Abstract
Background
HIV-1 subtype C has emerged as the most prevalent strain of HIV-1 worldwide, leading to speculation that subtype C may be more transmissible than other subtypes. We compared the risk of HIV-1 transmission for subtype C versus non-C subtypes (A, D, G and recombinant forms) among heterosexual African HIV-1 serodiscordant couples.
Methods
We conducted a nested case-control analysis using data from two prospective cohort studies of heterosexual HIV-1 serodiscordant couples from 6 countries in eastern and southern Africa. Cases (N=121) included incident HIV-1 transmissions that were established as linked within the serodiscordant partnership by viral sequencing; controls (N=501) were non-transmitting HIV-1 infected partners. Subtype was determined for partial env and gag genes. Multiple logistic regression controlled for age and gender of the HIV-1 infected partner and self-reported unprotected sex. Plasma and genital HIV-1 RNA concentrations were compared between subtype C and non-C subtypes using generalized estimating equations.
Results
HIV-1 subtype C was not associated with increased risk of HIV-1 transmission compared to non-C subtypes: env adjusted odds ratio (adjOR) 1.14 (95% confidence interval [CI] 0.74–1.75, p=0.6) and gag adjOR 0.98 (95% CI 0.63–1.52, p=0.9). Plasma and genital HIV-1 RNA levels did not differ significantly for subtype C versus non-C.
Conclusion
In a geographically diverse population of heterosexual African HIV-1 serodiscordant couples, subtype C was not associated with greater risk of HIV-1 transmission compared to non-C subtypes, arguing against the hypothesis that subtype C is more transmissible compared to other common subtypes.
Keywords: HIV-1 subtype, transmission, serodiscordant couples, Africa
Introduction
HIV-1 subtype C accounts for nearly half of all HIV-1 infections worldwide, primarily due to its predominance in sub-Saharan Africa, where the majority of HIV-1 infections occur 1, 2. The explosion of heterosexually transmitted HIV-1 throughout southern Africa in the 1990s was almost exclusively due to HIV-1 subtype C, leading some to hypothesize that subtype C might be more transmissible compared to other subtypes 3–6. Laboratory studies have suggested molecular and genetic characteristics of subtype C that could promote more efficient transmission 7–9. However, clear evidence for differential transmissibility of HIV-1 subtypes in population-level epidemiological studies has not been shown 10–12. HIV-1 genetic diversity, including subtype diversity, poses a challenge to the development of a globally-effective HIV-1 vaccine 13, and subtype-related differences in HIV-1 transmission, if present, would be a critical consideration in the selection of vaccine antigens 2, 14.
Epidemiologic studies directly measuring the relationship between HIV-1 subtype and heterosexual transmission risk have been challenging for two main reasons. First, prospective studies of HIV-1 transmission require following large numbers of HIV-1 infected persons and their uninfected sexual partners in order to identify rates of HIV-1 transmission occurring within the partnerships. Second, HIV-1 subtypes tend to be geographically specific, and thus studies must include populations from multiple regions in order to have sufficient subtype variation for comparison of transmission risk. Several studies of mother-to-child transmission have had mixed results when comparing vertical HIV-1 transmission by subtype15–18. Even fewer studies of subtype and transmission exist for heterosexual transmission. One HIV-1 serodiscordant couples study in Uganda found higher transmission risk for subtype A compared to D 19, but subtype C was not present in the study population. Another study of serodiscordant couples in Zambia found subtype C in 95% of genetically-linked transmissions20, but the Zambian epidemic is predominantly subtype C and thus comparing transmission rates to other subtypes was not possible in that study. In the present study, among a multinational population of heterosexual HIV-1 serodiscordant couples from eastern and southern Africa, our aim was to assess whether subtype C, compared with non-C subtypes, was associated with greater HIV-1 transmission risk.
Methods
Study Population
We conducted a nested case-control study using data from two prospective cohort studies of African HIV-1 serodiscordant couples. Between November 2004 and April 2007, 3408 heterosexual HIV-1 serodiscordant couples from 6 African countries (Botswana, Kenya, South Africa, Tanzania, Uganda, and Zambia) were enrolled into the Partners in Prevention HSV/HIV Transmission Study, a randomized, double-blind, placebo-controlled clinical trial of herpes simplex virus type 2 (HSV-2) suppressive therapy to reduce HIV-1 transmission, as previously described 21. Eligible couples were at least 18 years of age, reported at least three vaginal sex acts in the three months prior to enrollment, and intended to remain as a couple. At enrollment, all HIV-1 infected partners were HSV-2 seropositive, had CD4 counts ≥250 cells/μL (making them ineligible for antiretroviral therapy (ART) under the national guidelines of the study countries at that time), and were not currently taking ART. HSV-2 suppressive therapy was found not to reduce HIV-1 transmission within the partnerships22. In a parallel study at two sites (Kampala, Uganda and Soweto, South Africa), an additional 485 HIV-1 serodiscordant couples were enrolled into an observational study of immune correlates of HIV-1 protection (Couples Observational Study)23. Similar to the clinical trial cohort, participants were ≥18 years of age and sexually active and HIV-1 seropositive partners were not using ART. In both cohorts, initially HIV-1 uninfected participants were followed quarterly, with HIV-1 serologic testing.
