Abstract
Background
Molecular analysis has become important in colorectal carcinoma (CRC) evaluation. Alterations in KRAS, BRAF, or mismatch repair (MMR) genes may determine therapeutic response or define a hereditary cancer syndrome. Correlation of DNA studies with clinical findings will further clarify the clinical utility of these markers.
Patients and Methods
A retrospective study was performed on 111 paraffin-embedded tumor specimens submitted for microsatellite instability (MSI) testing based on clinical history or histologic examination, or both. DNA samples were screened for 7 KRAS mutations and the BRAF p.V600E mutation using fluorescent allele-specific polymerase-chain reaction (PCR) and capillary electrophoresis. Clinical data were collected through chart review.
Results
Fifty-eight male and 53 female patients were studied. The incidence of KRAS and BRAF mutations was 49.5% and 7.2%, respectively. Dideoxy sequencing verified KRAS mutation status in 46 of 49 specimens tested. There was a trend toward significance of individual KRAS mutations on survival (P = .003). Dually positive KRAS and MSI tumors exclusively demonstrated p.G12D and p.G13D mutations (G>A transitions). BRAF-mutated tumors were predominantly right-sided and associated with a borderline worse prognosis. Forty-eight percent of tumors with MSI were present in the left colon or rectum.
Conclusion
Allele-specific PCR is an accurate and convenient method to assess KRAS and BRAF mutations and may detect mutations not identified by dideoxy sequencing. KRAS mutation status, in conjunction with morphologic or clinical parameters, may be useful in determining whether a tumor should be tested for MSI. MSI testing should not be considered exclusively in right-sided lesions. BRAF analysis may not be useful in rectal adenocarcinomas and should be evaluated in larger studies.
Keywords: Allele specific, Missense, Mutation, Transversion, Biomarker
Introduction
Colorectal carcinoma (CRC) accounted for approximately 50,000 deaths in the United States in 2013.1 There are 2 primary molecular pathways for CRC carcinogenesis: the chromosomal instability (CIN) pathway (80%–85% of CRC) and the microsatellite instability (MSI) pathway (15%–20% of CRC).2–4 The CIN pathway consists of chromosomal deletions or duplications in addition to multiple mutations in various oncogenes and tumor suppressor genes. A well-characterized signaling pathway involved in the development of colorectal cancer through the CIN pathway is the epidermal growth factor receptor (EGFR) pathway. An initiating step in the stimulated EGFR pathway is activation of Kirsten ras (KRAS), a guanosine triphosphatase (GTPase) that is activated through binding of guanosine triphosphate (GTP), which subsequently stimulates the serine/threonine kinase BRAF, triggering the MAPK/ERK pathway.5 KRAS gene mutations are thought to occur early in the adenoma-carcinoma continuum and are estimated to be present in 30% to 50% of colon adenocarcinomas.2,6–9 Conserved missense mutations in codons 12, 13, and 61 result in prolonged binding of guanosine triphosphate (GTP) and constitutive activation of the KRAS protein. In the BRAF gene, 1 specific activating transversion mutation, c.1799T>A (p.V600E) accounts for most BRAF mutations associated with human cancers, including 10% to 40% of colon cancers.8,10–12 Activating mutations in KRAS or BRAF have recently been shown to render advanced CRCs insensitive to EGFR inhibitors, and KRAS and BRAF are now commonly evaluated for mutations before EGFR inhibitor therapy is initiated.9,13–15
Because KRAS and BRAF mutations have become increasingly important in prognosis and treatment, identifying rapid and sensitive methods to detect these mutations is critical. Several different approaches demonstrate utility, including Sanger sequencing (also known as dideoxy sequencing), pyrosequencing, high-resolution melting analysis, and allele-specific PCR.16,17 Many early studies used Sanger sequencing to identify KRAS mutations, a technique that is considered the gold standard but one that also requires the mutant allele to be present within the tumor at a rate of at least 15% to 20%. The sensitivity of allele-specific PCR is estimated to be 1% or better and allows for identification of target mutations in biopsy or pathologic specimens with low tumor burden. Next-generation sequencing platforms may eventually be useful for evaluating genes for somatic mutations in large panels; however, these platforms are currently not easily accessible or cost-effective for single gene assays and require sophisticated bioinformatics for analysis.
Like KRAS and BRAF, identifying MSI is important clinically to determine prognosis and counseling regarding inherited cancer risks. The presence of MSI in CRC may indicate a germline MMR defect, suggesting an autosomal dominant hereditary colon cancer syndrome—hereditary nonpolyposis colon cancer syndrome (HNPCC or Lynch syndrome).3,18 Alternatively, patients with sporadic MSI tumors caused by MLH1 gene silencing by biallelic promoter hypermethylation have a better prognosis but may have a decreased response to 5-fluorouracil.3,19–21 Although MSI high (MSI-H) tumors have an improved prognosis, CIN has been found to be associated with worse prognosis: In 1 meta-analysis of survival, the hazard ratio (HR) was 1.45 (95% confidence interval [CI], 1.35–1.55; P < .001)4 for a CIN (defined as polyploidy or aneuploidy)-related neoplasm.
To assess the utility of allele-specific PCR in patients with colorectal cancer, we used laboratory-developed allele-specific fluorescent PCR assays specific for KRAS and BRAF mutations to evaluate a cohort of patients diagnosed with colorectal neoplasms whose samples were submitted for MSI analysis. We evaluated the association between MSI and EGFR pathway mutations. An extensive chart review was also performed to relate mutation findings with patient parameters.
