Abstract
Post-translational protein modification is an important strategy for the regulation of the cell proteome independent of the need for new gene expression. Ubiquitin and ubiquitin-like modifiers mediate the regulation of protein levels, signaling pathways, vesicular trafficking, and many other cellular processes through their covalent conjugation to proteins. Interferon stimulated gene 15 (ISG15) is a ubiquitin-like modifier induced by type I interferon. In addition to conjugating to potentially hundreds of target proteins, ISG15 can be found in an unconjugated form both inside of the cell and released from interferon stimulated cells into the extracellular environment. Due to its robust expression after type I interferon stimulation and the broad panel of proteins that it targets, ISG15 has drawn much attention as a potential regulator of the immune response and has been shown to mediate protection in a number of different viral infection models. Here we will review the current state of the field of ISG15, the viruses against which ISG15 mediates protection, and the mechanisms by which ISG15 exerts antiviral activity.
Abbreviations: ISG15, interferon stimulated gene 15; siRNA, short interfering RNA; VSV, vesicular stomatitis virus; SeV, Sendai virus; NDV, Newcastle disease virus; ASLV, avian sarcoma leukosis virus; HPV, human papilloma virus; HIV-1, human immunodeficiency virus 1; VLP, virus-like particle; WNV, West Nile virus; WT, wild type; CHIKV, Chikungunya virus; HSV-1, herpes simplex virus 1; LCMV, lymphocytic choriomeningitis virus; ESCRT, endosomal sorting complexes required for transport; PKR, protein kinase R; IRF3, interferon regulatory factor 3; RIG-I, retinoic acid inducible gene 1; JNK, Jun N-terminal kinase; NK, natural killer; MEF, mouse embryonic fibroblast
Keywords: ISG15, interferons, viruses, innate immunity, ubiquitin-like protein
Graphical Abstract
Highlights
-
•
ISG15 is an interferon-induced ubiquitin-like modifier that plays an important role during host responses to viral infections.
-
•
ISG15 mediates these functions in a conjugation-dependent manner by targeting both host and viral proteins.
-
•
Unconjugated ISG15 can also regulate the host response to viral infection through distinct mechanisms of action.
Introduction
The study of ubiquitin conjugation has played a key role in furthering our understanding of the mechanisms by which proteins and biological pathways can be regulated at a post-translational level. Ubiquitin is an 8.5-kDa protein originally named for the nearly universal cross-immunoreactivity between the ubiquitin homologs throughout eukaryotic species. Ubiquitin covalently conjugates to target proteins forming an isopeptide bond between its C-terminal glycine and the amine groups of lysine residues in target proteins. Ubiquitination occurs through an ATP-dependent enzymatic pathway consisting of an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase [1]. One of the first functions ascribed to ubiquitin conjugation was the targeting of ubiquitinated proteins for proteasomal degradation [2]. However, ubiquitin has since been shown to affect many cellular processes independent of proteasomal degradation, including the regulation of vesicular trafficking, signaling pathways, and DNA damage repair [3].
Following the discovery of ubiquitin, numerous other proteins that also covalently conjugate to target proteins through an enzymatic pathway similar to that of ubiquitin conjugation were identified [4]. These ubiquitin-like modifiers share limited sequence homology to ubiquitin but contain one or more domains that adopt a β-grasp structure similar to that of ubiquitin. The first ubiquitin-like protein to be discovered was interferon stimulated gene 15 (ISG15). ISG15 was discovered independently by two different laboratories. Korant et al. identified ISG15 during a study characterizing proteins induced by type I interferons (IFN) [5]. Protein lysates from Daudi cells stimulated with IFN were analyzed by two-dimensional gel electrophoresis at different times after stimulation. ISG15 was found to be rapidly and robustly induced after IFN stimulation. ISG15 was also identified by a group evaluating how the pool of ubiquitin conjugates is affected by viral infection [6]. This study revealed that infecting A549 cells with encephalomyocarditis virus induced a 15-kDa protein that cross-reacted with an antibody raised against ubiquitin. Consequently, ISG15 was originally named ubiquitin cross-reactive protein by this group. Subsequent cloning and characterization of ISG15 revealed that it contained two ubiquitin-like domains, each with approximately 30% amino acid sequence homology to ubiquitin [6], [7], [8]. The structural similarity of ubiquitin and the ubiquitin-like domains of ISG15 was later confirmed when the crystal structure of ISG15 was solved [9]. Given the known regulatory potential of ubiquitin-like modifiers and the importance of type I IFN during viral infection, there has been great interest in understanding how ISG15 might regulate the immune response and virus replication.
ISG15 conjugation
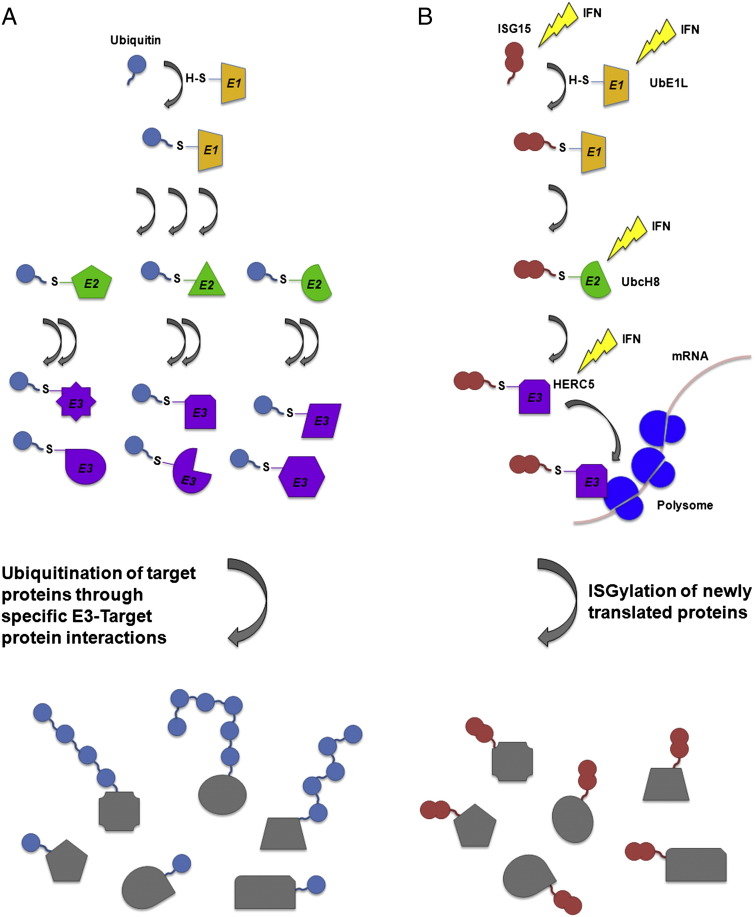
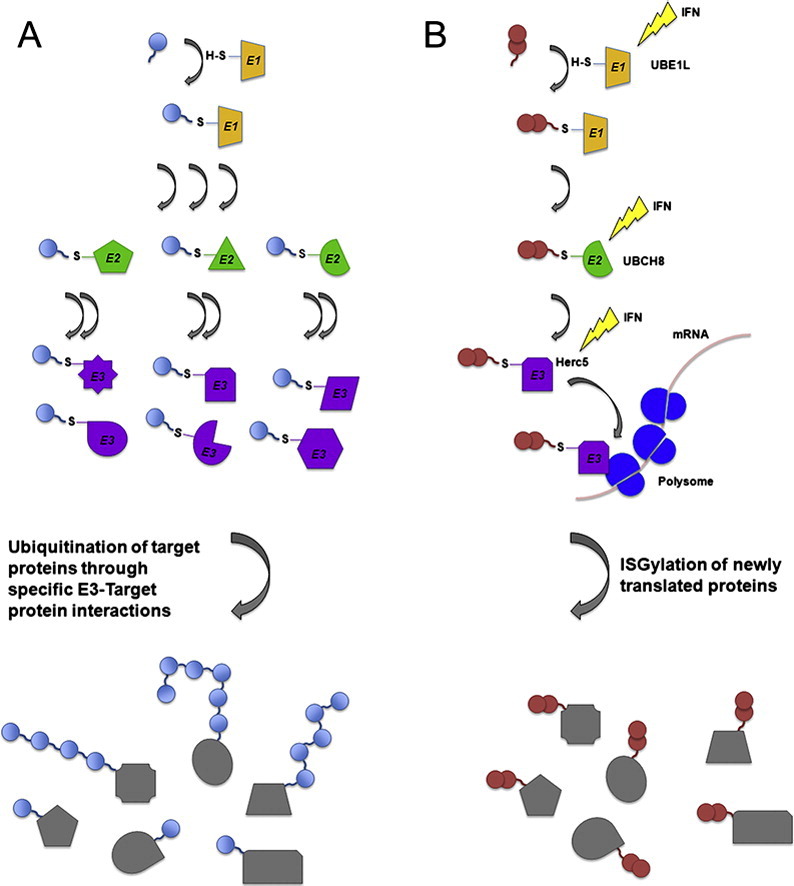
