efficiency and reducing the cytotoxicity of delivery carriers are required for successful gene delivery.
Recently, various bio-reducible polymers, which contain disulfide bonds, have been developed to overcome the cytotoxicity and low transfection efficiency of existing polymers [6, 7]. Disulfide bonds are more stable in physiological conditions than ester bonds, which are rapidly hydrolyzed by water. When the bio-reducible polymers are exposed to glutathione (GSH) in the cytosol, the incorporated disulfide bonds in the polymer undergo degradation by thiol-disulfide exchange [8, 9]. This intracellular degradation mechanism of bio-reducible polymers causes decreased cytotoxicity via prevention of polymer accumulation in the cells, and contributes to the enhancement of transfection efficiency [10]. Based on these advantages, several types of bio-reducible polymers have been developed as potential gene delivery carriers using PEI, polypeptide or PAMAM as a back bone [11-14].
Previously, our group developed a bio-reducible poly (cystamine bisacrylamide-diaminohexane, CBA-DAH), which had higher transfection efficiency and lower cytotoxicity than high molecular branched PEI (PEI25k) [13]. Additionally, poly (CBA-DAH) was used as the back-bone of the arginine-grafted bio-reducible poly (CBA-DAH) polymer (ABP) [15]. Arginine is a main component in cell penetrating peptides, therefore, arginine modified non-viral carriers would show improved intracellular uptake. In vitro and in vivo studies using ABP have demonstrated high transfection efficiency with low cytotoxicity in reductive conditions [16-18]. Despite its promising features, ABP forms loose complexes with pDNA, and the delivery of therapeutic genes requires large amounts of polymer due to its low molecular weight. These disadvantages limit its utility in vivo. To improve the drawbacks of ABP, four ABP residues were conjugated to PAMAM G0 resulting in a dendrimer type bio-reducible polymer, PAM-ABP [19]. Because of increased molecular weight, PAMAM conjugated ABP required a lower weight ratio than ABP to optimize the transfection efficiency and make well condensed nanoparticles with pDNA. In addition, PAM-ABP has been used to efficiently deliver genes into various types of cells while maintaining low cytotoxicity [20-22].
Cancer gene therapy has been widely studied and has continuously progressed as an alternative strategy to conventional treatments such as chemotherapy and radiotherapy. In recent years, many studies have focused on effective and safe cancer therapy by tumor targeting [23, 24]. An active targeting is achieved by coupling of specific tumor-homing ligands, such as antibodies and peptides to the gene carrier. Ligands of targeted nanocarriers bind to their receptors in tumor cells and then are uptaken into the cell by receptor-mediated endocytosis [25]. Specific and active targeting limits undesirable side effects to normal cells, and reduces the necessary dose for therapeutic efficacy. Therefore, selective targeting delivery to the tumor site is critical for effective cancer gene therapy.
Cancer cells express many different types of integrin. Integrins are adhesive molecules that mediate the progression of cancer stages such as tumor growth, invasion, and metastasis. Among them, αvβ3 and αvβ5 (αvβ3/5) are poorly expressed in non-angiogenic cells and strongly overexpressed in angiogenic vessels and tumor cells [26, 27]. Many cancer cells overexpress αvβ3/5 integrins, which can specifically recognize the short peptide motif Arg-Gly-Asp (RGD). Based on this biological feature of cancer, various polymers have been designed using the RGD motif cancer targeting delivery [28, 29]. A number of synthetic linear and cyclic RGD sequence containing peptides, such as RGD4C and RGDfK, have been shown to have high affinity for integrin αvβ3/5 [27]. For example, when the cationic polymer (PEI) was grafted with RGD moieties, cancer cell transfection was enhanced and the successful therapeutic efficacy of soluble VEGF receptor 1 (sFlt-1) gene was observed in a colon cancer animal model [30, 31]. Other types of nanocarriers using RGD peptides have been studied, such as targeted liposomes, chitosan, cationic polymers for genes or drugs targeted delivery [32-34].
Angiogenesis is a critical event in tumors and involves the formation of new blood vessels to fuel rapid cancer cell growth. A number of growth factors have been identified as regulators of angiogenesis such as fibroblast growth factor (FGF) and hepatocyte growth factor (HGF)[35, 36]. Most recently, vascular endothelial growth factor (VEGF) has been identified and been shown to be potent and specific for angiogenesis of endothelial cells in response to important angiogenic processes such as proliferation, and migration [37]. Therefore, inhibition of VEGF expression or disabling VEGF receptor function has been suggested as powerful strategy for cancer gene therapy by inhibition of tumor growth and metastasis [38-40].
The recently developed bio-reducible polymer PAM-ABP showed enhanced gene delivery efficiency with low cytotoxicity. However, lack of cell- targeting ability of PAM-ABP is a major obstacle to successful disease specific applications. For this reason, we developed tumor targeted bio-reducible polymers by coupling cyclic RGD peptides to the PAM-ABP, termed “PA-PEG1k-RGD”. The physiological properties, transfection efficiency and cytotoxiciy were investigated in detail. The ability of RGD peptides to mediate intracellular uptake was evaluated in cancer cells. Moreover, therapeutic efficacy was demonstrated using VEGF siRNA expressing plasmid (pshVEGF) as a therapeutic gene. PA-PEG1k-RGD/pshVEGF complex was examined to investigate its anti-cancer effects in human cancer cells (MCF7 and PANC-1, integrins αvβ3/5 overexpress) and normal cells (293, integrins αvβ3/5 negative).
2. Materials and methods
2.1. Materials
Hyperbranched poly(ethylenimine) (PEI25K,Mw=25 kDa), tert-butyl-N-(6-aminohexyl) carbamate (N-Boc-1,6-diaminohexane, N-Boc-DAH), poly(amidoamine) (PAMAM, ethylene diamine core, G0), 5-diphenyltetrazoliumbromide (MTT) and dithiothreitol (DTT) were purchased from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM), Dulbecco's phosphate-buffered saline (DPBS), Medium 200PRF, Low serum growth supplement (LSGS), phosphate-buffered saline (PBS), YOYO-1 iodide (1mM solution in DMSO), SYBR safe DNA gel stain, human VEGF ELISA kit, gel extraction kit, pSilencer 2.1-U6 neo kit and human umbilical vein endothelial cell (HUVEC) were purchased from Invitrogen (Carlsbad, CA). VEGF shRNA oligonucleotides were synthesized by University of Utah DNA/Peptide synthesis core facility. Cyclic Arg-Gly-Asp-D-Phe-Cys (RGDfC, M.W. 578.65) and Arg-Ala-Asp-D-Phe-Cys (RADfC, M.W. 592.68) peptides were obtained from Peptide International, Inc (Louisville, KY). Luciferase or green fluorescent expression plasmid, pCMV-Luc or pCMV-GFP, was purchased from Aldevron, Inc. (Fargo, ND). Maleimide polyethylene glycol succinimidyl ester (Mal-PEG-NHS, 1K Da) was purchased from Nanocs Inc. (New York, NY). N,N′-Cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). Traut's reagent, SPDP cross linker and BCA assay kit were purchased from pierce (Rockford, IL). Luciferase assay kit, 5× reporter lysis buffer, 5× passive lysis buffer and agarose were purchased from Promega (Madison, WI). MCF7 human breast cancer, PANC-1 human pancreatic cancer and 293 human embryonic kidney cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, VA).