Patients and Methods
Cohort Selection and Chart Review
A retrospective study was performed on 135 formalin-fixed, paraffin-embedded (FFPE) tumor specimens submitted to the Vanderbilt University Medical Center Clinical Molecular Diagnostic Laboratory between January 1, 2007 and March 31, 2009 for MSI testing. Samples were submitted by a clinician or anatomic pathologist for MSI testing loosely based on the Bethesda Criteria22: age < 50 years and family history or characteristic histologic features, or both. Twenty-four patient samples were excluded from additional studies because of insufficient DNA, noncolonic tumor testing, or outside referral, disqualifying these specimens from institutional review board approval. Chart reviews were performed on the remaining 111 patients. Parameters evaluated in the chart review included age, sex, age at diagnosis, pathologic stage, treatment (surgery, radiation, and chemotherapy), and course of disease. If germline MMR mutation status was known, it was also recorded. One patient had 2 synchronous tumors (1 in the right colon and 1 in the left colon) that were tested for all 3 markers. Only the right-sided MSI-H tumor was included in the study (the left tumor was microsatellite stable [MSS]). Fourteen cases were considered stage 0, which included 3 adenomas (no high-grade dysplasia), 4 adenomas with high-grade dysplasia, and 3 adenomas with carcinoma in situ. Two patients with stage 0 disease had no residual cancer after neoadjuvant therapy, and 1 patient had invasive carcinoma but did not undergo resection because of other comorbidities (no pathologic staging). In a final patient, an adenoma with high-grade dysplasia was tested, but the individual also had invasive carcinoma in a rectal adenoma that was not submitted for testing because it was outside material. Overall survival (OS) was defined as time from surgery to death from any cause or last follow-up and was determined using chart review and the Social Security Death Index. The study was approved by the Vanderbilt University Medical Center Institutional Review Board. Patient demographics and characteristics are identified in Table 1.
Table 1.
Patient Characteristics and Mutation Status (KRAS and BRAF)
| Variable | Overall n = 111 (%) |
No Mutation n = 48 (%) |
KRAS or BRAF n = 63 (%) |
P Value |
|---|---|---|---|---|
| Age (y) | 50 (44–61)a | 48 (42–56)a | 51 (45–61)a | .13b |
| < 40 | 16 (14.4) | 10 (20.8) | 6 (9.5) | |
| 40–49 | 39 (35.1) | 18 (37.5) | 21 (33.3) | |
| 50–59 | 27 (24.3) | 9 (18.8) | 18 (28.6) | |
| > 60 | 29 (26.1) | 11 (22.9) | 18 (28.6) | |
| Sex | .85c | |||
| Female | 53 (47.7) | 22 (45.8) | 31 (49.2) | |
| Male | 58 (52.3) | 26 (54.2) | 32 (50.8) | |
| MSI | .84c | |||
| MSS/MSI-L | 90 (81.1) | 39 (81.2) | 51 (81) | |
| MSI-H | 21 (18.9) | 9 (18.8) | 12 (19) | |
| Stage | .15d | |||
| 0 or adenoma | 14 (12.6) | 6 (12.5) | 8 (12.7) | |
| 1 | 19 (17.1) | 12 (25.0) | 7 (11.1) | |
| 2 | 17 (15.3) | 4 (8.3) | 13 (20.6) | |
| 3 | 33 (29.7) | 16 (33.3) | 17 (27.0) | |
| 4 | 28 (25.2) | 10 (20.8) | 18 (28.6) | |
| Location | .70c | |||
| Left Colon | 29 (26.1) | 12 (25.0) | 17 (27.0) | |
| Right Colon | 36 (32.4) | 14 (29.2) | 22 (34.9) | |
| Rectum | 46 (41.4) | 22 (45.8) | 24 (38.1) |
Abbreviations: MSI-H = microsatellite instability high; MSS/MSI-L = microsatellite stable/microsatellite instability low.
Median (interquartile range).
Wilcoxon rank-sum test.
χ2 test.
Fisher exact test.
DNA Extraction from Peripheral Blood and FFPE Specimens
FFPE tumor blocks were selected by the surgical pathologist for clinical testing. For MSI testing, separate tumor and normal FFPE blocks were sectioned. Only cases with 30% to 50% or more tumor present in the block were accepted for testing. Tissue was deparaffinized using xylene, ethanol washes, and acetone dehydration, and after cell lysis and proteinase K treatment, the DNA was extracted using the Puregene DNA Isolation or QIAquick PCR purification kit (QIAGEN, Inc. Valencia, CA). For certain MSI cases, when normal tissue was unavailable, peripheral blood was extracted using the EZ1 Biorobot automated extractor (QIAGEN, Inc. Valencia, CA) and used as normal DNA for comparison.
MSI Analysis
For MSI testing, 5 to 10 ng of each tumor and normal DNA was analyzed using the Promega MSI Analysis System (Promega Corp, Madison, WI) according to the manufacturer’s instructions. Amplicons were detected using capillary electrophoresis on an ABI 3130xl Genetic Analyzer and analyzed using GeneMapper Software, version 3.7 (Applied Biosystems/Life Technologies, Grand Island, NY). Nine patients with MSI-H tumors underwent further evaluation for specific MMR testing by immunohistochemical analysis or sequencing, or both, at an outside facility. Not all patients with MSI-H were tested because of patient preference, lack of adequate tissue, loss in follow-up, or BRAF p.V600E positivity.