ISG15 forms conjugates with a diverse pool of target proteins in a process referred to as ISGylation. ISGylation occurs through a series enzymatic reactions similar to the ubiquitin conjugation pathway (Fig. 1 ). ISG15 is encoded and expressed as a 17-kDa precursor protein that is immediately proteolytically processed at its C-terminus to expose a C-terminal LRLRGG amino acid sequence, identical with the C-terminal sequence of ubiquitin [10]. The cognate ISG15 E1 activating enzyme, UbE1L, initiates the ISG15 conjugation enzymatic pathway by forming a thioester bond between the C-terminal glycine of ISG15 and a cysteine residue of UbE1L [11], [12]. UbE1L is the only known E1 enzyme for ISG15, and it appears to be specific for ISG15 as it has not formed thioester bonds with other tested ubiquitin-like modifiers [11], [13]. Confirmation of the importance of UbE1L in this pathway has come from the analysis of mice deficient in UbE1L. These mice express unconjugated ISG15 but do not form ISG15 conjugates [14]. Once primed by UbE1L, ISG15 is then transferred to a cysteine residue of an E2 conjugating enzyme. Short interfering RNA (siRNA) knockdown of UbcH8 in human cells, and UbcM8 in mouse cells, results in nearly complete loss of ISG15 conjugate formation, suggesting that UbcH8 and UbcM8 serve as the predominant E2 enzymes in the ISG15 conjugation system [15], [16]. Additional experiments evaluating whether other proteins may serve as ISG15 E2 enzymes have found the transfer of ISG15 to UbcH5 and to UbcH7, the closest homologs of UbcH8, to be highly unfavorable [12], [17]. Finally, an E3 ligase facilitates the conjugation of ISG15 to target proteins. HERC5, HHARI, and Efp (TRIM25) have all been demonstrated capable of acting as E3 ligases for ISG15 [18], [19], [20], [21]. However, while HHARI and Efp have been shown to act as E3 ligases for specific target proteins, siRNA knockdown of the HECT E3 ligase HERC5 was shown to abrogate nearly all ISG15 conjugate formation, suggesting that it is the predominant E3 ligase for ISG15. Knockdown studies in mouse cells have similarly demonstrated that HERC6 is the predominant ISG15 E3 ligase in mice [22], [23], [24].
Fig. 1.
A comparison between the ubiquitin and ISG15 conjugation pathways.
Both ubiquitin and ISG15 are conjugated to target proteins by utilizing enzymatic cascades composed of E1, E2, and E3 enzymes. (a) In the case of ubiquitin, it achieves specificity in target protein modification through the use of multiple E2 and E3 enzymes, which can orchestrate a broad range of specific ubiquitin modifications. Ubiquitin can also conjugate to itself on multiple lysine residues to form polyubiquitin chains on target proteins that result in different downstream functional consequences for the targeted protein. (b) The ISG15 conjugation cascade is an IFN-induced cascade in which the vast majority of ISG15 conjugation utilizes UbE1L, UbcH8, and HERC5. Interaction between HERC5 and polysomes leads to the preferential ISGylation of newly translated proteins. Unlike ubiquitin, there is no evidence that ISG15 forms poly-ISG15 chains or targets proteins for degradation.
In an elegant study of ISG15 conjugation specificity, it was shown that almost any protein inserted into an expression vector can be ISGylated in a manner that appears to require little sequence specificity [25]. Truncation of target proteins leads to alteration in the ISGylation profile of the protein, further suggesting that the specificity of ISG15 conjugation is not strictly determined by the local amino acid sequence environment of its lysine residues. This study showed that ISGylation targets newly translated proteins, and this appears to be mediated in part by the interaction of HERC5 with polysomes. Thus, the specificity of ISG15 is in part derived from the temporal expression of target proteins. How, or if, ISG15 might discriminate between proteins within the pool of actively translated proteins is unclear.
Enzymes capable of deconjugating ISG15 have also been identified. UBP43, also known as USP18, has been shown to deconjugate ISG15 from ISGylated target proteins [26]. The biological function of UBP43 and the effect of increasing ISG15 conjugation were initially evaluated both in vitro and in UBP43-deficient mice [26], [27], [28]. However, the interpretations of these studies have been complicated by the fact that UBP43 has been found to bind to the interferon α/β receptor (IFNAR) 2 and inhibit IFN signaling independent of its deconjugating activity [29], [30]. USP2, USP5, USP13, and USP14 have also been shown to cleave peptide fused to the C-terminus of ISG15; however, further studies are needed to determine whether they can deconjugate ISG15 from ISGylated proteins and to understand what biological roles they may play in the regulation of ISGylation [31]. Characterization of these ISG15 deconjugases could advance our understanding of ISGylation, as deubiquitinating enzymes have been found to play an important role in ubiquitin-mediated regulation [32].
Proteomic studies have identified hundreds of proteins that either are ISGylated or interact with ISG15 after IFN stimulation [33], [34]. These potential target proteins are involved in all aspects of cellular biology. The functional consequence of ISGylation is still not well understood. Proteasome inhibition has been reported to increase the pool of ISG15 conjugates but does so by means of de novo conjugate formation, not inhibition of conjugate degradation [35]. In fact, there is no evidence that ISGylation results in the proteasome-mediated degradation of target proteins. One of the challenges of studying ISG15 has been that, for every target protein studied, only a small fraction of the total pool of target protein is actually modified by ISG15. This phenomenon is also observed with the small ubiquitin-like modifier (SUMO). SUMO modification is thought to be highly dynamic, but transient modification of transcription factors can lead to long-lasting downstream consequences through chromatin remodeling or recruitment of inhibitory complexes [36]. Whether a similar dynamic effect might mediate the functionality of ISGylation has yet to be determined.