2.2. Synthesis of RGD conjugated PAM-ABP
Arginine-grafted bio-reducible poly (cystaminebisacrylamide-diaminohexane, CBA-DAH) named “ABP” and dendrimer type poly (amido amine) (PAMAM) conjugated ABP named “PAM-ABP” were synthesized as described previously [13, 15, 19]. To synthesize the cyclic RGDfC (or RADfC) peptides conjugated PAM-ABP, PAM-ABP was dissolved in 0.1 M phosphate buffered saline (pH 7.2, 0.15 M NaCl, 2 mM EDTA). Mal-PEG1k-NHS was added to the PAM-ABP solution. The mixture was stirred for 1 h at room temperature, and then dialyzed against ultrapure water using a dialysis membrane (MWCO=3500 Da), followed by lyophilization. For the conjugation of target peptides, pegylated PAM-ABP (PA-PEG1k-Mal) was dissolved in 50 mM phosphate buffered saline (pH 7.2 0.15M NaCl, 10 mM EDTA). Two equivalents of cyclic RGDfC (or RADfC) peptides per maleimide groups of PA-PEG1k-Mal were added to the solution with stirring, and the mixture was further reacted for overnight. The product was dialyzed against ultrapure water using a dialysis membrane (MWCO=3500 Da). The final product was lyophilized and stored at -80 °C for further use.
2.3. Construction of VEGF siRNA expressing plasmid
Human VEGF siRNA (targeting bases position 189-207) with the following sequences were used: 5′-AAGGAGTACCCTGATGAGATC-3′ (sense), 5′-GATCTCATCTGGGTACTCC-3′ (antisense) [41]. Human VEGF specific siRNA expressing plasmid was designed and constructed using pSilencer 2.1-U6 neo kit. Two complementary oligonucleotides of siVEGF sequence containing hairpin structure were annealed and then ligated with BamHI and HindIII sites of linearized pSilencer 2.1-U6 neo vector, resulting inpshVEGF. Hairpin structure sequences as follows; gatccc AAGGAGTACCCTGATGAGATC ttcaagaga GATCTCATCAGGGTACTCCTT ttttttggaaa (sense) 5′-agcttttccaaaaaa AGGAGTACCCTGATGAGATC tctcttgaa GATCTCATCAGGGTACTCCTT g-3′ (antisense). VEGF target sequence is shown in uppercase letters and the loop sequence is underlined. Negative control plasmid that expresses a hairpin siRNA with limited homology to any known sequences in human was provided from pSilencer 2.1-U6 neo kit.
2.4. Agarose gel electrophoresis
For the gel retardation assay, PAM-ABP, PA-PEG1k-RGD or PA-PEG1k-RAD/pCMV-Luc complex was prepared at various weight ratios (0.25, 0.5, 1, 2, 3, 4 and 5) by mixing fixed amount of pDNA (0.5 μg) with increasing amounts of polymers in Hepes buffered saline (10 mM Hepes, 1 mM NaCl, pH 7.4). After 30 min incubation at room temperature, the samples were electrophoresed through a 1 % agarose gel containing a SYBR gel staining solution at 100V for 40 min. The migration of pDNA bands in agarose gel was visualized using a UV transilluminator (Bio-Rad, Hercules, CA). pDNA release from the complex under the reductive condition was confirmed. Polymers/pDNA complexes were prepared under optimal weight ratio condition. Complexes were incubated in the absence or presence of 5 mM DTT for 30 min at room temperature, and then the samples were electrophoresed through a 0.7 % agarose gel containing the SYBR solution at 100 V for 20 min.
2.5. Measurement of particle size and zeta-potential
PEI25k, PAM-ABP, PA-PEG1k-RGD or PA-PEG1k-RAD/pCMV-Luc complex was prepared at various weight ratios (1, 2, 3, 4, 5 and 6). The complexes were incubated 30 min at room temperature for complex formation. After incubation, the particle sizes and zeta potentials were measured by the Zetasizer Nano ZS system (Malvern Instruments, UK)
2.6. Cell culture and transfection
MCF7 human breast cancer cell, PANC-1 human pancreatic cancer cell and 293 human embryonic kidney cell lines were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum. Human umbilical vein endothelial cell (HUVEC) was maintained in Medium 200PRF supplemented with low serum growth supplement (LSGS). The cells were cultured at 37 °C in a 5% CO2 incubator. For the luciferase assay, the cells were seeded at a density of 5.0×104 cells/well in 12-well plates. PA-PEG1k-RGD/pCMV-Luc complex was prepared at various weight ratios. PEI25k or PAM-ABP/pCMV-Luc complex was prepared at a 1:1or 1:5 weight ratio, respectively, based on previous report [19]. The amount of plasmid DNA was fixed at 1 μg/well. Before transfection, the cells were washed twice with serum-free DMEM, and then 1 ml of serum-free fresh medium was added. Then, the polymers/pDNA complexes were added to the each well and incubated it for 4 hrs. After incubation, the transfection mixtures were removed, and 1 ml of fresh DMEM, containing 10% FBS, was added. The cells were incubated for an additional 48 hrs or 72 hrs.
For the fluorescence microscopy study, green fluorescent protein expression plasmid (pCMV-GFP) was transfected into MCF7 or PANC-1 cells. Cells were seeded at a density of 5.0×105 cells/well in 6-well plates. Polymers/pDNA complexes were prepared at the same ratio as described above. PA-PEG1k-RAD/pCMV-Luc complex was prepared at a 1:7 weight ratio. The amount of plasmid DNA was fixed at 2 mg/well. GFP intensity of the transfected cells was verified by fluorescence microscopy and a multimode microplate reader (Infinite M2000; Tecan Inc., Mannedorf, Switzerland). Excitation and emission peaks of GFP were fixed at 395nm and 509nm wavelength. GFP intensity was calculated according to the following equation: GFP intensity (% of control) = [(OD (sample)/mg protein)/(OD (control)/mg protein)]×100
2.7. Competition study with free RGD peptides
Competition study was performed in order to measure RGD ligand mediated cellular uptake of PA-PEG1k-RGD. 293 and PANC-1 cells were seeded at a density of 5.0×105 cells/well in 6-well plates. Before 15 min of complex treatment, a 10-fold excess of free RGDfC peptides in PBS was added to the cells [29]. After 48 hrs incubation, cells were washed and lysated in passive lysis buffer to measure the GFP intensity by microplate reader. The collected cell lysates were centrifuged and then the supernatants were harvested. The protein concentration of the extract was measured by BCA protein assay (Pierce, Rockford, IL).