Protection of Human Subjects
All participants received HIV-1 and risk-reduction counseling (both individually and as a couple), free condoms, and treatment for sexually transmitted infections (STIs), according to WHO guidelines. Written, informed consent was obtained from all participants. The study protocols were approved by the University of Washington Human Subjects Review Committee and ethical review committees at each of the study sites.
Selection of cases and controls
Cases were defined from our primary cohort studies as all HIV-1 infected partners of HIV-1 seroconverters, limited to those couples in which it was determined, through viral genetic linkage, that HIV-1 transmission occurred within the partnership (as opposed to from an outside partner) 24. A total of 121 cases were included: 106 from the Partners in Prevention HSV/HIV Transmission Study and 15 from the Couples Observational Study. Controls were selected randomly, in proportion to research site and gender distribution of each primary study, from non-transmitting HIV-1 infected partners to achieve a 1:4 case to control ratio. Since HIV-1 subtype was expected to be correlated with site, given the geographic association of HIV-1 subtypes in Africa, the proportional sampling of controls was used to select controls representative of the full cohort. In total, 501 controls were selected.
Laboratory Testing
HIV-1 seroconversion of initially HIV-1 uninfected partners was determined by quarterly serologic testing using dual rapid HIV-1 antibody tests with confirmatory HIV-1 enzyme immunoassay (EIA), Western blot, and plasma HIV-1 RNA detection. Plasma HIV-1 RNA levels for HIV-1 infected partners were quantified using the COBAS Ampliprep/COBAS TaqMan real-time HIV-1 RNA assay version 1.0 (Roche Diagnostics, Indianapolis, IN). Plasma HIV-1 RNA viral loads were assessed at enrollment and visit months 3, 6, 9, 12 and study exit for the Partners in Prevention HSV/HIV Transmission Study and at enrollment only for the Couples Observational Study. Genital HIV-1 RNA was quantified using the TaqMan assay from samples collected at a single study visit in the Partners in Prevention HSV/HIV Transmission Study: seminal plasma for HIV-1 infected men, collected at any visit ≥3 months after enrollment and endocervical swabs for HIV-1 infected women, collected at a visit 6 months after enrollment 25. All viral loads were log10 transformed, and results below the limit of quantification (<240 copies/mL) were assigned a value of half the limit.
Viral sequencing using blood plasma was performed on partial HIV-1 env (C2-V3-C3) and gag (p17–p24) genes using samples collected at the first post-seroconversion study visit for cases and at the last follow-up visit for controls. Genetic linkage of HIV-1 transmission events was based on phylogenetic analysis and posterior probability of linkage using pair-wise nucleotide distances between sequences 24.
Subtypes were determined by the REGA subtype tool version 2.0 (http://dbpartners.stanford.edu/RegaSubtyping/). Sequence data were provided to GenBank and accession numbers are pending.
Data analysis
We compared HIV-1 transmission risk in cases versus controls between subtype C and all non-C subtypes (including A, D, G, and recombinants) separately for both env and gag. All cases had subtype information available in gag, env or both gene regions, but among controls, 43/501 (8.6%) were missing all subtype data, including 34/332 (10.2%) from eastern African and 9/169 (5.3%) from southern Africa, due to low HIV-1 plasma viral loads preventing adequate viral amplification. To avoid bias because of control exclusion due to missing subtype data, we performed multiple imputation with 20 datasets imputed using Markov chain Monte Carlo methods 26.
To assess differences in HIV-1 transmission between subtype C to non-C subtypes, we performed a standard case-control analysis using logistic regression, analyzing the 20 imputed datasets and combining the results to produce standard estimates and 95% confidence intervals. All models were adjusted for gender and age of the HIV-1 infected partner and self-reported unprotected sex in the month prior to study enrollment. We assessed other variables for potential confounding, some of which may reflect regional differences, including circumcision status of male HIV-1 uninfected partners, duration of partnership, number of children, presence of sexually transmitted infections, any ART initiation during follow-up by HIV-1 infected partners, and CD4 count of HIV-1 infected partners; however, none of these factors substantially changed the effect estimates and thus were not included in the final models. In additional analyses, we further adjusted for baseline plasma HIV-1 RNA concentrations to assess the association of subtype C and HIV-1 transmission independent of plasma viral load. With the available sample size, we estimated we would have 80% power to detect a 1.85-fold increased odds of HIV-1 transmission for subtype C versus non-C at the alpha 0.05 level.
In addition to the nested case-control analysis, in order to incorporate changes in longitudinal covariates, including time-dependent covariates such as plasma HIV-1 RNA and unprotected sex, we also employed a case-cohort analysis, as a secondary analysis. We used Cox proportional hazards analyses, adjusted for gender, age of the HIV-1 infected partner, and longitudinal report of unprotected sex and plasma HIV-1 RNA, to compare transmission by HIV-1 subtype. Case-cohort analysis methods were used 27.