Though the biochemical conjugation of ISG15 parallels that of ubiquitination, there are several notable differences between the conjugation systems (Fig. 1). Firstly, like ISG15 itself, the predominant ISG15 conjugation and deconjugation enzymes UbE1L, UbcH8/UbcM8, HERC5/HERC6, and UBP43/USP18 are all induced by type I IFN stimulation. Secondly, the ubiquitin conjugation system is constituted by many E2 and E3 proteins [1]. It is the binding specificity of E2 to E3 and E3 to target proteins that allows for ubiquitin to affect such a vast number of biological processes with such specificity. In contrast, it seems that formation of the overwhelming majority of ISG15 conjugates occurs through one E2 enzyme and one E3 enzyme. Finally, there is no evidence that ISG15 conjugates to itself and forms ISG15 chains in the manner of polyubiquitin modification. While not fully understood, these differences between the ISG15 and ubiquitin pathways most likely reflect the specific function of ISG15 conjugation.
ISG15 has been implicated in regulating numerous biochemical pathways, and the generation and study of ISG15-deficient mice have revealed that it plays an important role during viral infection. Data collected to date have demonstrated that ISG15 can protect against a wide gamut of viruses, and the mechanisms by which it mediates this protection are varied. Here we will review the progress that has been made in the identification of viruses that are regulated by ISG15 and the mechanisms by which ISG15 mediates this protection.
Antiviral Activity of ISG15
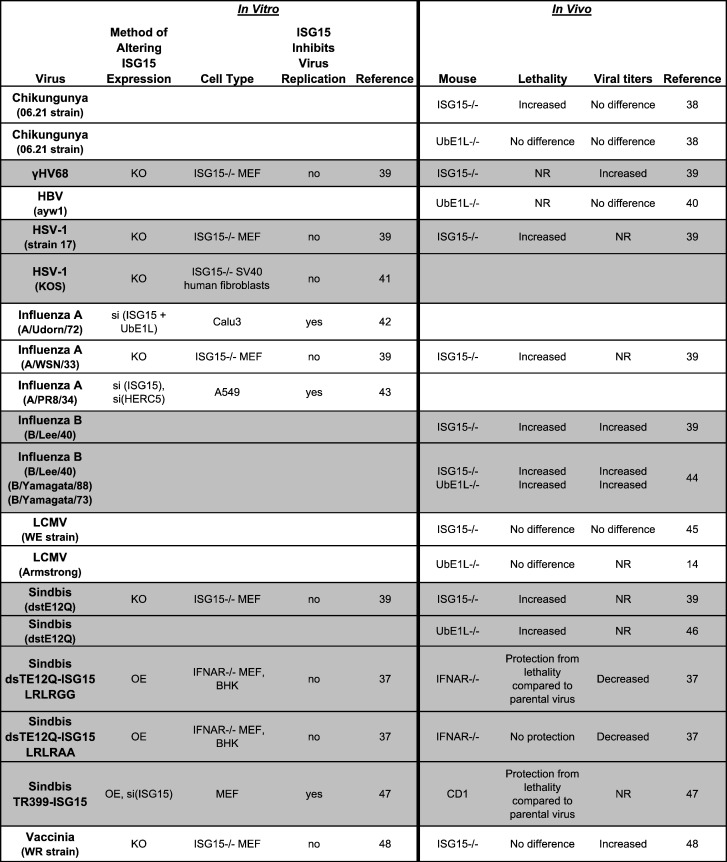
The antiviral activity of ISG15 was first observed in a screen designed to identify antiviral interferon stimulated genes (ISG) important during Sindbis virus infection [37]. Recombinant Sindbis viruses were engineered to express ISGs, and these ISGs were then evaluated for their ability to protect IFNAR −/− mice from lethality after infection. Mice infected with a Sindbis virus expressing ISG15 but not a control virus were protected from virus-induced lethality. This discovery was followed by numerous reports of ISG15 antiviral activity observed in tissue culture under conditions of ISG15 overexpression and siRNA knockdown (Table 1 ). Influenza virus, vaccinia virus, vesicular stomatitis virus (VSV), Sendai virus (SeV), Japanese encephalitis virus, Newcastle disease virus (NDV), avian sarcoma leukosis virus (ASLV), human papilloma virus (HPV), human immunodeficiency virus 1 (HIV-1), Ebola virus-like particles (VLPs), dengue virus, and West Nile virus (WNV) have all been reported to be modestly inhibited by ISG15 in vitro. For several viruses, contradictory results have been reported with regard to whether ISG15 antagonizes virus replication. These discrepancies are likely a result of different experimental conditions. In the case of influenza A virus, it has been reported that inhibition of replication by ISG15 is species and/or cell type specific [42].
Table 1.
Studies evaluating the impact of ISG15 on viral infection in human and mouse systems
γHV68, gamma herpes virus 68; HBV, hepatitis B virus; HCV, hepatitis C virus; JEV, Japanese encephalitis virus; OE, overexpression; si, siRNA knockdown; KO, genetic knockout; RP, ectopic treatment with recombinant protein; NR, not reported; MLF, mouse lung fibroblast; BHK, baby hamster kidney. Studies not discussed within text [40], [47], [53], [55], [56], [59].
aVirus packaging system or Gag VLP assay.
bProvirus DNA transfection.
Importantly, the generation of ISG15-deficient mice has allowed for the verification of the antiviral activity of ISG15 in vivo [45]. As predicted, ISG15 −/− mice infected with Sindbis virus were found to be more susceptible to lethality than wild-type (WT) mice [39]. The increased lethality observed in the ISG15 −/− mice could be rescued when the mice were infected with a recombinant virus expressing WT ISG15, but not when infected with a recombinant virus expressing an ISG15(LRLRAA) mutant incapable of forming conjugates [39]. This suggested that, in this viral model, the antiviral activity of ISG15 was dependent upon its ability to ISGylate proteins. A subsequent study confirmed the conjugation dependence of this protection by showing that UbE1L −/− mice are also more susceptible than WT mice to Sindbis virus infection [46].
Increased mortality was also observed in ISG15 −/− mice after infection with influenza A and influenza B viruses [39], [44]. During influenza B virus infection, viral loads in WT mice remained very low over the course of infection and the mice experienced minimal weight loss or other clinical signs of illness. In contrast, ISG15 −/− mice experienced dramatic weight loss and viral loads reached levels up to 1000-fold higher than in WT mice at late time points. These observations were found to be true for both the mouse-adapted influenza B/Lee/40 strain and a non-mouse-adapted influenza B/Yamagata/88 strain, indicating that ISG15 may contribute to the species tropism barrier for influenza B virus infection. UbE1L −/− mice infected with influenza B virus similarly exhibited increased lethality and increased viral burden compared to WT mice, suggesting that ISG15-mediated protection from influenza B virus is also dependent on the ability of ISG15 to form conjugates [44]. Bone marrow chimera studies suggest that the expression of ISG15 in a radioresistant cell line is responsible for mediating this protection; however, it remains unclear as to how ISG15 conjugation is affecting influenza B virus replication [44].