2.8. Flow cytometry assay for cellular uptake
MCF7 or PANC-1 cell was seeded at a density of 5×105 cells/well in 6-well plates. Polymers/pDNA complexes were prepared with YOYO-1 iodide (1 molecule of the dye per 20 base pairs of nucleotide) labeled pshVEGF at optimized weight ratios. Transfection was performed as described above. After 4 hrs of complexes treatment, the cells were harvested to fresh tubes. The cells were washed with cold PBS and then re-suspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 0.02% NaN3, 0.2% FBS). The degree of cellular uptake was immediately measured by BD FACScan II (BD, Biosciences Immunocytometry Systems, San Jose, CA, USA).
2.9. MTT assay
MCF7, PANC-1 and 293 cells were seeded at 1×104 cells/well in 96-well microassay plates. After 24 hrs of incubation, polymer/pDNA complexes were treated into cells. Complexes were prepared with optimized weight ratio respectively. pCMV-Luc or pshVEGF was used as a pDNA. The complexes were prepared at optimized ratios with fixed amount of pDNA (1 μg/well) and then transfected into cells as described above. After 48 hrs or 72 hrs of incubation, 20 μl (5 mg/ml) MTT solution in 1×PBS was added to each well. Cells were incubated for an additional 4 hrs at 37 °C. The MTT-containing medium was removed and 200 ml of DMSO was added to dissolve the formazan crystals formed by the live cells. Absorbance was measured at 570 nm using the microplate reader. Cell viability (%) was calculated according to the following equation: Cell viability (%) = (OD570 (sample)/OD570 (control)) × 100.
2.10. In vitro luciferase assay
Transfected cells were washed twice with PBS and 150μl of reporter lysis buffer was added to each well. After 15 min of incubation at room temperature, the cells were harvested and centrifuged. The supernatants were transferred to fresh tubes. The protein concentration of the extract was measured by BCA protein assay. Luciferase activity was measured in terms of relative light units (RLU) with the multimode micro-plate readers. The final values of luciferases were reported in terms of RLU/mg total protein.
2.11. VEGF ELISA
MCF7 and PANC-1 cells were seeded at 2 × 104 cells/well in 24 well plates and then polymers/pDNA complexes were transfected as described above. At 48 hrs and 72 hrs of incubation, each conditioned medium was harvested and the expression level VEGF was measured by human VEGF ELISA kit. VEGF levels were normalized relative to the total protein concentration in each sample.
3. Results
3.1. Synthesis of cyclic RGDfC peptides conjugated with PAM-ABP
In the present study, a tumor targeting bio-reducible polymer was developed by conjugating cyclic RGDfC(RGD) peptides to PAM-ABP(PA), named “PA-PEG1k-RGD” (Fig. 1A). The RGD peptides were used as the targeting ligand for αvβ3/5integrins on the cancer cell surface. RADfC peptides, with alanine substituted for glycine, were used as a negative control variant for RGDfC peptides. It has been observed that cyclic RGD peptides bind integrins with higher affinity than linear RGD peptides [42]. Bifunctional maleimide-polyethylene glycol-N-hydroxysulfosuccinimide (Mal-PEG-NHS) was used for the spacer between RGD peptides and PAM-ABP. All reactions were monitored by Thin-Layer Chromatography (TLC) with ninhydrin staining, UV spectroscopy and proton NMR. The structure of polymer was confirmed by proton NMR spectra (400 MHz, D2O), with unique peaks for PAMAM (3.5, 3.1, 2.7, 2.4 ppm), ABP (1.2–1.6 ppm), PEG (3.7 ppm), maleimide (7.0 ppm) and the phenylalanine of RGD peptide (7.2-7.4 ppm) (Fig. 1B). Briefly, poly (CBA-DAH) was synthesized by using N-Boc-DAH and CBA as repeated monomers. Based on the results of the proton NMR spectra, poly (CBA-DAH) was composed of eight repeating units. Then, 2 equivalents of Fmoc-Arg(pbf)-OH and HBTU, and 4 equivalents of DIPEA were used for arginine-graft reaction. 100% of primary amine of poly (CBA-DAH) was conjugated with arginine. For the chemical conjugation of PAMAM dendrimer and ABP, traut's reagent and SPDP cross linker were used. As a result, four ABPs had been conjugated to the four primary amines of PAMAM dendrimer. The molecular weight of PAM-ABP was measured using a gel permeation chromatography. The molecular weight of PAM-ABP was determined to be 19.2×103 Da/mole and its polydispersity value was 1.41. Chemical conjugation of PAM-ABP with RGD peptides was performed using PEG as a spacer. Finally, PA-PEG1k-RGD was synthesized by conjugation of one PEG and one RGD peptide with PAM-ABP. The overall synthesis scheme of PA-PEG1k-RAD was same as PA-PEG1k-RGD.
Fig. 1.
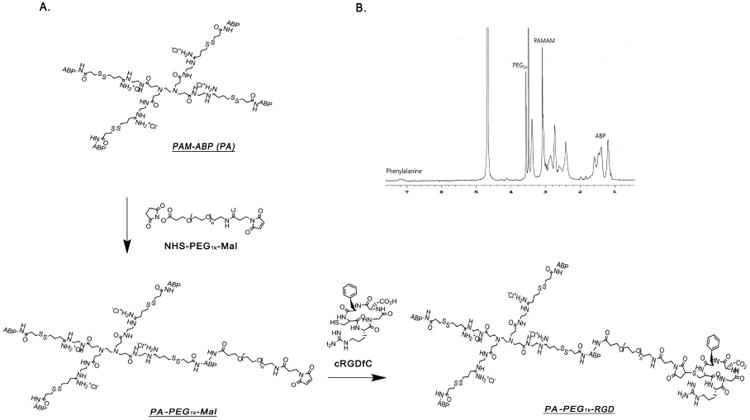
(A) Structure of PA-PEG1k-RGD Cyclic RGDfC peptides were conjugated to PAM-ABP using cross linker, Mal-PEG1k-NHS. (B) 1H NMR spectrum of PA-PEG1kRGD
3.2.Characterization of PA-PEG1k-RGD/pDNA complex
To confirm the complex formation of PA-PEG1k-RGD with plasmid DNA (pDNA), a gel retardation assay was performed. Polymers/pCMV-Luc complexes were prepared at various weight ratios of PA-PEG1k-RGD or PA-PEG1k-RAD with fixed amount of pDNA. PAM-ABP showed complete retardation above 1:1 (polymer: pDNA) weight ratio. PA-PEG1k-RGD and PA-PEG1k-RAD showed completely retardation of pDNA above 2:1 weigh ratio (Fig. 2A). Polymers with internal disulfide bonds were expected to be degraded in reducing environments such as cytoplasm. To confirm the pDNA release from the complex with PAM-ABP derivatives under the reductive condition, agarose gel electrophoresis was performed in the absence or presence of DTT. In the figure 2B, complexes with PAM-ABP derivatives showed pDNA release from the complex in the presence of DTT. However, non-degradable polymer, PEI, showed complete retardation both absence and presence of DTT condition. These results demonstrated that RGD peptides conjugated PAM-ABPs could rapidly degraded in reductive environment by cleavage of the disulfide bonds. The size and surface charge of the complex were measured at various weight ratios. The average mean diameter of the PA-PEG1k-RGD/pCMV-Luc complex was approximately 150 nm, and the zeta potential showed a positive charge with an average of 15 mV above 3:1 weight ratio (Fig. 2C and 2D). The PA-PEG1k-RAD/pCMV-Luc complex showed similar particle size and zeta-potential. PA-PEG1k-RGD showed a slightly higher complete retardation ratio with reduced surface charge than that of PAM-ABP, which may be due to a change in surface charge density by the peptides conjugation. However, PA-PEG1k-RGD had efficient condensing ability and was able to form nano-complexes with pDNAs.