Finally, we compared differences in plasma and genital HIV-1 RNA concentrations between subtype C and non-C subtypes for participants from the Partners in Prevention HSV/HIV Transmission Study. We assessed subtype differences related to longitudinal plasma HIV-1 RNA during study follow-up using repeated measures generalized estimating equations (GEE) models with unstructured correlation matrix, adjusting for gender, age of the HIV-1 infected partner, and unprotected sex. Participants were censored at ART initiation. Genital HIV-1 RNA levels were available at a single time point for 416/624 (66.7%) of the HIV-1 infected partners, and we assessed differences among subtypes using a multiple linear regression for endocervical and semen HIV-1 RNA levels, controlling for age of the HIV-1 uninfected partner, unprotected sex reported at enrollment, and plasma HIV-1 viral load.
All analyses were performed using SAS v.9.2 (SAS Institute, Inc., Cary, N.C.).
Results
Of the 622 HIV-1 infected study participants in the nested case-control cohort, subtype information was available for 579 (93.1%), including all 121 (100.0%) cases and 458/501 (91.4%) controls. The majority of participants were from eastern Africa: 80 (66.1%) cases and 332 (66.3%) controls (Table 1). Most couples (92.0%) were married. Age was similar between cases and controls: median age of cases was 30 years (IQR 26–35) and the median age of controls was 32 years (IQR 26–38). Cases were more likely to report unprotected sex in the month prior to enrollment (52.8% versus 36.2%, p=0.001) and less likely to be female (49.6% versus 65.5%, p<0.001). The median baseline HIV-1 plasma RNA was significantly higher in cases (4.8 log10 copies/mL, IQR 4.3–5.1) compared to controls (4.2 log10 copies/mL, IQR 3.6–4.8, p<0.001).
Table 1.
Characteristics of the study population
| Transmitting couples, N=121 | Non-transmitting couples, N=501 | Subtype C***, N=217 | Non-C Subtypes***, N=362 | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| N/median | %/IQR | N/median | %/IQR | N/median | %/IQR | N/median | %/IQR | |
| Demographic characteristics | ||||||||
| HIV-1 infected female | 60 | 49.6% | 328 | 65.5% | 149 | 68.7% | 208 | 57.5% |
| Age in years, HIV-1 infected partner | 30 | 26–35 | 32 | 26–38 | 32 | 27–39 | 30 | 26–37 |
| East African (vs. southern African) | 80 | 66.1% | 332 | 66.3% | 25 | 11.5% | 353 | 97.5% |
| Married/living together | 113 | 93.4% | 459 | 91.6% | 179 | 82.5% | 353 | 97.5% |
| Duration of partnership, years | 3.8 | 1.5–7.0 | 5 | 2.2–9.7 | 4.1 | 1.8–8.7 | 5.2 | 2.2–9.6 |
| Number of children within partnership | 1 | 0–2 | 1 | 0–2 | 1 | 0–2 | 1 | 0–2% |
| Unprotected sex | ||||||||
| One month prior to enrollment | 56 | 52.8% | 160 | 36.2% | 81 | 37.3% | 125 | 34.5% |
| Follow-up visits with unprotected sex** | 220/1057 | 20.8% | 643/9280 | 6.9% | 411/3365 | 12.2% | 420/6168 | 6.8% |
| Antiretroviral therapy initiated during follow-up | 1 | 0.8% | 67 | 13.4% | 6 | 2.8% | 54 | 14.9% |
| Baseline clinical characteristics | ||||||||
| CD4 count, cells/mm3 | 417 | 302–580 | 434 | 341–601 | 418 | 316–570 | 428 | 332–602 |
| HIV-1 plasma viral load, log10 copies/mL | 4.8 | 4.3–5.1 | 4.2 | 3.6–4.8 | 4.2 | 3.7–4.8 | 4.4 | 2.9–4.0 |
| Any genital tract infection (either partner)* | 23 | 20.4% | 74 | 15.9% | 51 | 23.5% | 43 | 11.9% |
| Circumcision (male HIV-1 uninfected partners) | 39 | 65.0% | 239 | 72.9% | 84 | 38.7% | 175 | 48.3% |
Includes Neisseria gonorrhoeae, Chlamydia trachomatis, Trichomonas vaginalis
Numerator=all followup visits with unprotected sex reported, denominator=total follow-up visits
Subtype data shown for env; gag results were similar (N=43 without subtype information)
IQR=interquartile range
Bold indicates statistical significan at the p<0.05 level, comparing transmitting to non-transmitting couples and subtype C to non-C subtypes
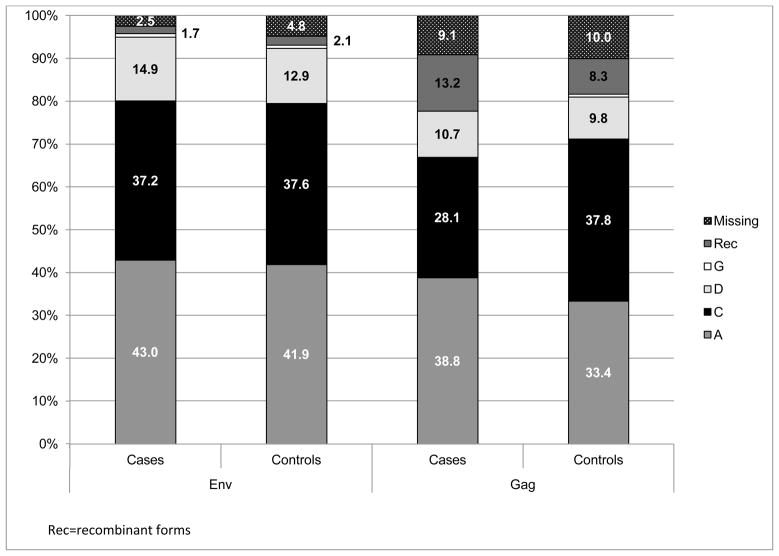
The most common subtypes were A (env 44.0%, gag 38.3%) and C (env 39.2%, gag 39.7%), followed by D (env 13.9%, gag 11.1%), and G or recombinant subtypes (env 2.9%, gag 10.9%). Subtype was missing in env for 25 (4.3%) and in gag for 57 (9.8%). For participants with both env and gag subtypes available, concordance between genes was 82.5%, with concordance of 95.5% for subtype C env and gag. Nearly all participants from southern Africa were infected with subtype C (env 98.5%, gag 99.5%). In eastern Africa, the predominant subtypes were subtype A (env 67.7%, gag 59.6%) and subtype D (env 21.5%, gag 17.4%). The distribution of subtype among cases and controls is shown in Figure 1.