These initial studies established the precedent of ISG15 antiviral activity in vivo. Subsequent studies have identified additional viruses that ISG15 mediates protection against in vivo (see Table 1). Increased susceptibility has been seen with Chikungunya virus (CHIKV), herpes simplex virus 1 (HSV-1), gamma herpes virus 68, and vaccinia virus infections. However, despite this diverse array of viruses that appear to be affected by ISG15, ISG15 −/− mice have not displayed increased susceptibility to all viruses tested. No phenotype was observed following infection of ISG15 −/− mice with lymphocytic choriomeningitis virus (LCMV) or VSV [45]. Our laboratory also has not observed increased susceptibility in ISG15 −/− mice after WNV infection (D.L., unpublished results).
After the discovery of ISG15, much attention was invested in studying ISG15 conjugation. The initial discoveries of conjugation-mediated protection against Sindbis virus and influenza B virus seemed to corroborate the importance of ISGylation. Recently, however, it has been reported that, while ISG15 −/− mice are more susceptible to CHIKV infection, UbE1L −/− mice display similar susceptibility to CHIKV as WT mice, suggesting a conjugation-independent role for ISG15 during virus infection [38]. This phenomenon has also been observed with Ross River virus infection (D.L., unpublished results). In vitro and in vivo studies have shown that the antiviral activity of ISG15 targets viruses from diverse families ranging from retroviruses (HIV-1, ASLV) to large DNA viruses (vaccinia virus, HSV-1) to both positive and negative sense RNA viruses (Sindbis virus and influenza A virus, respectively) [39], [48], [51], [57]. The mechanisms through which ISG15 mediates protection are likely to be just as diverse. We will cover below what is currently known about the mechanisms through which ISG15 exerts its protective effects.
Mechanisms of ISG15 Activity
Inhibition of virus release
Several studies have found that ISG15 can impact the viral life cycle at the stage of virus release. This has been seen with the retroviruses HIV-1 and ASLV, as well as with Ebola virus. Despite impacting the release of all of these viruses, detailed studies have identified different mechanisms through which ISG15 is acting. The first evidence of ISG15 inhibiting virus release was observed when characterizing the effect of ISG15 on HIV-1 replication. The replication of HIV-1 in 293T cells transfected with HIV-1 proviral DNA was found to be inhibited when cotransfected with a plasmid expressing ISG15 [57]. While ISG15 inhibited HIV-1 release into the supernatant, it did not affect HIV-1 protein production. Expression of ISG15 was shown to inhibit the ubiquitination the HIV-1 Gag protein and the interaction between Gag and Tsg101. Both of these events are important for mediating HIV-1 budding and release [60]. Interestingly, while cotransfection of UbE1L enhanced HIV-1 inhibition, the ectopic expression of ISG15 alone could inhibit the Gag–Tsg101 interaction, raising the possibility that the unconjugated form of ISG15 may be partly mediating this inhibition. A subsequent study has shown that HERC5-mediated ISG15 conjugation can inhibit HIV-1 budding from the plasma membrane [58]. Unlike the initial HIV-1 study, this group found that the HIV-1 Gag protein is ISGylated by HERC5. It was also shown that overexpression of ISG15 without HERC5 resulted in an accumulation of Gag in cytoplasmic clusters, whereas coexpression of ISG15 with HERC5 resulted in an accumulation of Gag at the plasma membrane. These results suggest that unconjugated ISG15 and ISGylation may act to block HIV-1 budding by two distinct mechanisms.
ISG15 was also shown to inhibit Ebola VLP budding by a similar mechanism. Expression of the Ebola matrix protein, VP40, alone is sufficient to produce budding and release of VP40 VLPs [61]. This release is regulated by the ubiquitin E3 ligase Nedd4 through a mechanism dependent on its ligase activity [62]. Two studies demonstrated that the coexpression of ISG15 with VP40 can inhibit VP40 VLP release by inhibiting Nedd4 ligase activity and, thus, inhibiting ubiquitination of VP40 [49], [54]. Like the HIV-1 studies, overexpression of ISG15 alone was able to disrupt Ebola VLP release, suggesting that unconjugated ISG15 may be capable of inhibiting Nedd4 ligase activity [49]. Malakhova et al. was able to show that purified ISG15 protein was able to disrupt the ability of Nedd4 to bind to ubiquitin E2 conjugating enzymes, thus preventing ubiquitin from being transferred to the catalytic cysteine residue of Nedd4 [54]. This study further showed that ISG15 could inhibit other Nedd4-like E3 ligases from augmenting VP40 VLP release. The ISG15 disruption of ubiquitin-mediated regulation during both Ebola virus and HIV-1 release raises an intriguing model for general ISG15 regulation through ubiquitin antagonism. It is possible that unconjugated ISG15 could compete with ubiquitin by binding to ubiquitin interacting domains in proteins or by forming thioester bonds with ubiquitin E2 or E3 enzymes. Additionally, ISG15 conjugation to Ubc13 on a lysine residue that is normally ubiquitinated has been reported to inhibit the E2 conjugating activity of Ubc13 [63], [64]. Thus, competing for target lysine residues is yet another mechanism by which ISG15 can antagonize ubiquitin.
Studies of the retrovirus ASLV revealed yet another mechanism by which ISG15 can inhibit virus budding [51], [52]. ASLV budding and release is dependent upon Gag binding to Nedd4 [60]. Despite the observed effects of ISG15 on Nedd4 in the Ebola studies, cotransfection of ISG15 with the ASLV Gag protein inhibited ASLV VLP release without disrupting the interaction between Gag and Nedd4. The disruption of Gag–Tsg101 interaction in the HIV-1 study suggested that ISG15 is disrupting an early stage in the endosomal sorting complexes required for transport (ESCRT) pathway. Chimeric HIV-1 or ASLV Gag proteins fused to downstream ESCRT proteins can bypass early stages of the ESCRT pathway to facilitate VLP release [60]. However, in a study by Pincetic et al., it was found that ASLV Gag-ESCRT chimeras were still sensitive to ISG15-mediated inhibition [51]. ISG15 was found to prevent VLP release by inhibiting the recruitment of the AAA-ATPase Vps4 to the ESCRT pathway. This inhibition of Vps4 recruitment was dependent on charged multivesicular body protein (CHMP) 5 and correlated with ISGylation of CHMP5. However, other CHMP proteins were also found to be ISGylated in this study, and the mechanism by which ISG15 mediates this modulation of the ESCRT pathway was not conclusively shown.