KRAS Mutational Analysis and Sequencing
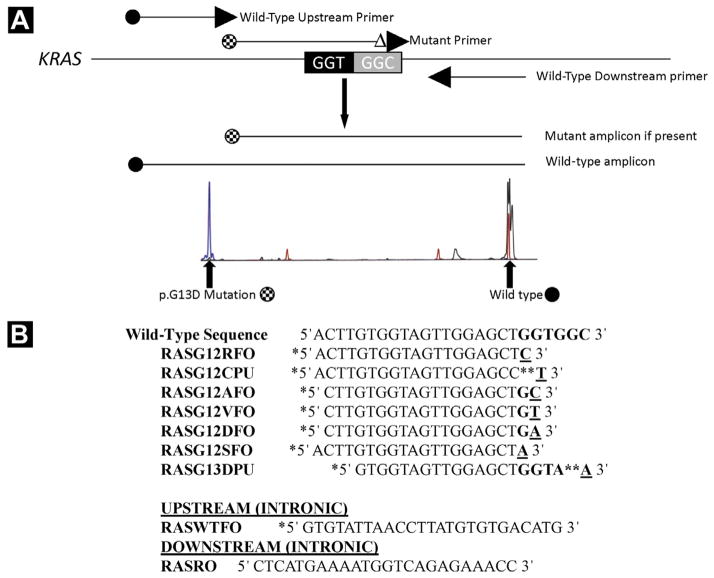
Mutations in KRAS codons 12 and 13 in exon 2 were identified using allele-specific PCR with primer sequences adapted from primers previously reported by Takeda et al or Lecomte et al (Figure 1).23,24 The 7 most common mutations in colon cancer were evaluated and included p.G12D, p.G12V, p.G12A, p.G12C, p.G12S, p.G12R, and p.G13D. Seven separate reactions were performed, each containing a fluorescently labeled upstream wild-type primer (RASWTFO), an unlabeled downstream primer (RASRO), and 1 differentially labeled fluorescent mutation-specific primer. Each primer was used at a final concentration of 0.25 μM. The reaction conditions included 1 cycle at 95°C for 5 minutes; 40 cycles at 95°C for 30 seconds, 60°C for 2 minutes, 70°C for 2 minutes; and a final extension at 70°C for 10 minutes. Amplicons were detected using capillary electrophoresis on an ABI 3130xl Genetic Analyzer (Applied Biosystems/Life Technologies, Grand Island, NY) and analyzed using GeneMapper Software (Applied Biosystems/Life Technologies, Grand Island, NY).
Figure 1.
(A) KRAS Assay Design. Seven Separate Reactions are Performed, Each Containing a Fluorescently Labeled Upstream Wild-Type Primer, an Unlabeled Downstream Primer, and 1 Differentially Labeled Fluorescent Mutation-Specific Primer. Amplicons are Detected by Capillary Electrophoresis at Approximately 176 to 180 Base Pairs (Checkered circle) if the Respective Mutation is Present (Left Peak is p.G13D). Wild-Type Amplicons (Black circle) are Detected at 267 Base Pairs in All Reactions (Right peak). (B) Primer Sequences used in The Assay (Adapted from References 23 and 24), Including 7 Mutant Allele-Specific Primers and 2 Wild-Type Primers (Upstream and Downstream). * = Fluorochrome Label; ** = Penultimate Substitution of Nucleotide to Increase Specificity
To validate our KRAS mutation–specific assay, a subset of specimens (n = 49) were sequenced using the ABI BigDye Terminator v3.1 system (Applied Biosystems/Life Technologies, Grand Island, NY) according to the manufacturer’s instructions.
BRAF p.V600E Mutational Analysis
The c.1799T>A (p.V600E) mutation was identified using allele-specific fluorescent PCR and capillary electrophoresis detection. Five to 20 ng of genomic DNA extracted from FFPE tumor was amplified using the following primers: BRAF-FO-5′ TGTTT TCCTTTACTTACTACACCTCAGA 3′ (0.05 μM final concentration), BRAF-RO 5′ HEX-CAGTGGAAAAATAGCCTCAATTCT 3′ (0.05 μM final concentration), and BRAF-Rmt 5′ FAM-A CCCACTCCATCGAGATTTCT 3′ (0.1 μM final concentration). The reaction conditions included 1 cycle at 94°C for 5 minutes; 40 cycles at 94°C for 30 seconds, 56°C for 45 seconds, 72°C for 45 seconds; and a final extension cycle at 72°C for 10 minutes. Amplicons were detected using capillary electrophoresis on an ABI 3130xl Genetic Analyzer and analyzed using GeneMapper Software (Applied Biosystems/Life Technologies, Grand Island, NY).
Statistical Analysis
Demographic and pathologic characteristics were summarized and compared across mutation group (KRAS or BRAF present or absent). Continuous characteristic age was summarized by median with 25th and 75th percentiles (interquartile ratio) and compared with the Wilcoxon rank-sum test. Frequencies and percentages were shown for the remaining categorical variables, which were compared with the χ2 test or the Fisher exact test when appropriate. Frequencies of mutation type were also provided.
Overall survival is summarized by individual categorical demographic and pathologic variables with the Kaplan-Meier method and compared with the log-rank test. Hazard ratios from the Cox proportional hazards model were presented with P values from the likelihood ratio test. An exploratory multivariate model to assess the associations between OS and risk factors was performed. This model investigates the OS difference in patients with and without mutations while accounting for differences in age, sex, MSI, stage (0–2 vs. 3–4), and tumor location (colon vs. rectum). Our sample size was small, and thus in our multivariable analysis we did not attempt to assess the differences in survival among specific different types of mutation. All statistical inferences were assessed at a 2-sided 5% significance level, and all summary statistics, graphics, and survival models were generated using R statistical software, version 2.13.25
Results
Patient Characteristics
Female patients composed 47.7% of our cohort and male patients composed 52.3%. The median age at diagnosis was 50 years, with 73.8% of patients < 60 years at diagnosis, consistent with referral for MSI testing loosely based on the Bethesda criteria. Sixty-one patients (54.9%) had stage III or IV disease, 36 (32.4%) patients had stage I or II disease, and 14 (12.6%) patients had stage 0 disease at diagnosis (defined in Patients and Methods section). Location of tumor was defined anatomically: right colon, from cecum through transverse colon (n = 36; 32.4%), left colon, from splenic flexure through sigmoid colon (n = 29; 26.1%), and rectum, beyond the sigmoid (n = 46; 41.4%). Clinical characteristics are presented in Table 1.