Fig. 2. Physical characterization of PA-PEG1k-RGD/pDNA complexes.
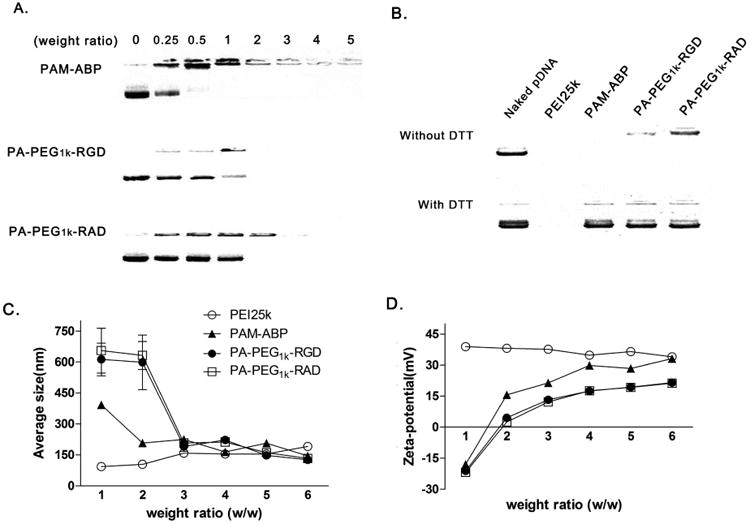
(A) Gel retardation assay Polymers/pDNA complexes were prepared at various weight ratios and analyzed by 1% agarose gel electrophoresis. (B) Agarose gel electrophoresis in the absence and in the presence of 5 mM DTT. PEI25k or PAM-ABP/pCMV-Luc complex was prepared at 1:1 or 1:5 weight ratio, respectively. PA-PEG1k-RGD or PA-PEG1k-RAD/pCMV-Luc complex was prepared 1:7 (pDNA: polymer) weight ratio condition. (C) Size and (D) zeta potential Complexes were prepared with various weight ratios polymers and pDNA. Data are expressed as mean values (±standard deviation) of triplicate experiments.
3.3. Transfection efficiency of PA-PEG1k-RGD
Transfection condition of PA-PEG1k-RGD was optimized at various weight ratios. The PA-PEG1k-RGD/pCMV-Luc complex was prepared and transfected into PANC-1 pancreatic cancer cells. The highest transfection efficiency of PA-PEG1k-RGD was observed at a 7:1 (PA-PEG1k-RGD: pCMV-Luc) weight ratio. Therefore, the complex weight ratio was fixed at a 7:1 in further studies (Fig. 3A). Transfection efficiency of PA-PEG1k-RGD was determined in αvβ3 integrin positive HUVEC and αvβ3/5 integrin negative 293 cells to confirm the RGD mediated delivery. For the luciferase assay, complexes were prepared at the optimized weigh ratio and then treated to cells. After 48 hrs incubation, PA-PEG1k-RGD transfected HUVEC showed strong luciferase activity compared to PEI25k, PAM-ABP and PA-PEG1k-RAD. Otherwise, there was no significant difference between the RGD and RAD conjugated PAM-ABP polymers and PA-PEG1k-RGD had lower transfection efficiency than PAM-ABP and PEI25K in 293 cells (Fig. 3B and 3C). In breast and pancreatic tumors, αvβ3/5 integrins are overexpressed and associated with tumor growth and metastasis [27]. Thus, MCF7 breast cancer and PANC-1 pancreatic cancer cells were used as model cancer cells. Transfection efficiency of PA-PEG1k-RGD was evaluated by GFP expressing plasmid delivery into the MCF7 or PANC-1cells. Fluorescence microscopy images demonstrated that PA-PEG1k-RGD/pCMV-GFP showed higher GFP expression compared with the PEI25k, unmodified PAM-ABP and PA-PEG1k-RAD in both cancer cells (Fig. 4A and 4B). In the measurement of GFP intensity, the PA-PEG1k-RGD/pCMV-GFP complex transfected MCF7 or PANC-1 cells elucidated 290% and 340% stronger GFP intensity respectively than untreated controls. In addition, PA-PEG1k-RGD showed significantly higher GFP expression levels than PEI25k or PAM-ABP (Fig. 4C and 4D). These results demonstrated that gene delivery efficiency of RGD conjugated PAM-ABP was regulated by binding of RGD with αvβ3/5 integrins on the cell surface. Therefore, PA-PEG1k-RGD could deliver therapeutic genes into cancer cells with limited side effects in normal cells.
Fig. 3. Transfection efficiency of PA-PEG1k-RGD.
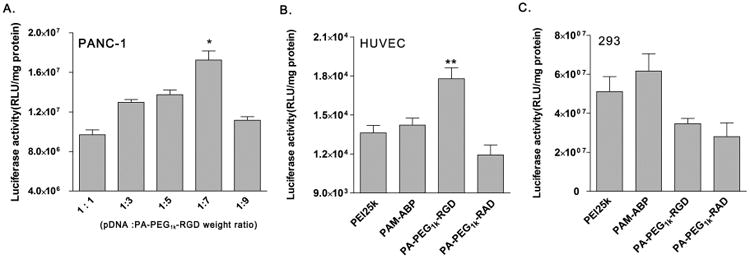
PA-PEG1k–RGD/pDNA complex was prepared at various weight ratios and transfected into (A) PANC-1 pancreatic cancer cells. PEI25k, PAM-ABP, PA-PEG1k-RGD or PA-PEG1k-RAD/pCMV-Luc complex was transfected into (B) HUVEC αvβ3 positive and (C) 293 αvβ3/5 negative normal cells. Each polymer/pDNA complex was prepared under optimal condition. After 48 hrs of incubation, luciferase activity was measured. The data are expressed as mean values (±standard deviation) of triple experiments. *P<0.01 as compared with weight ratio of 1:5 (pDNA: PA-PEG1k–RGD).**P<0.003 as compared with PAM-ABP.