It is not clear whether the ISG15 manipulation of the ESCRT pathway represents a general strategy for the inhibition virus propagation. Notably, both LCMV and HSV-1 have been shown to utilize the ESCRT pathway for virus budding, yet ISG15 has not been found to affect LCMV pathogenesis in vivo or inhibit HSV-1 replication in vitro [39], [45], [65], [66], [67]. It is possible that these viruses are capable of antagonizing ISG15 and its effects on the ESCRT pathway. More studies will be needed to more precisely define the mechanisms by which ISG15 regulates viral budding. It will also be interesting to determine if ISG15 alteration of the ESCRT pathway represents specific strategy for targeting viruses or if this might be reflective of a broader role that ISG15 plays in vesicular trafficking after IFN stimulation.
ISGylation of viral proteins
The recombinant Sindbis virus studies not only demonstrated that ISG15 could act as an antiviral molecule but also showed that it could do so when expressed only from within the virus genome, exerting its activity from within a virally infected cell [37], [39]. Similarly, knockdown of ISG15 in human cells was shown to increase influenza A virus replication, further suggesting that ISG15 can directly antagonize virus replication and sparking subsequent studies exploring the possibility of ISG15 acting by directly conjugating to viral proteins [42]. Indeed, there is now evidence that, during some infections, viral proteins are targeted for ISGylation and this contributes to the type I IFN-mediated inhibition of virus replication.
The first viral protein shown to be modified by ISG15 was the NS1 protein of influenza A virus. Two studies showing that influenza A virus NS1 protein can be ISGylated by HERC5 have been published, and different functional consequences have been attributed to this modification [43], [68]. Both studies found that NS1 is ISGylated on more than one lysine residue. Zhao et al. found that the lysine residue at position K41 was the dominant site for ISGylation [68]. Modification at this site was shown to have no impact on the ability of NS1 to bind to double-stranded RNA but did inhibit its ability to interact with importin α, an interaction involved in the nuclear translocation of NS1. A recombinant influenza A virus containing the K41A mutation in its NS1 protein to reduce ISGylation of NS1 was less susceptible to inhibition of replication by IFN as compared to a WT virus. Tang et al. found that seven lysines in NS1 had to be mutated to generate a non-ISGylated NS1 [43]. This study found that ISGylated NS1 could not interact with the N-terminus of protein kinase R (PKR), the RNA binding domain of NS1, U6snRNA, or double-stranded RNA. On a functional level, ISGylation of NS1 inhibited the ability of NS1 to antagonize the induction of ISGs by SeV infection, suggesting that ISGylation of NS1 would render influenza A virus more susceptible to the antiviral response. Differences in the findings of these two studies could be due to the different strains of influenza A virus that were used in each study. However, both studies further support the hypothesis that, by modifying viral proteins, ISG15 can directly antagonize virus replication.
The HPV capsid protein has also been shown to be ISGylated by HERC5 [25]. Using an HPV pseudovirus packaging system, Durfee et al. demonstrated that the expression of the ISG15 conjugation system led to the ISGylation of the HPV capsid protein, which then incorporated into released virus. ISGylation of HPV capsid protein correlated with a decrease both in the amount of virus released and in the subsequent infectivity of the virus produced in the presence of ISG15. This observation led to a model in which ISGylated capsid protein is incorporated into virions but results in an alteration of the geometry of the viral capsid. It is hypothesized that this change in the capsid structure then inhibits the infectivity of the released virus. As mentioned previously, Woods et al. also found that HIV-1 Gag protein can also be ISGylated by HERC5 [58]. It is important to note that direct causality of virus replication inhibition through viral protein ISGylation, as opposed to ISGylation of host proteins, was not verified with non-ISGylatable HPV capsid or HIV-1 Gag mutants. Nonetheless, these observations help to further an attractive model for ISG15 function that could help to explain how ISGylation of a small fraction of the total pool of a protein could lead to a large downstream effect on virus fitness. Homooligomerization of viral proteins is a universal necessity in viral genomes to facilitate virion formation and genome packaging. Future studies will be needed to determine if this model can be applied to other viral proteins such as matrix proteins or ribonucleoproteins.
Modification of host proteins
Since ISGylation has been shown to target newly translated proteins, ISGs would be predicted to be a population of proteins rife with targets for ISG15 conjugation following IFN stimulation. In one of the first high-throughput studies identifying ISGylated proteins, both Jak1 and STAT1 were shown to be ISGylated in human thymus tissue [69]. In subsequent proteomic studies, many ISGs have been identified as ISG15 targets [33], [34]. Among these, three antiviral effectors, interferon regulatory factor 3 (IRF3), retinoic acid inducible gene 1 (RIG-I), and PKR, have been analyzed in significant detail.
ISGylation of IRF3 has been reported to stabilize activated IRF3, thus positively regulating the type I IFN response [50], [70]. Shi et al. showed that, after SeV infection, HERC5 can bind to and ISGylate IRF3 on multiple lysine residues. ISGylation of IRF3 inhibits the interaction of IRF3 with PIN1 thus inhibiting the ubiquitination and degradation of IRF3, resulting in a more robust IFN response [50]. This study went on to show that siRNA knockdown of HERC5 or ISG15 resulted in increased replication of VSV, SeV, and NDV. However, it is unclear from these studies whether inhibition of virus replication was dependent on ISGylation of IRF3 or if ISGylation of other target proteins mediated these antiviral effects.
RIG-I was found to be ISGylated after transfection of COS-7 cells with ISG15, UbE1L, and UbcH8 expression plasmids [71]. Overexpression of the ISG15 conjugation system also resulted in decreased levels of non-ISGylated RIG-I, though this decrease was not dependent on proteasome activity. This reduction of RIG-I levels was not seen in UbE1L −/− mouse embryonic fibroblasts (MEFs) further implicating the importance of ISG15 conjugation to mediate this phenomenon. The reduction of RIG-I protein levels correlated with a reduction in the intensity of the IFN response triggered by NDV [71]. In this study, the residues modified by ISG15 were not identified and mutated; thus, it could not be ruled out that the decrease in RIG-I expression was an indirect result of the ISGylation of another protein. Whether directly a result of RIG-I ISGylation or not, this study suggests that ISG15 might not always engender a more intense IFN response but, rather, it may act to dampen the IFN response down to an appropriate level in certain scenarios.
PKR was also identified in proteomic analysis as being a target of ISGylation [33], [34]. Okumura et al. recently verified that PKR is ISGylated after IFN or LPS stimulation [72]. Mutational analysis revealed that PKR is ISGylated at K69 and K159. In lung fibroblasts, PKR was found to be activated even in the absence of viral RNA; however, this activation was dependent on ISG15. A non-ISGylatable K69R, K159R PKR mutant did not exhibit constitutive activation demonstrating that this RNA-independent activation of PKR was due to ISGylation of PKR. This increased activation of PKR was also observed when ISG15 was fused to the N-terminus of PKR resulting in increased phosphorylation of eIF2α and decreased protein synthesis. These findings suggest that ISG15 may have a broader impact on protein translation prior to viral infection.