Allele-Specific PCR for KRAS and BRAF
All 111 patients with CRC were tested for KRAS codon 12 and codon 13 mutations and BRAF p.V600E mutations. In our population, we found an incidence of 49.5% KRAS mutations, slightly higher than that noted in previous publications.26–32 The most common KRAS mutations identified in our cohort were c.35G>A (p.G12D; 16.1%), c.35G>T (p.G12V; 12.5%), and c.38G>A (p.G13D; 10.7%) (Table 2). The c.34G>C (p.Gly12Arg) mutation was not identified in any patients in our cohort. One patient was identified with concomitant KRAS mutations (c.38G>A [p.G13D] and c.35G>C [p.G12A]); analysis of the normal tissue of this patient did not identify a mutation, indicating that both mutations were sporadic. KRAS mutations were not found to be significantly associated with location, either at 3 levels (left vs. right vs. rectum; P = .49) or 2 levels (colon vs. rectum; P = .70) (Table 3). To validate our allele-specific PCR laboratory test, 49 of the specimens were also examined by dideoxy sequencing, with KRAS mutation status (mutated or wild type) verified in 46 of 49 specimens by dideoxy sequencing. Three mutations identified by allele-specific PCR could not be confirmed by observing the dideoxy sequencing curve amplitudes alone.
Table 2.
Molecular Markers: MSI Status, KRAS Mutations, and BRAF p.V600E Mutation
| Molecular Markers | n (%) |
|---|---|
| MSI Status (n = 111)a,b | |
| High | 21 (18.9) |
| Stable | 90 (81.1) |
| KRAS Mutations (n = 111) | |
| Positive | 55 (49.5) |
| Negative | 56 (50.5) |
| Specific KRAS Mutations (n = 112)c | |
| c.35G>C (p.G12A) | 5 (4.5) |
| c.34G>T (p.G12C) | 4 (3.6) |
| c.35G>A (p.G12D) | 18 (16.1) |
| c.34G>A (p.G12S) | 3 (2.7) |
| c.35G>T (p.G12V) | 14 (12.5) |
| c.38G>A (p.G13D) | 12 (10.7) |
| Wild type | 56 (50.0) |
| BRAF c.1799T>A; p.V600E (n = 111) | |
| Positive | 8 (7.2) |
| Negative | 103 (92.8) |
| MSI-H Tumors (n = 21 of 111) | |
| KRAS positive | 8 of 21 (38.1) |
| p.G12D | 4 |
| p.G13D | 4 |
| KRAS negative | 13 of 21 (61.9) |
| BRAF positive | 4 of 21 (19.0) |
| BRAF negative | 17 of 21 (81.0) |
| MSS (n = 90 of 111) | |
| KRAS positive | 47 of 90 (52.2) |
| KRAS negative | 43 of 90 (47.8) |
| BRAF positive | 4 of 90 (4.4) |
| BRAF negative | 86 of 90 (95.6) |
Microsatellite instability low (MSI-L) and microsatellite stable (MSS) tumors were considered as a single entity in the statistical analysis.
One patient had synchronous tumors: 1 MSI-H and 1 MSS. Both were BRAF p.V600E mutation positive and KRAS mutation negative. He was considered MSI-H/BRAF mutation positive for statistical analysis.
One patient had 2 KRAS mutations (p.G12A and p.G13D). For statistical analysis, this patient was counted as having a p.G13D mutation because this is the more common mutation and was the more predominant mutation by dideoxy sequencing.
Table 3.
Association of Tumor Location and Presence of Molecular Markers (n = 111)
| Marker | Right (32.4%, n = 36) | Left (26.1%, n = 29) | Rectum (41.4%, n = 46) |
|---|---|---|---|
| MSI Statusa | |||
| MSS, n = 90 (%) | 25 (27.8) | 25 (27.8) | 40 (44.4) |
| MSI-H, n = 21 (%) | 11 (52.4) | 4 (19.0) | 6 (28.6) |
| KRAS (n = 111)b | |||
| Wild type, n = 56 (%) | 21 (37.5) | 13 (23.2) | 22 (39.3) |
| Positive, n = 55 (%) | 15 (27.3) | 16 (29.1) | 24 (43.6) |
| BRAF c.1799T>A (p.V600E)c | |||
| Wild type, n = 103 (%) | 29 (28.2) | 28 (27.2) | 46 (44.7) |
| Positive, n = 8 (%) | 7 (87.5) | 1 (12.5)d | 0 (0.0) |
Abbreviations: MSI = microsatellite instability; MSI-H = microsatellite instability high; MSS = microsatellite stable. All comparisons were done by Fisher exact test.
P = .11 at 3 locations: .22 colon vs. rectum.
P = .49 (any KRAS mutation at 3 locations); .70 (any KRAS mutation colon vs. rectum).
P = < .001 at 3 locations; P = .02 colon vs. rectum.
One patient had synchronous tumors, 1 in the right colon and 1 in the left colon. Only data for the right (MSI-H) was included in statistical analysis. Both tumors were BRAF p.V600E positive. The single BRAF p.V600E mutation (shown here) in the left colon occurred in a different patient.
Eight patients (7.2%) had BRAF p.V600E mutations, similar to the incidence in previously published reports (~8%).33,34 A majority of BRAF mutations were identified in tumors of the right colon (7 of 8) compared with the left colon (1 of 8) or rectum (0 of 8), which was statistically significant (P < .001) (Table 3). There were no concomitant BRAF and KRAS mutations, consistent with previous data suggesting that these mutations are typically mutually exclusive.8,34,35
Microsatellite Status and KRAS/BRAF Mutations
Microsatellite status was defined in accordance with the Bethesda guidelines: MSI-H was defined as 2 of 5 markers positive for MSI and low microsatellite instability (MSI-L) was defined as 1 of 5 markers positive for MSI.22 Twenty-one patients (18.9%) were identified as MSI-H and 1 patient as MSI-L. Given the lack of conclusive distinction between MSI-L and MSS tumors, we combined MSI-L and MSS for further analyses (Table 2).3 MSI status was not associated with location, at either 3 levels (left vs. right vs. rectum; P = .11) or 2 levels (colon vs. rectum; P = .22) (Table 3).