Fig. 4. Green fluorescent protein expression levels by PA-PEG1k-RGD.
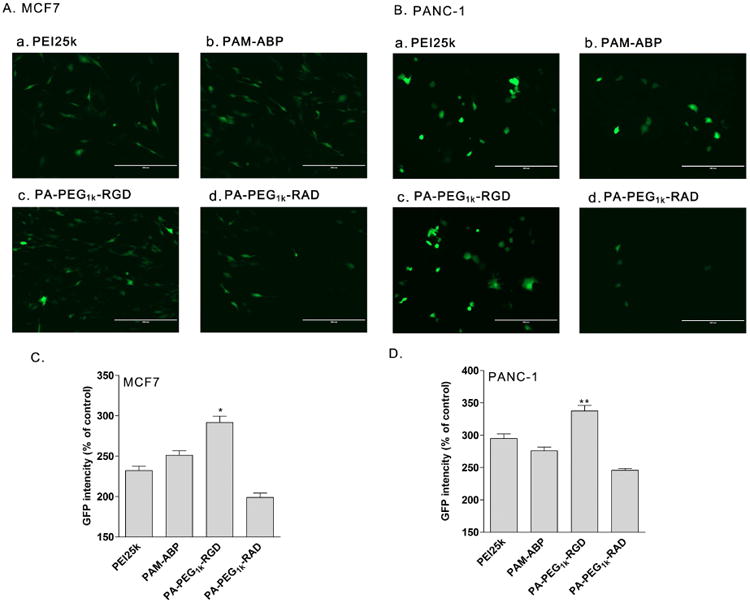
Polymers/pCMV-GFP complexes were transfected into MCF7 and PANC-1 cancer cells. After 48 h of incubation, GFP expression level was evaluated by (A and B) fluorescence microscopy and (C and D) fluorescence intensity measurement. The data are expressed as mean values (±standard deviation) of triplicate experiments. *P<0.003 as compared with PAM-ABP. **P<0.001 as compared with PEI25k.
3.4. Cytotoxicity of PA-PEG1k-RGD
In order to evaluate cytotoxicity, polymers/pDNA complexes were transfected into MCF7 or PANC-1 cancer cells and cell viability was measured by MTT assay after 48 hrs of incubation. PA-PEG1k-RGD and pDNA complexes were prepared with increasing weight ratios of PA-PEG1k-RGD. At the transfection optimized weight ratio (1:7), cells showed over 85% viability. Even polymer ratio increased up to 1:15, cells showed approximately 70% viability (Fig. 5A). In figure 5B, cytotoxicity of PA-PEG1k-RGD was compared with various polymers; PEI25k, PAM-ABP, and PA-PEG1k-RAD. Each polymer was mixed with pDNA at optimizing weight ratio condition and naked pDNA was used as a control. PEI25k transfected MCF7 and PANC-1 showed less than 70% cell viability even at low weight ratio (1:1). On the other hand, PAM-ABP and peptides (RGD or RAD) conjugated PAM-ABP groups showed higher (over 80%) cell viability than that of PEI25k. In addition, similar cell viability at optimal ratios of modified and unmodified PAM-ABP indicated that modification of PAM-ABP with RGD peptides ligand did not affect the cytotoxicity of PAM-ABP.
Fig. 5. Cytotoxiciy of RGD peptides conjugated PAM-ABP.
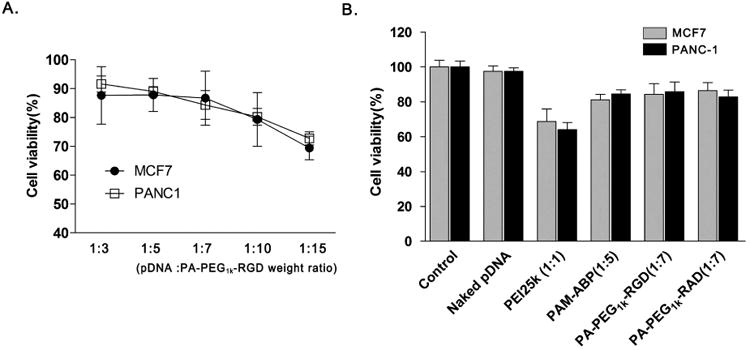
(A) Cell viability of PA-PEG1k-RGD at various weight ratios PA-PEG1k-RGD/pCMV-Luc complexes were transfected into MCF7 and PANC-1 cells with increasing weight ratios of PA-PEG1k-RGD polymer. (B) Comparison of polymer cytotoxicity PEI25k, PAM-ABP, PA-PEG1k-RGD and PA-PEG1k-RAD were prepared with pDNA at optimal conditions for complex formation. After 48 hrs, cytotoxicity was evaluated by MTT assay. The complex ratio was presented by pDNA: polymer weight ratio. The data are expressed as mean values (±standard deviation) of quadruplicate experiments.
3.5. Competition assay with free cyclic RGDfC peptides
The main advantage of PAM-ABP conjugated RGD is selective internalization into the cancer cells through receptor-mediated or clathrin-dependent endocytosis [43, 44]. To determine the transfection efficiency dependance on RGD peptides, a competition assay was performed. Transfection efficiency was measured by expressed GFP intensity and PA-PEG1k-RAD was used as a control. Excess amount of free RGD peptides were added to the cells 15 min prior to the complex treatment. After 48 hrs incubation, PA-PEG1k-RGD transfected PANC-1 cells showed significantly decreased GFP expressing levels (∼10%) with free RGD peptides. However, transfection efficiency of PA-PEG1k-RAD was not influenced by free RGD peptides (Fig. 6A). In addition, 293 non-cancer cells did not show significant difference between with and without free RGD peptides in each polymer group (Fig. 6B). These results suggest that PA-PEG1k-RGD had specificity to αvβ3/5 integrins expressing cancer cells and this targeting ability would be achieved by conjugation of RGD peptides to PAM-ABP.
Fig. 6. Competition assay with free cyclic RGDfC peptides.