While the ISG15-mediated regulation of antiviral effectors has received much attention, it is also important to understand how ISGylation of other “non-antiviral effector” proteins might affect virus replication and pathogenesis. For example, as previously discussed, ISGylation of CHMP5 has been correlated with the ISG15-dependent inhibition of HIV-1 budding [52]. ISG15 has also been implicated in the regulation of IFN-induced apoptosis through modification of filamin B [73]. In addition to cross-linking actin filaments, filamin B also serves as a scaffold protein for the Jun N-terminal kinase (JNK) signaling pathway. Jeon et al. has shown that ISGylation of filamin B disrupts its ability to bind to RAC-1, MEKK1, and MEKK4, thereby inhibiting the signaling cascade leading to JNK-mediated apoptosis [73]. In this study, coexpression of UbcH8 and filamin B resulted in co-localization of the two proteins in actin-rich membrane ruffles. Thus, while only a small fraction of total filamin B is ISGylated, the fraction of ISGylated filamin B within the local microenvironment of a membrane ruffle might be quite high. This might explain how ISGylation of a seemingly small fraction of filamin B can have a large impact on JNK signaling.
It is important to note that, despite the number of studies evaluating the regulation of ISGs by ISG15, no studies in ISG15 −/− mice or ISG15 −/− cells have demonstrated ISG15 conjugation as having a role in the regulation of the type I IFN response. Analysis of cells from ISG15 −/− and UbE1L −/− mice did not reveal any misregulation of STAT1 activation or ISG induction after stimulation with IFNβ, LPS, or combinations of both stimuli [14], [45]. PolyI:C injections of mice had no effect on cell proliferation, and UbE1L −/− cells were found to undergo similar levels of apoptosis after IFN stimulation. There could be several explanations for these discrepancies including (1) the differences in utilization of overexpression, knockdown, and knockout systems; (2) differences between the function of ISGylation in humans and mice; or (3) differences in experimental conditions and types of stimulations. Additional studies will be required to better understand these discrepancies.
Unconjugated ISG15
In addition to existing in its conjugated form, ISG15 is present in an unconjugated form both intracellularly and released into the extracellular space. Recent evidence in both murine and human models has indicated that unconjugated ISG15 may also play an important role in the host response to infections.
A recent study by Werneke et al. was the first in vivo study to suggest that unconjugated ISG15 has a biologically relevant role during infection [38]. In this study, ISG15 −/− neonatal mice were found to be highly susceptible to infection with the re-emerging viral pathogen, CHIKV. However, UbE1L −/− mice did not display increase lethality, indicating that the expression of unconjugated ISG15 was sufficient to protect these mice from CHIKV-induced lethality. The protective mechanism of ISG15 in this model was distinct from previous models that have been studied in that ISG15 did not appear to function as an antiviral molecule. Viral loads in all organs examined were the same in WT, UbE1L −/−, and ISG15 −/− mice. Instead, the ISG15 −/− mice had significantly higher levels of multiple cytokines in their serum, and death of the animals occurred in a manner consistent with a cytokine storm. Together, these data suggested that unconjugated ISG15 protected these neonatal mice from CHIKV-induced lethality through its ability to regulate the production of proinflammatory cytokines and chemokines. The mechanism through which ISG15 regulates these responses and the cell types responsible for the increased cytokine production in this model are not known. A recombinant CHIKV engineered to express ISG15 was no less virulent than the WT virus in ISG15 −/− mice. This suggests that unconjugated ISG15 may need to be expressed by non-infected cells or expressed before virus infection in order to mediate protection. It is also not certain whether intracellular unconjugated ISG15 or extracellular unconjugated ISG15 is responsible for the regulation of cytokines in this model. Future studies will be needed to address these important questions.
ISG15 has long been implicated as having a role as a cytokine-like molecule. Though ISG15 has no canonical signal peptide for release, T-cells, B-cells, monocytes, and epithelial cells have all been shown to release ISG15 after IFN stimulation in vitro [74]. In studying the potential role for this released ISG15, it was found that treatment of peripheral blood lymphocytes with recombinant ISG15 resulted in specific proliferation of natural killer (NK) cells [75]. This expansion of NK cells was found to be due to the induction of IFNγ by T-cells. Recently, a study by Bogunovic et al. confirmed a role for unconjugated ISG15 in the stimulation of IFNγ production and correlated this function with increased disease susceptibility in humans [41]. In this study, ISG15 deficiency was identified as a potential predisposition for Mendelian susceptibility to mycobacterial disease. These authors found that recombinant ISG15 induced IFNγ production by both NK cells and other lymphocytes. They went on to show that the levels of IFNγ produced after stimulation with recombinant ISG15 were greatly enhanced by co-stimulation of cells with ISG15 and IL-12. Whole blood cells from ISG15-deficient individuals produced decreased levels of IFNγ compared to cells from ISG15-sufficient individuals after stimulation with either mycobacteria alone or in combination with IL-12. Treatment of ISG15-deficient cells with recombinant ISG15 in addition to mycobacteria and IL-12 enhanced their ability to produce IFNγ to levels near ISG15-sufficient cells. These data indicate that released ISG15 may play an important role in IFNγ production during mycobacterial infection.
Together, these studies suggest that ISG15 contributes to the host response to infection not only through its modification of target proteins but also through the actions of unconjugated ISG15, perhaps functioning as a cytokine. Many questions still remain, including the identity of a receptor that can mediate these biological properties. Given the therapeutic potential for administration or inhibition of cytokines during viral infection, it is of great interest to further define the role of ISG15 in cytokine regulation.
Viral countermeasures
Viral antagonism of antiviral effectors is a phenomenon so frequent and inevitable that the study of mutant viruses is routinely a means by which novel host biology is elucidated. Discoveries of viral antagonism of ISG15 have further highlighted the importance of ISG15 as a part of the host antiviral response. Before the antiviral activity of ISG15 had been demonstrated, a role for ISG15 during influenza B virus infection was eluded to by the discovery of the interaction between ISG15 and the influenza B virus NS1 protein (B/NS1) [11]. Influenza B/NS1, but not influenza A/NS1, was found to bind ISG15 and inhibit it from interacting with UbE1L, thereby preventing the formation of ISG15 conjugates. A detailed review covering the interaction between B/NS1 and ISG15 has recently been published [76]. Since this discovery of B/NS1 interacting with ISG15, several other viruses that encode proteins that can interact with and potentially antagonize ISG15 have also been identified.
The vaccinia E3L protein initially masked the sensitivity of vaccinia virus to ISG15 [48]. A WT vaccinia strain replicated to similar levels in ISG15 −/− and WT MEFs after 48 h of infection, and ISG15 −/− mice did not exhibit increased lethality compared to WT mice. However, after infection with WT vaccinia virus, it was noted that ISG15 was induced but did not form conjugates. Infection of cells with a ΔE3L mutant vaccinia virus did result in ISG15 conjugate formation. The E3L protein was shown to bind to ISG15, suggesting that the E3L protein directly antagonizes ISG15 conjugation, though the exact mechanism by which this occurs is not known. ISG15 −/− mice exhibited increased lethality compared to WT mice when infected with this ΔE3L mutant vaccinia virus, and the mutant virus exhibited a ~ 25-fold increase in virus replication in ISG15 −/− MEFs compared to WT MEFs. Therefore, similar to influenza B virus, vaccinia virus appears to have evolved a mechanism to disrupt ISG15 conjugation and its antiviral activity.