Figure 1. Distribution of env and gag subtype among cases and controls.
The percentage distribution of HIV-1 subtypes by cases (HIV-1 infected partner in transmitting couples, determined to be linked by viral sequencing) and controls (HIV-1 non-transmitting controls) for both the env and gag gene regions. Letters refer to the subtype for that gene region (RF=recombinant forms)
Subtype C and HIV-1 Transmission Risk
In the nested case-control multivariate logistic regression analysis, subtype C was not associated with an increased risk of HIV-1 transmission compared to non-C subtypes, both when considering subtype based on env sequencing (adjusted odds ratio [adjOR] 1.14, 95% confidence interval [CI] 0.74–1.75, p=0.6) and gag sequencing (adjOR 0.98, 95% CI 0.63–1.52, p=0.9) (Table 2). Additionally, separate comparisons of subtype C to individual subtypes showed no statistically significant differences in the odds of HIV-1 transmission risk with subtype A (env adjOR 1.17, p=0.5 and gag adjOR 1.09, p=0.7) or subtype D (env adjOR 1.39, p=0.3 and gag adjOR 1.79, p=0.08). Separate comparisons between subtype C and subtype G or recombinant forms was not possible due to the small number of participants with these subtypes. Further adjusting these same regression models for plasma HIV-1 RNA did not to substantially change these results. Additionally, when we compared HIV-1 transmission for subtype A compared to subtype D, we did not find significant differences in env (adjOR 1.25, 95%CI 0.66–2.36, p=0.5) or gag (adjOR 0.89, 95%CI 0.48–1.67, p=0.7).
Table 2.
Adjusted multivariate models for the nested case-control and sensitivity case-cohort analyses comparing HIV-1 transmission for subtype C versus non-C subtypes
| Nested case-control | Case-Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Odds ratio (95%CI) adjusted for gender, age and unprotected sex | Odds ratio (95%CI) adjusted gender age, unprotected sex plus plasma HIV-1 RNA | Hazard ratio (95%CI) adjusted for gender, age and unprotected sex | Hazard ratio (95%CI) adjusted gender age, unprotected sex plus plasma HIV-1 RNA | |||||
|
| ||||||||
| env | gag | env | gag | env | gag | env | gag | |
| C vs. non-C | 1.14 (0.74–1.75) p=0.6 |
0.98 (0.63.1.52) p=0.9 |
1.18 (0.76–1.83) p=0.5 |
1.03 (0.66–1.60) p=0.9 |
1.56 (0.89–2.76) p=0.1 |
0.92 (0.51–1.67) p=0.8 |
1.58 (0.86–2.90) p=0.1 |
0.97 (0.52–1.80) p=0.9 |
|
| ||||||||
| C vs. A | 1.17 (0.74–1.84) p=0.5 |
1.09 (0.68–1.75) p=0.7 |
1.25 (0.78–1.99) p=0.3 |
1.13 (0.70–1.84) p=0.6 |
1.22 (0.65–2.29) p=0.5 |
0.88 (0.45–1.72) p=0.7 |
1.26 (0.65–2.44) p=0.5 |
0.91 (0.45–1.83) p=0.8 |
|
| ||||||||
| C vs. D | 1.39 (0.76–2.56) p=0.3 |
1.79 (0.93–3.47) p=0.08 |
1.27 (0.68–2.38) p=0.5 |
1.60 (0.82–3.13) p=0.2 |
2.19 (0.95–5.06) p=0.07 |
1.29 (0.49–3.37) p=0.6 |
2.29 (0.95–5.51) p=0.07 |
1.28 (0.45–3.62) p=0.6 |
Adjusted for gender and age of HIV-1 infected partner
Unprotected sex and plasma HIV-1 RNA assessed at baseline in nested case-control model and longtitudinally for case-cohort model
In the case-cohort analysis, which permitted adjustment for unprotected sex as a time-varying covariate, results were similar to those in the nested case-control approach: subtype C was not significantly associated with increased HIV-1 transmission compared to non-C subtypes, in env (adjHR 1.56, 95% CI 0.89–2.76, p=0.1) or gag (adjHR 0.92, 95% CI 0.51–1.67, p=0.8). In separate comparisons of HIV-1 transmission risk between subtype C and subtypes A and D, there were also no statistically significant differences for env or gag. These results were similar with the addition of time-dependent plasma HIV-1 RNA to the models.