Within our MSI-H tumor population, 38.1% had KRAS mutations (8 of 21), whereas 19% (4 of 21) had a BRAF c.1799T>A (p.V600E) mutation (Table 2). Of the MSS/MSI-L tumors, 52.2% (47 of 90) had KRAS mutations and 4.4% (4 of 90) had a BRAF mutation (Table 2). Interestingly, all KRAS mutations in MSI-H tumors were a G>A transition within the second base pair of either codon 12 or 13 (c.35G>A [p.G12D]: GGT>GAT; c.38G>A [p.G13D]: GGC>GAC) (Table 2). Based on the data presented in this study, there was strong evidence (P = .006) to reject the null hypothesis that MSI-H and KRAS mutations c.35G>A or c.38G>A were independent, suggesting a possible correlation between the type of KRAS mutation and the presence of MSI.
Clinicopathologic Correlations
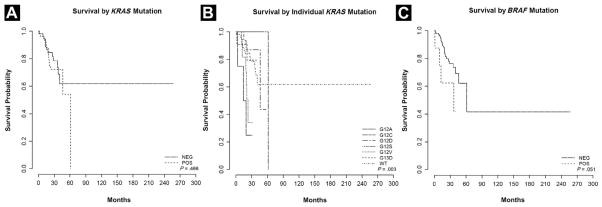
Median OS was 46.3 months. Only stage was found to be significantly associated with survival in both univariate and multivariate analyses (Table 4). Sex, age at surgery, and tumor location were not found to be associated with significant differences in survival in univariate or multivariable analysis. MSI status did not show a significant association with OS by univariate analysis (HR, 0.85; 95% Cl, 0.32–2.23; P = .73) or multivariate analysis (HR, 1.44; 95% CI, 0.52–4.00; P = .49). The presence or absence of any KRAS mutation was not found to be significantly associated with OS on univariate analysis (P = .47) (Table 4A, Figure 2A). However, when the mutations were stratified by amino acid substitution, there was a trend in effect of a specific mutation on survival (P = .003) (Figure 2B), as has been suggested by larger studies of KRAS mutation using other detection methods.36 Specifically, all 4 patients with isolated p.G12A mutations appeared to exhibit aggressive disease with either metastasis or extensive recurrence (Table 4A, Figure 2B). The fifth patient with a p.G12A mutation also had a p.G13D mutation. On dideoxy sequencing, the p.G13D mutation, a more common KRAS mutation, was the more predominant mutant allele, and this patient was classified as having a p.G13D mutation for these reasons (Table 2). This patient did not undergo resection because of other comorbidities (no pathologic staging). BRAF-mutated tumors trended toward worse OS than did KRAS-mutated tumors (P = .051), similar to previous studies (Table 4A, Figure 2C).33,34,37
Table 4A.
Univariate Survival Analysis
| Variable | % Alive after 3 Yearsa |
95% CI | Log-Rank P Value |
HR | 95% CI | LR testb P Value |
|---|---|---|---|---|---|---|
| Age | 0.99 | 0.96–1.03 | .74 | |||
| Sex | .77 | .78 | ||||
| Female | 75.4 | 63.5–89.7 | 1 | |||
| Male | 67.2 | 51.6–87.7 | 1.12 | 0.53–2.35 | ||
| Stage | <.0001 | <.0001 | ||||
| 0–2 | 90.8 | 82.6–99.9 | 1 | |||
| 3–4 | 57.0 | 42.9–75.8 | 5.03 | 1.9–13.35 | ||
| Location | .27 | .26 | ||||
| Colon | 71.9 | 60.9–84.7 | 1 | |||
| Rectum | 72.4 | 55.3–94.8 | 0.63 | 0.27–1.43 | ||
| MSI | .74 | .73 | ||||
| MSS or MSI-L | 73.1 | 63.4–84.2 | 1 | |||
| MSI-H | 72 | 50.1–100 | 0.85 | 0.32–2.23 | ||
| KRAS | .47 | .47 | ||||
| Negative | 73.6 | 60.2–89.9 | 1 | |||
| Positive | 72.2 | 60.3–86.4 | 1.32 | 0.63–2.78 | ||
| KRASa | .0030 | |||||
| Wild type | 84.5 | 75.2–95.0 | 1 | |||
| p.G12A | 25.0 | 4.6–100.0 | 6.51 | 1.78–23.73 | .0046 | |
| p.G12C | 100.0 | 100.0–100.0 | NA | No deaths | ||
| p.G12D | 87.1 | 71.8–100.0 | 0.76 | 0.21–2.67 | .66 | |
| p.G12S | 100.0 | 100.0–100.0 | 0.70 | 0.09–5.63 | .74 | |
| p.G12V | 34.1 | 12.5–92.9 | 3.60 | 1.30–9.99 | .01 | |
| p.G13D | 79.6 | 57.7–100.0 | 0.81 | 0.18–3.65 | .79 | |
| BRAF | .05 | .10 | ||||
| Negative | 73.5 | 63.6–84.8 | 1 | |||
| Positive | 62.5 | 36.5–100 | 2.79 | 0.95–8.17 | ||
| KRAS or BRAF | .10 | .10 | ||||
| Negative | 74.9 | 59.8–93.8 | 1 | |||
| Positive | 70.7 | 59.5–84.0 | 1.93 | 0.87–4.28 |
Abbreviations: CI = confidence interval; HR = hazard ratio; MSI = microsatellite instability: MSI-L = microsatellite instability low level; MSS = microsatellite stable; NA = not applicable.