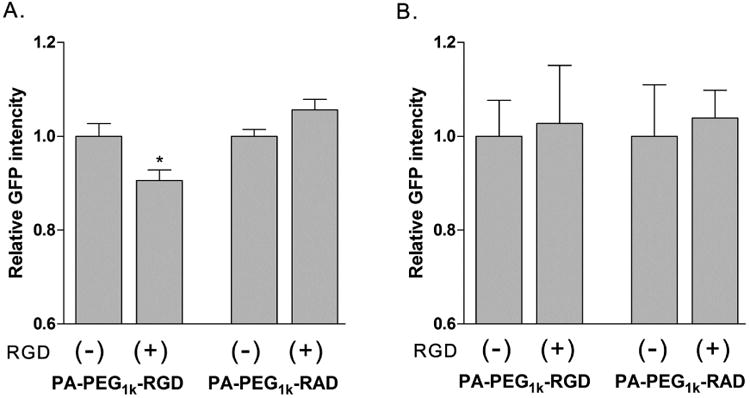
PA-PEG1k-RGD/pCMV-GFP or PA-PEG1k-RAD/pCMV-GFP complex was prepared 1:7 (pDNA: polymer) weight ratio condition and then transfected into (A) PANC-1 and (B) 293 cells. The free cyclic RGDfC peptides were pretreated before 15 min of complex treatment. After 48 hrs incubation, expressed GFP intensity (OD (sample)/mg protein) was measured and presented by relative GFP activity (% of free RGD untreated sample in each polymer) *P<0.014 as compared with free RGD untreated (-) PA-PEG1k-RGD.
3.6. Cellular binding and uptake of PA-PEG1k-RGD/pshVEGF complexes
Inhibition of VEGF expression or disabling VEGF receptor function has been used as a potent strategy to inhibit tumor growth and metastasis in a variety of animal tumor models [38, 39]. Therefore, VEGF siRNA expressing plasmid, pshVEGF, was constructed as a therapeutic gene. The efficacy of cancer treatment of the developed cancer targeted bio-reducible polymer, PA-PEG1k-RGD, could be clearly elucidated by this therapeutic gene. Also, the VEGF siRNA expressing plasmid could have longer lasting expression than delivering siRNA directly. The pshVEGF contained the hairpin structure of VEGF siRNA encoding oligonucleotides (Fig. 7A). Detailed construction was described in the Materials and Methods section. Prior to evaluating the therapeutic gene efficacy, the binding and uptake of PA-PEG1k-RGD/pshVEGF nanoparticles into MCF7 and PANC-1 cancer cells was evaluated by FACS analysis using YOYO-1-labeled shVEGF plasmid. PA-PEG1k-RGD showed the highest cellular uptake rate into MCF7 or PANC-1 cells (65.1% or 72.5%) compared with other carriers. Otherwise, non-targeting ligand conjugated polymer, PA-PEG1k-RAD showed much lower cellular uptake rate into MCF7 or PANC-1 cells (8% or 13.3%) than that of PEI25k, PAM-ABP and PA-PEG1k-RGD (Fig. 7B). The decreased uptake rate of PA-PEG1k-RAD might be due to that the conjugated non-targeting ligands have no cellular binding ability and also they block the endocytosis entering ability of bio-reducible polymer backbone. These results suggest that the PAM-ABP bio-reducible polymer acquired cancer targeting efficiency by conjugation of RGD peptides.
Fig. 7. Cellular binding and uptake efficiency of PA-PEG11k-RGD/pshVEGF complex.
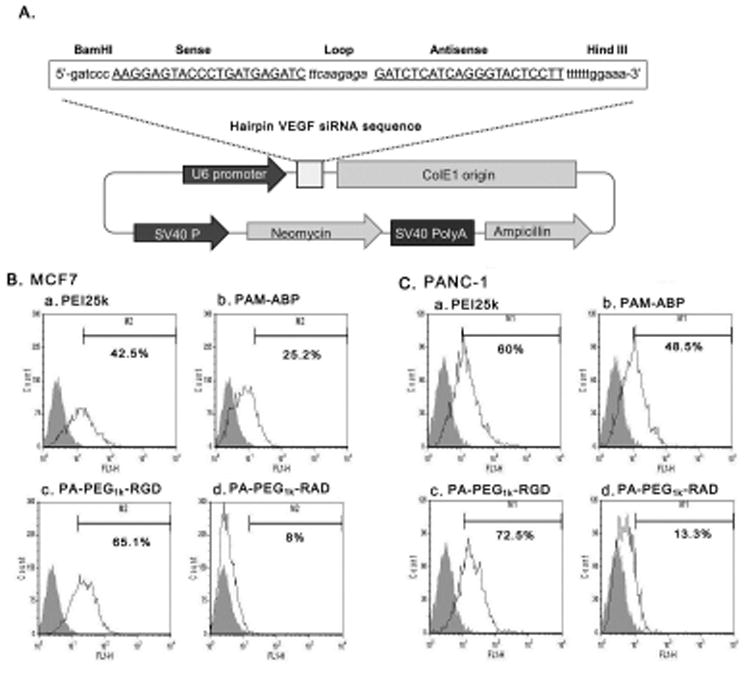
(A) Structure of pshVEGF VEGF siRNA sequence containing hairpin structure was inserted into pSilencer plasmid to constructed VEGF siRNA expressing plasmid, pshVEGF. Polymers/YOYO-1 dye stained pshVEGF complexes were prepared at optimized weight ratio, respectively. The complexes were treated into (B) MCF7 and (C) PANC-1 cells for 4 hrs and then, cells were analyzed by FACS. a; PEI25k, b; PAM-ABP, c; PA-PEG11k-RGD and d; PA-PEG11k-RAD. Gray area indicated untreated cells.
3.7. Cancer cell growth inhibition efficacy by PA-PEG1k-RGD/shVEGF delivery
The PA-PEG1k-RGD carrier was evaluated for cancer cell specific delivery of VEGF siRNA expressing plasmid (pshVEGF). VEGF gene expression levels related with gene silencing efficacy was measured using VEGF ELISA after transfection of polymer/pDNA complexes (Fig. 8). In the results, expression level of endogenous VEGF was significantly reduced in MCF7 and PANC-1 cells transfected with PA-PEG1k-RGD/pshVEGF complex. Compared to non-treated control cells, the PA-PEG1k-RGD/pshVEGF transfected group showed 67% or 71% reduction of VEGF levels in MCF7 or PANC-1 cell after 48 hrs incubation (Fig. 8A and 8B). In addition, gene silencing efficiency of PA-PEG1k-RGD/pshVEGF was continued with 51% or 60% reduction of VEGF expression until 72 hrs incubation (Fig. 8C and 8D). PA-PEG1k-RGD/pshVEGF complex transfected cells showed significantly lower VEGF expression than PEI25k, PAM-ABP and PA-PEG1k-RAD. In addition, scrambled siRNA expressing plasmid (pscRNA) was delivered using PA-PEG1k-RGD to confirm the constructed VEGF siRNA expressing plasmid (pshVEGF) silencing efficiency. The PA-PEG1k-RGD/pscRNA complex transfected group did not show a decrease in VEGF expression as similar to non-treated control cells (Fig. 8). The anti-proliferative effect of PA-PEG1k-RGD/pshRNA on the cancer cells was evaluated by same experimental condition as with VEGF ELISA. After 72 hrs of incubation, inhibition rates of proliferation and cell growth was measured by MTT method [45]. Similar to the VEGF expressing comparison results, PA-PEG1k-RGD/pshVEGF complex showed less than 43% or 35% cell viability in MCF7 or PANC-1 cells and had lower cell viability than PA-PEG1k-RGD/pscRNA complex (Fig. 9A and 9B). However, cell viability of 293 cells was not affected by PA-PEG1k-RGD/pshVEGF treatment (Fig. 9C). In the results, cell viability demonstrated VEGF gene silencing effect and indirectly indicated the inhibition of cell proliferation and growth rates by PA-PEG1k-RGD/pshVEGF complex treatment. Although PEI25K transfected cells showed lower cell viability than PAM-ABP and PA-PEG1k-RAD, these were not a VEGF siRNA silencing effect but were a result of the cytotoxicity of the PEI25K polymer (Fig. 9). Overall, VEGF siRNA delivery by PA-PEG1k-RGD maintained a long-term silencing effect; resulting in a decrease of the effective VEGF gene expression. Also, treatment with PA-PEG1k-RGD/pshVEGF caused decreased cancer cell viability. However, it had no significant effect on viability of non-cancer cells. These results clearly showed the targeting efficiency of the RGD peptide conjugated bio-reducible polymer.