Subtype C and HIV-1 Concentrations in Plasma and Genital Secretions
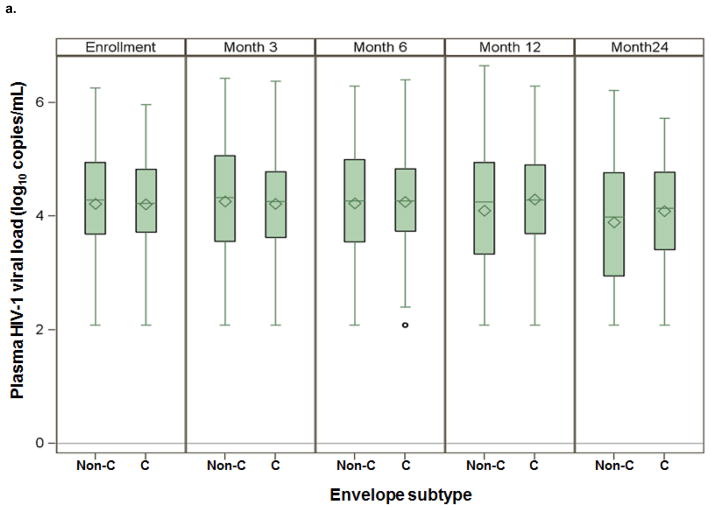
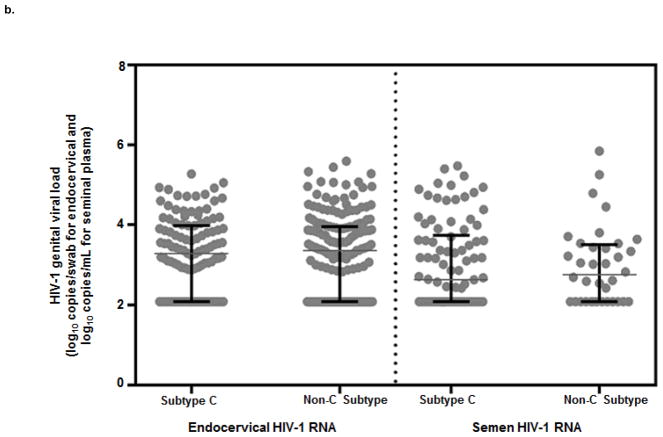
The median plasma HIV-1 RNA during the study was 4.3 log10 copies/mL (IQR 3.7–4.8) among those with env subtype C and 4.2 log10 copies/mL (IQR 3.4–4.9) among those with a non-C env subtype (Figure 2a; p=0.2). The median endocervical HIV-1 RNA for env subtype C was 3.3 log10 copies/mL (IQR 2.5–4.0) and for non-C env subtypes was 3.4 log10 copies/mL (IQR 2.5–4.0, p=0.9) (Figure 2b). The median semen HIV-1 RNA was 2.8 log10 copies/mL (IQR 2.1–3.5) for env subtype C and 2.6 log10 copies/mL (IQR 2.1–3.7) for non-C env subtypes. Individuals with env subtype C did not differ significantly from non-C subtypes by genital viral load in either endocervical fluid (p=0.9) or semen plasma (p=0.6). Results for gag subtype were similar to env (data not shown).
Figure 2. Median plasma and genital HIV-1 RNA by env subtype C and non-C subtypes.
a) Box plot distribution of log10 plasma HIV-1 RNA for env subtypes C and non-C subtypes by study month. Mean values denoted by diamonds and median values denoted by bars.
b) Median and interquartile range (IQR) for endocervical and semen HIV-1 RNA concentrations for those with env subtype C and non-C subtypes. Individual HIV-1 RNA concentrations plotted with median HIV-1 RNA concentration denoted and IQR denoted by black bars.
Discussion
In this analysis comparing transmitting and non-transmitting HIV-1 serodiscordant couples from eastern and southern Africa, we did not find evidence that subtype C was associated with increased HIV-1 transmission risk, compared with non-C subtypes. Our study population included a wide geographic region with sufficient subtype variation (primarily A, C and D) in order to perform the analyses, and genetic linkage information improved the precision of the results. Previous studies of subtype and HIV-1 transmission have either lacked the subtype diversity to compare subtype C to non-C subtypes or been based on ecological data of prevalent trends in subtype. To our knowledge, this is the first study to provide direct evidence for the question of whether subtype C is associated with increased heterosexual transmission risk compared to other non-C subtypes common in sub-Saharan Africa. Our results do not support the hypothesis that HIV-1 subtype C has greater transmissibility compared with other subtypes.