For KRAS with 7 groups, the reported time is 2-year survival.
Likelihood ratio test from Cox proportional hazard model.
Figure 2.
(A) Survival Stratified by KRAS Mutation–Positive Tumors vs. KRAS Wild-Type Tumors. (B) Survival Stratified by Individual KRAS Mutations. The P Value Indicates That the Difference in Overall Survival Among Different KRAS Mutation Groups is Statistically Significant. (C) Survival Stratified by BRAF Mutation–Positive Tumors vs. BRAF Wild-Type Tumors. NEG = Negative; POS = Positive; WT = Wild Type
Discussion
As we enter an era of personalized cancer medicine, identifying mutations within patient tumors is no longer just an academic pursuit. In colorectal cancer, multiple studies have demonstrated the prognostic impact of mutations of MAP-kinase pathway components KRAS and BRAF.9,13–15,33,34 KRAS mutation analysis in patients with CRC delineates those patients who respond to EGFR inhibition, providing an additional level of treatment individualization. More recently, drug development efforts focusing on key “driver” mutations led to BRAF inhibitors that have shown impressive results in melanoma; trials of similar inhibitors in BRAF-mutant CRC are ongoing. Historically, mutation detection required Sanger sequencing. Sequencing is remarkably faster and cheaper than it was even a decade ago, but it is still hindered by the need for an adequate sample, at least 20% to 30% of which must be tumor for accurate detection. Given our growing awareness of tumor heterogeneity,38 Sanger sequencing may also miss small mutant clonal populations within tumors that could, if undetected, become the dominant population.
With the rising number of tumors that must be tested, the need for methods beyond sequencing led to the development of several other methods of mutation detection. Among these, allele-specific PCR mutation detection is a rapid, cost-effective, and specific technique that has the capacity to detect mutations from samples with only a small percentage tumor.16,17 In this study, we evaluated a cohort of patients identified by screening for MSI—not for potential KRAS or BRAF mutations—and demonstrated that allele-specific PCR identified mutations at rates close to those identified in large studies. The incidence of KRAS mutations in our study was slightly higher than seen in previous studies—approximately 50% in the current study vs. 30% to 45% in previous studies.7,9,14,26–32,34,39,40 This may result from the increased sensitivity of allele-specific PCR over that of dideoxy sequencing, or it may also suggest variation in tumor sampling or sample bias. The stratification of KRAS mutations in our cohort (most common mutations: p.G12D, p.G12V, and p.G13D) mirrors that previously reported in the literature.7,26,27 Corroborating sequencing results indicate that our test is accurate and reliable. The 3 mutations that could not be detected by dideoxy sequencing were likely present at allelic percentages less than the dideoxy sequencing limit of detection, further supporting the enhanced sensitivity of allele-specific PCR. Although all specimens were microscopically examined for adequate tumor (30%–50%), tumor heterogeneity within the block may account for either decreased tumor burden in the remaining tissue that was analyzed or smaller clonal populations containing KRAS-mutated cells.
As seen in this study, allele-specific PCR is an accurate and convenient method to assess KRAS and BRAF mutations and is known to have enhanced sensitivity relative to dideoxy sequencing, which is potentially important in identifying mutations in the context of tumor heterogeneity. Allele-specific PCR is widely used in clinical molecular diagnostic laboratories for the detection of KRAS mutations, and recently an allele-specific–based assay was approved by the US Food and Drug Administration for KRAS mutation detection (QIAGEN, Inc, Valencia, CA). However, although allele-specific PCR is highly sensitive for mutation detection (as opposed to Sanger sequencing), there are certain limitations to its use that should be noted. First, allele-specific PCR will detect only the specific mutations for which the primers were designed. The assay presented in this study was designed to detect 7 of the most common and clinically relevant mutations in codons 12 and 13, as is the current US Food and Drug Administration–approved assay. Other variant mutations that may be present in codons 12, 13, or 61 or anywhere in exons 2 or 3 of the KRAS gene would not be detected by allele-specific PCR. Conversely, these would be detected using the gold-standard Sanger sequencing of KRAS exons 2 and 3, as long as the mutant allele was present at > 15% to 20% of the mutant allele population. Another issue that can occur with allele-specific PCR is nonspecificity of the mutant primer for a nonmutated site, resulting in the presence of an artifactual mutant peak in a wild-type specimen and a false-positive result. Primers can be designed in a way that minimizes this issue, including a penultimate substitution mismatch at the 3′ end as was done with 2 primers in this assay (Figure 1).
Additionally, this study provides an opportunity to investigate further the clinical significance of KRAS mutations in the setting of MSI and MSS tumors. Eight of 21 MSI-H tumors (38%) in this cohort had concomitant KRAS mutations. Previous reports have indicated a variable percentage (6%–40%) of KRAS mutations in MSI-H–positive or HNPCC-related tumors.29,30,41 Specifically, 1 study demonstrated KRAS mutations in 40% of HNPCC-associated tumors, 22% of sporadic MSI-H tumors, and overall 31% of MMR-deficient tumors.30 Thus, KRAS mutations can be quite common in MSI tumors.