Multiple viruses have evolved deubiquitinating enzymes capable of antagonizing host cell biology. Several of these viral proteins are capable of deconjugating ISG15 from ISGylated target proteins. Crimean-Congo hemorrhagic fever virus and equine arteritis virus encode L proteins containing ovarian tumor (OTU) domains. In host proteins, OTU domains can contain ubiquitin deconjugation activity [32]. When transfected into cells, these viral L proteins were able to reduce the total pool of both ubiquitin and ISG15 conjugates [77]. Expression of the Crimean-Congo hemorrhagic fever virus L protein OTU domain by recombinant Sindbis virus in parallel with expression of ISG15 abolished the protection provided by ISG15 during infection of IFNAR −/− mice [77]. Similarly, SARS Coronavirus encodes a papain-like protease that has been shown to cleave K48-linked ubiquitin chains, as well as ISG15 fusion proteins [78], [79], [80]. While these examples highlight another potential mechanism of circumventing ISG15, direct evidence for ISG15 antagonism by these proteins remains to be demonstrated during viral infection.
The convergent evolution of viral proteins antagonizing the ISG15 conjugation system demonstrates the broad antiviral activity of ISG15. It will be interesting to investigate whether the lack of a role for ISG15 during infections with certain viruses, such as VSV and LCMV, might also be due to viral antagonism of ISG15 that has yet to be discovered.
Concluding Remarks
ISG15 is a type I IFN stimulated effector protein that has been shown to play an important role during infection with a broad range of viruses. Studies of ISG15-deficient mice have clearly established that ISG15 mediates protection from virus-induced lethality; however, the molecular mechanisms mediating this protection are still not well understood. Protection has been shown to occur through a variety of mechanisms involving both its conjugation to host and viral proteins and the function of unconjugated ISG15 in the regulation of the immune response. We must note that, while this review has focused on the role of ISG15 in humans and mice, many other vertebrates including fish, sheep, and cows have been shown to encode ISG15 homologs [8]. The fish ISG15 homolog has been shown to inhibit virus replication in vitro, and a secreted form of fish ISG15 has been shown to have cytokine-like properties, thus highlighting the conserved antiviral functions of ISG15 homologs between species [81], [82].
The ISG15 conjugation system has been intimately tied to protein translation, preferentially targeting newly translated proteins for ISG15 modification [25]. This cotranslational model for ISG15 conjugation supports the hypothesis that ISG15 modification of viral proteins and ISGs will play an important role during infection. To date, influenza A virus NS1, influenza A virus M1, HPV capsid, and HIV-1 Gag have all been shown to be ISGylated [25], [43], [58], [68]. Likewise, multiple studies have demonstrated a role for ISGylation of key antiviral ISGs, suggesting that ISG15 conjugation is important for proper regulation of the host antiviral response. Understanding this relationship between ISGylation and protein translation may play a key role in future discoveries of ISG15 functions. However, this observation also raises an important concern about the context within which ISG15 is studied. When possible, functional characterizations of ISG15 conjugation should be verified under conditions of endogenous protein expression. If new protein translation of target proteins truly is essential for ISGylation, phenotypes generated in overexpression systems with ectopic expression of target proteins may be real but also biologically artifactual.
While the ISGylation of viral proteins has been reported and, in at least some systems, may contribute to the antiviral activity of ISG15, virus replication has not been altered in several models where ISG15 is known to play a role during virus infection in vivo. For example, ISG15 −/− mice display increased lethality following Sindbis virus infection; however, no defect in virus replication was observed when WT and ISG15 −/− MEFs were infected with Sindbis virus [39]. Similarly, ISG15 −/− mice are more susceptible to HSV-1 infection; however, no difference in HSV-1 replication has been observed in ISG15 −/− MEFs or in fibroblasts from human patients deficient for ISG15 [39], [41]. Discrepancies between in vivo protection and cell culture phenotypes might be a result of the particular cell types chosen for study. However, it is also possible that ISG15-mediated protection from lethality might not be a result of direct antagonism of virus replication, as has been demonstrated for CHIKV infection [38]. To date, very little has been reported on the role of ISG15 conjugation in the regulation of the immune response. The initial characterization of ISG15 −/− mice revealed no difference in the virus-specific T-cell response after infection of WT and ISG15 −/− mice with LCMV [45]. Importantly, however, no lethality phenotype was noted between WT and ISG15 −/− mice after LCMV infection. Given the discrepancies between certain in vivo survival phenotypes and cell culture replication phenotypes, it may be informative to more thoroughly examine the role of ISG15 conjugation in the immune response in the context of an infection in which ISG15 −/− mice display increased susceptibility.
Finally, recent findings support the need for further investigation into the function of the unconjugated form of ISG15. Released ISG15 has been shown to modulate IFNγ production by multiple cell types and has been suggested to play a role in defense against mycobacteria [41], [75]. Unconjugated ISG15 also appears to regulate cytokine production after certain viral infections, though it is unclear if this is through the extracellular or intracellular form of ISG15 [38]. In addition to providing evidence that ISG15 may function to regulate the immune response, both of these activities suggest the presence of ISG15 specific binding partners. In the case of extracellular ISG15, the identification of a putative cellular receptor will be critical in better understanding the biological significance of extracellular ISG15. Intracellular ISG15 may also interact with other unidentified intracellular proteins independent of conjugation. Recent work in the ubiquitin field has demonstrated that unanchored ubiquitin chains can bind to innate signaling molecules such as RIG-I and members of the NF-κB signaling cascade to influence signaling through these important pathways [83], [84]. This raises the possibility that other ubiquitin-like proteins could function in a similar manner. The body of research on non-covalent binding partners of ISG15 is quite small compared to the conjugation-dependent interactions, but these interactions may influence important biological processes. Uncovering potential binding partners for ISG15 will be instrumental in understanding its molecular mechanism of action.