We conducted both a nested case-control analysis and a longitudinal analysis using a case-cohort study design to assess whether subtype C was associated with an increased risk for HIV-1 transmission. We adjusted for age, gender and reported unprotected sex, and we determined that other factors (e.g., male circumcision status) were not confounding. We did not initially include plasma HIV-1 RNA in our initial models because we hypothesized that if HIV-1 transmission differed by subtype, it could be mediated by subtype-related differences in viral load. However, after finding no association between subtype C and HIV-1 transmission, we further adjusted our models to control for plasma HIV-1 RNA and continued to see no significant relationship between subtype C and HIV-1 transmission risk, compared to non-C subtypes. In both the nested case-control and case-cohort analyses, we also compared subtype C and subtypes A and D separately and found no statistically significant difference in HIV-1 transmission risk.
A limited number of studies have found individuals with subtype C to have higher HIV-1 DNA or RNA concentrations in plasma and genital secretions, which could indicate higher transmission risk 15, 28, 29; however, not all studies have found increased HIV-1 concentrations associated with subtype C infection 30. In the present study, we assessed whether subtype C was associated with higher plasma and genital HIV-1 RNA concentrations, as a proxy for infectiousness and potential onward transmission. We found no statistically significant differences in plasma and genital HIV-1 RNA levels in participants with subtype C compared to non-C subtypes, further supporting the results of our nested case-control and case-cohort transmission analyses.
The rapid expansion of HIV-1 subtype C throughout sub-Saharan Africa has led some to hypothesize a causal relationship between subtype C and increased HIV-1 transmissibility. However, a combination of other factors may be as likely to contribute to the swift growth of HIV-1 subtype C. A founder effect, which has been hypothesized to explain the dominance of specific subtypes throughout Africa, could be relevant 31, 32. Additionally, Tatem et al. recently provided evidence to suggest that regions with greater accessibility allowing for increased mobility, such as in southern Africa, are associated with clusters of similar subtypes throughout the transportation infrastructure 33. Another potential explanation is that subtype C has shown lower viral fitness, and therefore may result in slower disease progression compared to other subtypes 11, 34–36; individuals with a slower progressing disease not only add person-years to prevalence estimates, but also have more opportunity to transmit their infection over a longer period of time. Finally, subtype C may be more prevalent in sexual networks with behavioral and demographic characteristics leading to higher risk for HIV-1 transmission 31, 37, 38.
Our analyses have limitations. First, it is likely that most HIV-1 infected partners in our study had chronic, as opposed to acute, HIV-1 infection. Some have speculated that subtype C is associated with higher viremia during acute infection that may contribute to increased transmission 39, 40. However, in a separate analysis of seroconverters from our studies, we found no significant association between subtype C and plasma HIV-1 RNA levels during early HIV-1 infection 30. Second, as subtypes are geographically distributed, there may be unmeasured differences across study sites that could potentially confound the results, in spite of our assessment of a number of behavioral, demographic, and clinical factors for potential confounding. In the primary cohorts from which our case-control sample derived, there was higher incidence of HIV-1 transmissions within couples in southern Africa (3.7 per 100 person-years, 95% CI 2.6–4.8) compared to couples in eastern African (2.2 per 100 person-years, 95% CI 1.7–2.7), a difference that was statistically significant in a proportional hazards model adjusted for age, gender, circumcision status and unprotected sex (adjusted HR 1.65, 95%CI 1.14–2,38, p=0.007); however, our results suggest that this difference is not explained by subtype. The selection of controls from our analysis was based on gender and geographic distribution of the primary cohort to ensure a representative population of controls from the entire cohort.
In summary, we found no statistically significant differences in risk of heterosexual HIV-1 transmission associated with HIV-1 subtype C infection, nor was subtype C significantly associated with higher HIV-1 plasma and genital concentrations. A better understanding the impact of viral diversity on HIV-1 transmission and pathogenicity is important to HIV-1 prevention efforts, including treatment and vaccine development. The role of subtype on HIV-1 disease progression and pathogenicity should continue to be evaluated, particularly to inform the development of a globally applicable cross-protective vaccine.
Acknowledgments
Funding: US National Institutes of Health (P30 A127757 to the University of Washington Center for AIDS Research, P01 057005, and K08 AI074424) and the Bill and Melinda Gates Foundation (grant 26469).
We gratefully acknowledge the invaluable contributions of the HIV-1 serodiscordant couples who participated in this study. We also thank Airin L. Lam for her work sequencing the non-transmitting couples. E.M.K. and J.M.B. designed the study and wrote the first draft of the manuscript. E.M.K. performed the statistical analysis. M.C. and J.I.M. performed viral sequencing and provided interpretation of sequence data. J.L., D.D., C.C., R.O., A.M., K.F., N.M., and S.K. contributed critical revisions to the analysis and interpretation. All authors contributed to the writing of the final draft.