In the current study, all 8 KRAS mutations in patients with MSI-H were transition mutations (G>A) in the second position of codon 12 or 13, which was significantly more likely to occur than would be expected by chance (P = .006). Four of 21 (19.0%) MSI-H specimens were positive for p.G12D mutations and 4 of 21 (19.0%) MSI-H specimens were positive for p.G13D mutations. Comparatively, 14 of 90 (15.6%) MSS and 8 of 90 (9%) MSS specimens were positive for the p.G12D and p.G13D mutations, respectively. If one adjusts for the number of wild-type specimens and the presence of alternative KRAS mutations, the exclusivity of the combined p.G12D and p.G13D KRAS mutations becomes apparent (22 of 47 [46.8%] MSS KRAS-positive tumors are p.G12D or p.G13D mutated vs. 8 of 8 [100%] positive for a p.G12D or p.G13D mutation in MSI-H KRAS-positive tumors; Fisher exact test P = .006). In a large study evaluating KRAS mutations in MSI CRC, 81% of HNPCC-related CRC had G>A transition mutations and 76% of MSI-H CRC had G>A transition mutations, more than would be expected based on known percentages of KRAS mutations.30 Two other smaller studies also demonstrated 5 MSI-H KRAS-mutated tumors harboring exclusively p.G12D mutations.29,41 Early studies in Escherichia coli demonstrated that mutS-encoded protein complexes have high affinity for G-T mismatches.42 Such mismatches, if not repaired properly, would result in a G>A transition. The current study is small and, although not definitive, supports other findings suggesting a potential link between MSI status and specific KRAS mutations. Additional studies evaluating KRAS mutations in MSI-H tumors will be important in identifying whether the presence of a G>A transition mutation (p.G12D or p.G13D) may be useful in algorithms to identify MSI-H tumors.
Multiple studies have tried to identify the prognostic significance of individual KRAS mutations and the general presence of KRAS mutations, with contradictory results. Similar to our findings, many studies have not been able to correlate the presence of any KRAS mutation with a worse prognosis.9,34,43 Despite the lack of survival effect from KRAS mutations as a whole in this study, analysis of individual KRAS mutations suggests that specific KRAS mutations may be heterogeneous in their phenotype. Heterogeneity in behavior of specific KRAS mutations has been recently reported by De Roock et al with regard to EGFR response.44 In the current study, all 4 patients with an isolated p.G12A mutation (no other KRAS mutation) had aggressive disease (stage III or IV and extensive metastatic or recurrent disease) and 3 of the 4 patients (75%) died of their disease during the study. Similar to our studies, Span et al demonstrated that all p.G12A mutations (n = 15) were associated with metastatic disease.6 Another study found that both G>T and G>C transversions in codon 12 (p.G12V and p.G12A) were identified in metastatic tumors.45 Finally, a recent analysis of a previous study comparing panitumumab and best supportive care (BSC) vs. BSC alone revealed the p.G12A mutation to be a negative prognostic factor for OS in the BSC arm.46 Because KRAS p.G12A mutations are less common, a meta-analysis of the predictive importance of p.G12A mutations would be useful in further determining the prognostic significance of this mutation.
As in previous studies, a BRAF mutation in this cohort was associated with shorter OS.33 Additionally, we showed that BRAF mutations are overwhelmingly associated with right-sided colon cancers and are incredibly rare in rectal cancer, again confirming results presented elsewhere and supporting the validity of this data set.34,37 In 2 large rectal cancer trials, the Dutch TME trial and a retrospective study of first primary rectal cancers, BRAF was mutated in only 2.1% and 2.8% of patients, respectively.47,48 Several smaller studies, like this one, failed to identify any BRAF p.V600E mutations among rectal cancers.49,50 These data collectively suggest that BRAF mutation is a rare event in rectal carcinogenesis, and testing for BRAF mutations reflexively in the setting of KRAS negativity in rectal cancers should be carefully considered and further studied.
Finally, we found that 10 of 21 of our MSI-H tumors (48%) were present in the left colon or rectum (Table 3). Seven of 9 of our patients with documented MMR defects and a clinical history suggestive of HNPCC had tumors located in the left colon or rectum (Table 5). MSI is thought to be associated with right-sided neoplasms, a criteria that is currently used in MSI scoring analysis systems51 and is often used in deciding whether to test for MSI. Additionally, certain publications have suggested that right-sidedness should be listed as a criteria in the revised Bethesda criteria based on their findings.52 Hoogerbrugge et al found a very low incidence of MSI-positive rectal adenocarcinomas.53 Although sporadic MSI-H tumors are more often found in the right colon, our study is a reminder that MSI-H adenocarcinomas can and do occur in the left colon and rectum, often in the setting of Lynch syndrome. In a screening study for Lynch syndrome, 10 of 23 patients with mutated MMR enzymes presented with tumors in the left colon or rectum.18 Although our population was enhanced for younger patients because of specimen collection methods, the number of patients with MSI-H tumors in the left colon or rectum suggests that reflex testing for only right-sided tumors may miss a significant number of MSI-H tumors.
Table 5.
Defects in MMR-Associated Genes With KRAS Status and Tumor Location (n = 9)a
| Patient | IHC Loss | Sequencing Resultc | KRAS Status | Tumor Location |
|---|---|---|---|---|
| 1 | MLH1 and MSH2 | MSH2 c.490G>A (p.G164R) | Wild type | Rectum |
| 2 | MSH6 | MSH6 1634delAA | p.G13D | Left colon |
| 3 | MLH1, MSH2 MSH6, PMS2 | MSH2 del exons 1–7 | Wild type | Rectum |
| 4 | NA | MSH2 p.N127S MSH6 p.L585P | p.G12D | Right colon |
| 5b | MLH1/PMS2 | NA | Wild type | Rectum |
| 6 | MSH2/MSH6 | No MSH6 mutation detected | p.G12D | Rectum |
| 7 | NA | MSH6 p.R911X | p.G13D | Left colon |
| 8 | NA | MLH1 1489 insC | p.G12D | Rectum |
| 9 | NA | MSH2 del exons 1–3 | Wild type | Right colon |
Abbreviations: IHC = immunohistochemistry; MMR = mismatch repair; NA = not available.