Edited by E. Freed and M. Gale
References
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Tracing the history of the ubiquitin proteolytic system: the pioneering article. Biochem Biophys Res Commun. 2009;387:1–10. doi: 10.1016/j.bbrc.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 3.Komander D., Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 5.Korant B.D., Blomstrom D.C., Jonak G.J., Knight E. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J Biol Chem. 1984;259:14835–14839. [PubMed] [Google Scholar]
- 6.Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 7.Blomstrom D.C., Fahey D., Kutny R., Korant B.D., Knight E. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem. 1986;261:8811–8816. [PubMed] [Google Scholar]
- 8.Dao C.T., Zhang D.E. ISG15: a ubiquitin-like enigma. Front Biosci. 2005;10:2701–2722. doi: 10.2741/1730. [DOI] [PubMed] [Google Scholar]
- 9.Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 10.Potter J.L., Narasimhan J., Mende-Mueller L., Haas A.L. Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J Biol Chem. 1999;274:25061–25068. doi: 10.1074/jbc.274.35.25061. [DOI] [PubMed] [Google Scholar]
- 11.Yuan W., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krug R.M., Zhao C., Beaudenon S. Properties of the ISG15 E1 enzyme UbE1L. Methods Enzymol. 2005;398:32–40. doi: 10.1016/S0076-6879(05)98004-X. [DOI] [PubMed] [Google Scholar]
- 13.Chiu Y.H., Sun Q., Chen Z.J. E1-L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Kim K.I., Yan M., Malakhova O., Luo J.K., Shen M.F., Zou W. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol. 2006;26:472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C., Beaudenon S.L., Kelley M.L., Waddell M.B., Yuan W., Schulman B.A. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K.I., Giannakopoulos N.V., Virgin H.W., Zhang D.E. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durfee L.A., Kelley M.L., Huibregtse J.M. The basis for selective E1-E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou W., Zhang D.E. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 19.Dastur A., Beaudenon S., Kelley M., Krug R.M., Huibregtse J.M. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 20.Wong J.J., Pung Y.F., Sze N.S., Chin K.C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okumura F., Zou W., Zhang D.E. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21:255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ketscher L., Basters A., Prinz M., Knobeloch K.P. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem Biophys Res Commun. 2012;417:135–140. doi: 10.1016/j.bbrc.2011.11.071. [DOI] [PubMed] [Google Scholar]
- 23.Oudshoorn D., van Boheemen S., Sanchez-Aparicio M.T., Rajsbaum R., Garcia-Sastre A., Versteeg G.A. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One. 2012;7:e29870. doi: 10.1371/journal.pone.0029870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Versteeg G.A., Hale B.G., van Boheemen S., Wolff T., Lenschow D.J., Garcia-Sastre A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J Virol. 2010;84:5423–5430. doi: 10.1128/JVI.02395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durfee L.A., Lyon N., Seo K., Huibregtse J.M. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malakhov M.P., Malakhova O.A., Kim K.I., Ritchie K.J., Zhang D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie K.J., Hahn C.S., Kim K.I., Yan M., Rosario D., Li L. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 28.Malakhova O.A., Yan M., Malakhov M.P., Yuan Y., Ritchie K.J., Kim K.I. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knobeloch K.P., Utermohlen O., Kisser A., Prinz M., Horak I. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol Cell Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catic A., Fiebiger E., Korbel G.A., Blom D., Galardy P.J., Ploegh H.L. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannakopoulos N.V., Luo J.K., Papov V., Zou W., Lenschow D.J., Jacobs B.S. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 34.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M., Li X.L., Hassel B.A. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J Biol Chem. 2003;278:1594–1602. doi: 10.1074/jbc.M208123200. [DOI] [PubMed] [Google Scholar]
- 36.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 37.Lenschow D.J., Giannakopoulos N.V., Gunn L.J., Johnston C., O'Guin A.K., Schmidt R.E. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werneke S.W., Schilte C., Rohatgi A., Monte K.J., Michault A., Arenzana-Seisdedos F. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 2011;7:e1002322. doi: 10.1371/journal.ppat.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.H., Luo J.K., Zhang D.E. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J Immunol. 2008;181:6467–6472. doi: 10.4049/jimmunol.181.9.6467. [DOI] [PubMed] [Google Scholar]
- 41.Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiang T.Y., Zhao C., Krug R.M. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol. 2009;83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y., Zhong G., Zhu L., Liu X., Shan Y., Feng H. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol. 2010;184:5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- 44.Lai C., Struckhoff J.J., Schneider J., Martinez-Sobrido L., Wolff T., Garcia-Sastre A. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osiak A., Utermohlen O., Niendorf S., Horak I., Knobeloch K.P. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giannakopoulos N.V., Arutyunova E., Lai C., Lenschow D.J., Haas A.L., Virgin H.W. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol. 2009;83:1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Burke C.W., Ryman K.D., Klimstra W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guerra S., Caceres A., Knobeloch K.P., Horak I., Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okumura A., Pitha P.M., Harty R.N. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H.X., Yang K., Liu X., Liu X.Y., Wei B., Shan Y.F. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pincetic A., Kuang Z., Seo E.J., Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84:4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuang Z., Seo E.J., Leis J. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol. 2011;85:7153–7161. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai J., Pan W., Wang P. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J. 2011;8:468. doi: 10.1186/1743-422X-8-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malakhova O.A., Zhang D.E. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim M.J., Yoo J.Y. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J Immunol. 2010;185:4311–4318. doi: 10.4049/jimmunol.1000098. [DOI] [PubMed] [Google Scholar]
- 56.Broering R., Zhang X., Kottilil S., Trippler M., Jiang M., Lu M. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut. 2010;59:1111–1119. doi: 10.1136/gut.2009.195545. [DOI] [PubMed] [Google Scholar]
- 57.Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woods M.W., Kelly J.N., Hattlmann C.J., Tong J.G., Xu L.S., Coleman M.D. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology. 2011;8:95. doi: 10.1186/1742-4690-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsiao N.W., Chen J.W., Yang T.C., Orloff G.M., Wu Y.Y., Lai C.H. ISG15 over-expression inhibits replication of the Japanese encephalitis virus in human medulloblastoma cells. Antiviral Res. 2010;85:504–511. doi: 10.1016/j.antiviral.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Pincetic A., Leis J. The mechanism of budding of retroviruses from cell membranes. Adv Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timmins J., Scianimanico S., Schoehn G., Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda J., Nakao M., Kawaoka Y., Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou W., Papov V., Malakhova O., Kim K.I., Dao C., Li J. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun. 2005;336:61–68. doi: 10.1016/j.bbrc.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 64.Takeuchi T., Yokosawa H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun. 2005;336:9–13. doi: 10.1016/j.bbrc.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 65.Perez M., Craven R.C., de la Torre J.C. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crump C.M., Yates C., Minson T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol. 2007;81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pawliczek T., Crump C.M. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol. 2009;83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao C., Hsiang T.Y., Kuo R.L., Krug R.M. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci USA. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malakhov M.P., Kim K.I., Malakhova O.A., Jacobs B.S., Borden E.C., Zhang D.E. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 70.Lu G., Reinert J.T., Pitha-Rowe I., Okumura A., Kellum M., Knobeloch K.P. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol Biol (Noisy-le-grand) 2006;52:29–41. [PubMed] [Google Scholar]
- 71.Kim M.J., Hwang S.Y., Imaizumi T., Yoo J.Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okumura F., Okumura A.J., Uematsu K., Hatakeyama S., Zhang D.E., Kamura T. Activation of double-stranded RNA-activated protein kinase (PKR) by interferon-stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem. 2013;288:2839–2847. doi: 10.1074/jbc.M112.401851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeon Y.J., Choi J.S., Lee J.Y., Yu K.R., Kim S.M., Ka S.H. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009;10:374–380. doi: 10.1038/embor.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D'Cunha J., Ramanujam S., Wagner R.J., Witt P.L., Knight E., Borden E.C. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- 75.D'Cunha J., Knight E., Haas A.L., Truitt R.L., Borden E.C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao C., Collins M.N., Hsiang T.Y., Krug R.M. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang W., Zhang M., Xiao Z.Z., Sun L. Cynoglossus semilaevis ISG15: a secreted cytokine-like protein that stimulates antiviral immune response in a LRGG motif-dependent manner. PLoS One. 2012;7:e44884. doi: 10.1371/journal.pone.0044884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langevin C., van der Aa L.M., Houel A., Torhy C., Briolat V., Lunazzi A. Zebrafish ISG15 exerts a strong antiviral activity against RNA and DNA viruses and regulates the interferon response. J Virol. 2013;87:10025–10036. doi: 10.1128/JVI.01294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia Z.P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]