Fig. 8. Down regulation of VEGF expression by pshVEGF delivery into cancer cells 48hrs incubated.
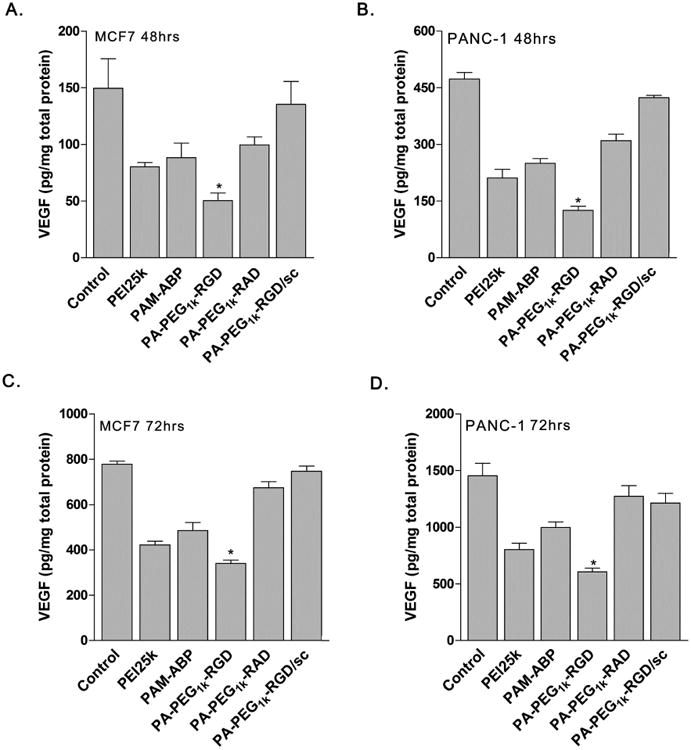
(A) MCF7 and (B) PANC-1, 72 hrs incubated of (C) MCF7 and (D) PANC-1 cells. Polymers/pshVEGF complexes were trasnfected into MCF7 and PANC-1 cells. After 48 or 72 hrs incubation, VEGF expression level of each conditioned medium was measured by VEGF ELISA. Control;untreated cells, PA-PEG1k-RGD/sc; scrambled shRNA expressing plasmid (pscRNA) was use for complex formation with PA-PEG1k-RGD. The data are expressed as mean values (±standard deviation) of quadruplicate experiments. *P<0.003 as compared with PEI25k.
Fig. 9. Cell growth inhibition.
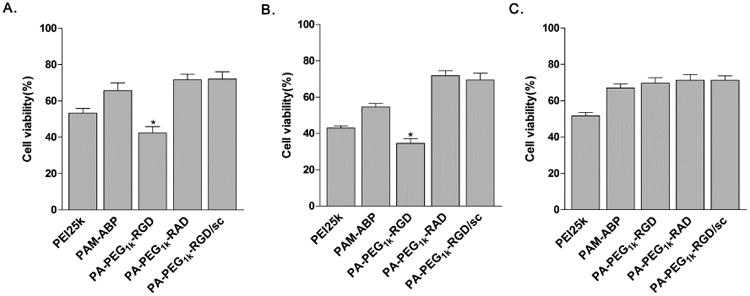
Polymers/pshVEGF complexes were transfected into (A) MCF7, (B) PANC-1 (C) 293 cells. After 72 hrs of incubation, cell growth inhibition efficiency was indirectly measured by MTT assay. Scrambled shRNA (pscRNA) was used in PA-PEG1k-RGD/sc complex. The data are expressed as mean values (±standard deviation) of triplicate experiments. *P<0.005 as compared with PEI25k.
4. Conclusion
In this study, we developed cyclic RGDfC peptides conjugated bio-reducible polymer, PA-PEG1k-RGD for tumor targeted therapeutic gene delivery. PA-PEG1k-RGD was efficiently delivered plasmid DNA into αvβ3/5 integrin expressing cancer cells, MCF7 and PANC-1, with low cytotoxicity. Competition assay with free RGD peptides and cellular uptake assay showed RGD mediated gene delivery efficiency of PA-PEG1k-RGD into MCF7 and PANC-1 cells. To confirm the anti-cancer therapeutic effect, the VEGF siRNA expressing plasmid, pshVEGF, was delivered to MCF7, PANC-1 or 293 cells. PA-PEG1k-RGD/pshVEGF complexes showed 51-71% decreased VEGF expression by silencing effect and reduced the viability of cancer cells less than 43%. Therefore, the gene therapy system with the tumor targeted bio-reducible polymer, PA-PEG1k-RGD and VEGF siRNA expressing plasmid may be a strong candidate for cancer gene therapy.