Partners in Prevention HSV/HIV Transmission Study Team
University of Washington Coordinating Center, Seattle, USA: Connie Celum (principal investigator), Anna Wald (protocol co-chair), Jairam Lingappa (medical director), Amalia Magaret, James P. Hughes (protocol statisticians), Lawrence Corey, Jared M. Baeten, M. Juliana McElrath (co-investigators)
Study sites and site principal investigators
Cape Town, South Africa (University of Cape Town): David Coetzee; Eldoret, Kenya (Moi University, Indiana University): Kenneth Fife, Edwin Were; Gaborone, Botswana (Botswana Harvard Partnership): Max Essex, Joseph Makhema; Kampala, Uganda (Infectious Disease Institute, Makerere University): Elly Katabira, Allan Ronald; Kisumu, Kenya (Kenya Medical Research Institute, University of California San Francisco): Elizabeth Bukusi, Craig Cohen; Kitwe, Zambia (Rwanda Zambia HIV Research Group, and Emory University): Susan Allen, William Kanweka; Moshi, Tanzania (Kilimanjaro Christian Medical College, Harvard University): Saidi Kapiga, Rachel Manongi; Nairobi, Kenya (University of Nairobi, University of Washington): Carey Farquhar, Grace John-Stewart, James Kiarie; Ndola, Zambia (Rwanda Zambia HIV Research Group, and Emory University): Susan Allen, Mubiana Inambao; Orange Farm, South Africa (Reproductive Health Research Unit, University of the Witwatersrand): Sinead Delany-Moretlwe, Helen Rees; Soweto, South Africa (Perinatal HIV Research Unit, University of the Witwatersrand): Guy de Bruyn, Glenda Gray, James McIntyre; Thika, Kenya (University of Nairobi, University of Washington): Nelly Rwamba Mugo
Data management was provided by DF/Net Research, Inc. (Seattle, USA) and site laboratory oversight was provided by Contract Lab Services (University of the Witwatersrand, Johannesburg, South Africa).
Footnotes
Conflict of Interest: None of the authors have commercial or other conflicts of interest related to the content of this manuscript.
Previous Presentation: A portion of this work was presented at the XIX International AIDS Conference, 22–27 July 2012, Washington, DC, USA. Abstract TUAC0101.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Isolation W-UNfH Characterisation Global trends in molecular epidemiology of HIV-1 during 2000–2007. Aids. 2011;25(5):679–89. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: a decade in review. AIDS reviews. 2012;14(2):83–100. [PubMed] [Google Scholar]
- 3.Renjifo B, Chaplin B, Mwakagile D, et al. Epidemic expansion of HIV type 1 subtype C and recombinant genotypes in Tanzania. AIDS research and human retroviruses. 1998;14(7):635–8. doi: 10.1089/aid.1998.14.635. [DOI] [PubMed] [Google Scholar]
- 4.Hunter DJ. AIDS in sub-Saharan Africa: the epidemiology of heterosexual transmission and the prospects for prevention. Epidemiology. 1993;4(1):63–72. [PubMed] [Google Scholar]
- 5.Van Harmelen JH, Van der Ryst E, Loubser AS, et al. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS research and human retroviruses. 1999;15 (4):395–8. doi: 10.1089/088922299311376. [DOI] [PubMed] [Google Scholar]
- 6.Engelbrecht S, Laten JD, Smith TL, van Rensburg EJ. Identification of env subtypes in fourteen HIV type 1 isolates from south Africa. AIDS research and human retroviruses. 1995;11(10):1269–71. doi: 10.1089/aid.1995.11.1269. [DOI] [PubMed] [Google Scholar]
- 7.Walter BL, Armitage AE, Graham SC, et al. Functional characteristics of HIV-1 subtype C compatible with increased heterosexual transmissibility. Aids. 2009;23(9):1047–57. doi: 10.1097/QAD.0b013e32832a1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters M, Vincent R, Perret JL, et al. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. Journal of acquired immune deficiency syndromes and human retrovirology : official publication of the International Retrovirology Association. 1999;20(2):115–21. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 9.Iversen AK, Learn GH, Skinhoj P, Mullins JI, McMichael AJ, Rambaut A. Preferential detection of HIV subtype C′ over subtype A in cervical cells from a dually infected woman. Aids. 2005;19(9):990–3. doi: 10.1097/01.aids.0000171418.91786.ad. [DOI] [PubMed] [Google Scholar]
- 10.Hu DJ, Buve A, Baggs J, van der Groen G, Dondero TJ. What role does HIV-1 subtype play in transmission and pathogenesis? An epidemiological perspective. Aids. 1999;13(8):873–81. doi: 10.1097/00002030-199905280-00002. [DOI] [PubMed] [Google Scholar]
- 11.Rainwater S, DeVange S, Sagar M, et al. No evidence for rapid subtype C spread within an epidemic in which multiple subtypes and intersubtype recombinants circulate. AIDS research and human retroviruses. 2005;21(12):1060–5. doi: 10.1089/aid.2005.21.1060. [DOI] [PubMed] [Google Scholar]
- 12.Morison L, Buve A, Zekeng L, et al. HIV-1 subtypes and the HIV epidemics in four cities in sub-Saharan Africa. Aids. 2001;15 (Suppl 4):S109–16. doi: 10.1097/00002030-200108004-00012. [DOI] [PubMed] [Google Scholar]
- 13.Taylor BS, Hammer SM. The challenge of HIV-1 subtype diversity. The New England journal of medicine. 2008;359(18):1965–6. doi: 10.1056/NEJMc086373. [DOI] [PubMed] [Google Scholar]
- 14.Lal RB, Chakrabarti S, Yang C. Impact of genetic diversity of HIV-1 on diagnosis, antiretroviral therapy & vaccine development. The Indian journal of medical research. 2005;121(4):287–314. [PubMed] [Google Scholar]