Acknowledgments
This work was financially supported by NIH Grants CA107070.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu F, Huang L. Development of non-viral vectors for systemic gene delivery. J Control Release. 2002;78:259–66. doi: 10.1016/s0168-3659(01)00494-1. [DOI] [PubMed] [Google Scholar]
- 2.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery—past, present, and future. Prog Polym Sci. 2007;32:799–837. [Google Scholar]
- 3.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–31. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 4.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem rev. 2008;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 5.Lim YB, Kim SM, Suh H, Park JS. Biodegradable, endosome disruptive, and cationic network-type polymer as a highly efficient and nontoxic gene delivery carrier. Bioconjugate Chem. 2002;13:952–7. doi: 10.1021/bc025541n. [DOI] [PubMed] [Google Scholar]
- 6.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J Control Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly (disulfide amine) s for gene delivery. Biomaterials. 2009;30:5804–14. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjugate Chem. 2001;12:989–94. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 9.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, et al. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J Am Chem Soc. 2004;126:2355–61. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 10.Kim HJ, Kwon MS, Choi JS, Yang SM, Yoon JK, Kim K, et al. Highly effective and slow-biodegradable network-type cationic gene delivery polymer: Small library-like approach synthesis and characterization. Biomaterials. 2006;27:2292–301. doi: 10.1016/j.biomaterials.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Kim TI, Kim SW. Bioreducible polymers for gene delivery. React Funct Polym. 2011;71:344–9. doi: 10.1016/j.reactfunctpolym.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn CH, Chae SY, Bae YH, Kim SW. Biodegradable poly (ethylenimine) for plasmid DNA delivery. J Control Release. 2002;80:273–82. doi: 10.1016/s0168-3659(01)00547-8. [DOI] [PubMed] [Google Scholar]
- 13.Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Novel biodegradable poly (disulfide amine) s for gene delivery with high efficiency and low cytotoxicity. Bioconjugate Chem. 2008;19:626–33. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Novel bioreducible poly (amido amine) s for highly efficient gene delivery. Bioconjugate Chem. 2007;18:138–45. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 15.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly (disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–64. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, McGinn AN, Olsen CD, Nam K, Lee M, Shin SK, et al. Human erythropoietin gene delivery for cardiac remodeling of myocardial infarction in rats. J Control Release. 2013;171:24–32. doi: 10.1016/j.jconrel.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim PH, Lee M, Kim SW. Delivery of two-step transcription amplification exendin-4 plasmid system with arginine-grafted bioreducible polymer in type 2 diabetes animal model. J Control Release. 2012;162:9–18. doi: 10.1016/j.jconrel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim PH, Kim J, Kim Ti, Nam HY, Yockman JW, Kim M, et al. Bioreducible polymer-conjugated oncolytic adenovirus for hepatoma-specific therapy via systemic administration. Biomaterials. 2011;32:9328–42. doi: 10.1016/j.biomaterials.2011.08.066. [DOI] [PubMed] [Google Scholar]
- 19.Nam HY, Nam K, Lee M, Kim SW, Bull DA. Dendrimer type bio-reducible polymer for efficient gene delivery. J Control Release. 2012;160:592–600. doi: 10.1016/j.jconrel.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 20.Kim HA, Nam K, Lee M, Kim SW. Hypoxia/hepatoma dual specific suicide gene expression plasmid delivery using bio-reducible polymer for hepatocellular carcinoma therapy. J Control Release. 2013;171:1–10. doi: 10.1016/j.jconrel.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Won YW, Lee M, Kim HA, Nam K, Bull DA, Kim SW. Synergistically Combined Gene Delivery for Enhanced VEGF Secretion and Antiapoptosis. Mol Pharm. 2013;10:3676–83. doi: 10.1021/mp400178m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Won YW, McGinn AN, Lee M, Nam K, Bull DA, Kim SW. Post-translational regulation of a hypoxia-responsive VEGF plasmid for the treatment of myocardial ischemia. Biomaterials. 2013;34:6229–38. doi: 10.1016/j.biomaterials.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis ME, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–82. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 24.Byrne JD, Betancourt T, Brannon-Peppas L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv Drug Deliv Rev. 2008;60:1615–26. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Prokop A, Davidson JM. Nanovehicular intracellular delivery systems. J Pharm Sci. 2008;97:3518–90. doi: 10.1002/jps.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danhier F, Breton AL, Préat Vr. RGD-based strategies to target alpha (v) beta (3) integrin in cancer therapy and diagnosis. Mol Pharm. 2012;9:2961–73. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- 28.Park J, Singha K, Son S, Kim J, Namgung R, Yun C, et al. A review of RGD-functionalized nonviral gene delivery vectors for cancer therapy. Cancer gene ther. 2012;19:741–8. doi: 10.1038/cgt.2012.64. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Jarzyna PA, van Tilborg GA, Nguyen VA, Cormode DP, Klink A, et al. RGD peptide functionalized and reconstituted high-density lipoprotein nanoparticles as a versatile and multimodal tumor targeting molecular imaging probe. FASEB J. 2010;24:1689–99. doi: 10.1096/fj.09-139865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suh W, Han SO, Yu L, Kim SW. An angiogenic, endothelial-cell-targeted polymeric gene carrier. Mol Ther. 2002;6:664–72. [PubMed] [Google Scholar]
- 31.Kim WJ, Yockman JW, Jeong JH, Christensen LV, Lee M, Kim YH, et al. Anti-angiogenic inhibition of tumor growth by systemic delivery of PEI-g-PEG-RGD/pCMV-sFlt-1 complexes in tumor-bearing mice. J Control Release. 2006;114:381–8. doi: 10.1016/j.jconrel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 32.Cai LL, Liu P, Li X, Huang X, Ye YQ, Chen FY, et al. RGD peptide-mediated chitosan-based polymeric micelles targeting delivery for integrin-overexpressing tumor cells. Int J Nanomed. 2011;6:3499–508. doi: 10.2147/IJN.S26670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J, Yang Sj, Wang Jc, Yang Lj, Xu Zz, Yang T, et al. Sequential treatment of drug-resistant tumors with RGD-modified liposomes containing siRNA or doxorubicin. Eur J Pharm Biopharm. 2010;76:170–8. doi: 10.1016/j.ejpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Nam HY, Kim Ti, Kim PH, Ryu J, Yun CO, et al. Active targeting of RGD-conjugated bioreducible polymer for delivery of oncolytic adenovirus expressing shRNA against IL-8 mRNA. Biomaterials. 2011;32:5158–66. doi: 10.1016/j.biomaterials.2011.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seghezzi G, Patel S, Ren CJ, Gualandris A, Pintucci G, Robbins ES, et al. Fibroblast growth factor-2 (FGF-2) induces vascular endothelial growth factor (VEGF) expression in the endothelial cells of forming capillaries: an autocrine mechanism contributing to angiogenesis. J Cell Biol. 1998;141:1659–73. doi: 10.1083/jcb.141.7.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 37.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–9. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 38.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–4. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 39.Inai T, Mancuso M, Hashizume H, Baffert F, Haskell A, Baluk P, et al. Inhibition of vascular endothelial growth factor (VEGF) signaling in cancer causes loss of endothelial fenestrations, regression of tumor vessels, and appearance of basement membrane ghosts. Am J Pathol. 2004;165:35–52. doi: 10.1016/S0002-9440(10)63273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrara N. VEGF as a therapeutic target in cancer. Oncology. 2005;69:11–6. doi: 10.1159/000088479. [DOI] [PubMed] [Google Scholar]
- 41.Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer res. 2004;64:3365–70. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 42.Juan HF, Wang I, Huang TC, Li JJ, Chen ST, Huang HC. Proteomics analysis of a novel compound: Cyclic RGD in breast carcinoma cell line MCF-7. Proteomics. 2006;6:2991–3000. doi: 10.1002/pmic.200500435. [DOI] [PubMed] [Google Scholar]
- 43.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58:467–86. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145:182–95. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]