- 15.John-Stewart GC, Nduati RW, Rousseau CM, et al. Subtype C Is associated with increased vaginal shedding of HIV-1. The Journal of infectious diseases. 2005;192(3):492–6. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Li M, Newman RD, et al. Genetic diversity of HIV-1 in western Kenya: subtype-specific differences in mother-to-child transmission. Aids. 2003;17(11):1667–74. doi: 10.1097/01.aids.0000060412.18106.d4. [DOI] [PubMed] [Google Scholar]
- 17.Koulinska IN, Villamor E, Msamanga G, et al. Risk of HIV-1 transmission by breastfeeding among mothers infected with recombinant and non-recombinant HIV-1 genotypes. Virus research. 2006;120(1–2):191–8. doi: 10.1016/j.virusres.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Eshleman SH, Guay LA, Mwatha A, et al. Comparison of mother-to-child transmission rates in Ugandan women with subtype A versus D HIV-1 who received single-dose nevirapine prophylaxis: HIV Network For Prevention Trials 012. Journal of acquired immune deficiency syndromes. 2005;39(5):593–7. [PubMed] [Google Scholar]
- 19.Kiwanuka N, Laeyendecker O, Quinn TC, et al. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. Aids. 2009;23(18):2479–84. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trask SA, Derdeyn CA, Fideli U, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. Journal of virology. 2002;76 (1):397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lingappa JR, Kahle E, Mugo N, et al. Characteristics of HIV-1 discordant couples enrolled in a trial of HSV-2 suppression to reduce HIV-1 transmission: the partners study. PloS one. 2009;4(4):e5272. doi: 10.1371/journal.pone.0005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Celum C, Wald A, Lingappa JR, et al. Acyclovir and transmission of HIV-1 from persons infected with HIV-1 and HSV-2. The New England journal of medicine. 2010;362(5):427–39. doi: 10.1056/NEJMoa0904849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingappa JR, Petrovski S, Kahle E, et al. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PloS one. 2011;6(12):e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell MS, Mullins JI, Hughes JP, et al. Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PloS one. 2011;6(3):e16986. doi: 10.1371/journal.pone.0016986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baeten JM, Kahle E, Lingappa JR, et al. Genital HIV-1 RNA predicts risk of heterosexual HIV-1 transmission. Science translational medicine. 2011;3(77):77ra29. doi: 10.1126/scitranslmed.3001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulla ZD, Seo B, Kalamegham R, Nuwayhid BS. Multiple imputation for missing laboratory data: an example from infectious disease epidemiology. Annals of epidemiology. 2009;19(12):908–14. doi: 10.1016/j.annepidem.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. Journal of clinical epidemiology. 1999;52(12):1165–72. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 28.Quinn TC, Wawer MJ, Sewankambo N, et al. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. The New England journal of medicine. 2000;342(13):921–9. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 29.Coombs RW, Reichelderfer PS, Landay AL. Recent observations on HIV type-1 infection in the genital tract of men and women. Aids. 2003;17(4):455–80. doi: 10.1097/00002030-200303070-00001. [DOI] [PubMed] [Google Scholar]
- 30.Campbell MS, Kahle EM, Celum C, et al. Plasma Viral Loads During Early HIV-1 Infection Are Similar in Subtype C- and Non-Subtype C-Infected African Seroconverters. The Journal of infectious diseases. 2013;207(7):1166–70. doi: 10.1093/infdis/jit015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalai SC, de Oliveira T, Harkins GW, et al. Evolution and molecular epidemiology of subtype C HIV-1 in Zimbabwe. Aids. 2009;23(18):2523–32. doi: 10.1097/QAD.0b013e3283320ef3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayehunie S, Johansson B, Sonnerborg A, et al. New subtype of HIV-1 in Ethiopia. Lancet. 1990;336(8720):942. doi: 10.1016/0140-6736(90)92312-6. [DOI] [PubMed] [Google Scholar]
- 33.Tatem AJ, Hemelaar J, Gray RR, Salemi M. Spatial accessibility and the spread of HIV-1 subtypes and recombinants in sub-Saharan Africa. Aids. 2012 doi: 10.1097/QAD.0b013e328359a904. [DOI] [PubMed] [Google Scholar]
- 34.Tscherning C, Alaeus A, Fredriksson R, et al. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241(2):181–8. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 35.Abraha A, Nankya IL, Gibson R, et al. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. Journal of virology. 2009;83(11):5592–605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arien KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. Journal of virology. 2005;79(14):8979–90. doi: 10.1128/JVI.79.14.8979-8990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buonaguro L, Tornesello ML, Buonaguro FM. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. Journal of virology. 2007;81(19):10209–19. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson E, Engelbrecht S. Molecular characterization of non-subtype C and recombinant HIV-1 viruses from Cape Town, South Africa. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9(5):840–6. doi: 10.1016/j.meegid.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Novitsky V, Ndung’u T, Wang R, et al. Extended high viremics: a substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection. Aids. 2011;25 (12):1515–22. doi: 10.1097/QAD.0b013e3283471eb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Centlivre M, Sala M, Wain-Hobson S, Berkhout B. In HIV-1 pathogenesis the die is cast during primary infection. Aids. 2007;21(1):1–11. doi: 10.1097/QAD.0b013e3280117f7f. [DOI] [PubMed] [Google Scholar]