As expected in patients with Lynch syndrome, all patients were negative for the BRAF p.V600E mutation.
Sequencing was not performed to our knowledge. The patient had 3 metachronous tumors over 30 years, the first before the age of 50 years.
These results were from outside reports, and therefore standard nomenclature could not always be used.
This study has several limitations. Relative to the RASCAL (The Kirsten Ras In-Colorectal-Cancer Collaborative Group) and similar studies, our cohort is small and is unique in its young average age at diagnosis. The small cohort makes definitive conclusions regarding individual KRAS mutations difficult; however, the observation of G>A transitions in patients with both MSI and KRAS mutations and the prognostic significance of the p.G12A mutation is intriguing. Because the cohort was identified for MSI testing to evaluate for inherited colon cancer syndromes, selection bias is possible but should not impact evaluation for KRAS/BRAF mutations. We attempted to minimize sample heterogeneity by limiting inclusion to cases with 30% to 50% or greater tumor present in the block.
In conclusion, we found that allele-specific PCR is an accurate method to identify KRAS and BRAF mutations in tumors submitted for MSI analysis from patients with CRC. Novel techniques such as allele-specific PCR are increasingly relevant to rapidly identify these mutations for clinical decision making. We observed that patients with MSI and concomitant KRAS mutations exclusively had a G>A mutation in either codon 12 or 13, which in the context of other studies, warrants further investigation regarding mechanism and impact on classification, algorithmic testing for MSI, and disease outcomes. Additionally, BRAF mutations were absent in rectal ad-enocarcinomas in this study. We hypothesize that clinical testing for BRAF p.V600E may not need to be done in rectal adenocarcinomas, but this requires further validation. Conversely, MSI-H is commonly present in left-sided tumors and therefore MSI testing should not be limited to right-sided lesions. These findings may be useful in improving algorithmic approaches to molecular analysis of CRCs and laying the groundwork for future studies on the prognostic significance of specific KRAS mutations in MSS and MSI-H tumors.
Conclusion
In this study, we identified patterns of KRAS and BRAF mutations, using allele-specific PCR, in both microsatellite-stable and -unstable CRCs that may have clinical utility in molecular diagnostics laboratories and in determining behavior of CRCs. First, we identified a rate of KRAS mutations that is higher than that often described in the literature, indicating the need for highly sensitive methods to detect KRAS mutations, such as allele-specific PCR. Although the presence of any KRAS mutation was not associated with poorer survival, similar to other studies, we identified that individual mutations may have an impact on survival, especially the p.G12A mutation. Although the presence of a KRAS mutation was not dependent on location, the p.V600E BRAF mutation was common in right-sided colon cancers, rare in left-sided colorectal cancers, and absent in rectal cancers. This raises the issue of whether testing for the BRAF mutation in rectal adenocarcinomas to identify resistance to EGFR inhibitors is beneficial or cost-effective, and requires further study. Finally, as identified in other studies, the presence of the BRAF p.V600E mutation is associated with poorer OS.
With regard to microsatellite-unstable tumors, concomitant KRAS mutations were exclusively p.G12D and p.G13D mutations, which are both G>A transition mutations in the second position of the codon. These findings, in conjunction with findings in a small number of previous studies, suggest that the mechanism of KRAS mutation is related to defective MMR enzymes. The presence of either of these 2 mutations in CRC may be useful in algorithms used to identify carcinomas that should be tested for MSI. KRAS mutations in microsatellite-unstable carcinomas were also identified in a higher percentage of MSI tumors than seen in previous studies. Finally, we found that almost half of the MSI-H tumors studied were in the left colon or rectum, indicating the need to test for MSI in both right- and left-sided neoplasms.
This study does have several limitations, including relatively small sample size and bias with regard to population studied (specimens submitted for MSI analysis). Because of this, important findings in this study should be confirmed by larger prospective studies or meta-analysis. These findings include significance of individual KRAS mutations, especially p.G12A, and the significance of recurrent KRAS G>A mutations in MSI-H tumors. Verification of these findings through additional research may help clinicians triage patients for more aggressive therapies or select individuals with certain KRAS mutations for MSI analysis.
Clinical Practice Points
Alterations in KRAS, BRAF, or MMR genes may determine therapeutic response or define a hereditary cancer syndrome. MSI resulting from defective MMR enzyme function has been shown to occur more commonly in right-sided CRCs. It is known that KRAS- and BRAF-mutated CRCs do not respond to EGFR inhibitor therapy, and mutations in these genes can be present in MSI-H tumors. However, the significance of KRAS mutations in MSI tumors is unknown, as is prognostic information of certain KRAS mutations in CRC.
We have further identified specific G>A KRAS mutations that are present in MSI-H tumors, indicating defective MMR as a possible mechanism of KRAS mutation in MSI-H tumors. We have also proposed that certain individual KRAS mutations may have prognostic significance and should be further studied in larger populations. Finally, MSI is commonly present in left-sided neoplasms, whereas BRAF mutations are not, as suggested by a few other studies. Based on these findings, algorithms used to assess whether a CRC should be tested for MSI or MMR enzyme loss should be reviewed and evaluated to consider including KRAS transition mutations G>A in their assessment. MSI or MMR status should be assessed in both right- and left-sided neoplasms, and testing for the p.V600E BRAF mutation in rectal carcinomas should be carefully considered when determining response to EGFR inhibition.
Table 4b.
Multivariable Survival Analysis
| Characteristic | HR | 95% CI | P Value |
|---|---|---|---|
| Age (Inczrement of 1 y) | 1.01 | 0.97–1.05 | .71 |
| Sex (Male vs. Female) | 1.35 | 0.62–2.90 | .45 |
| Stage (III–IV vs. 0–II) | 5.69 | 1.98–16.34 | .001 |
| Location (Rectum vs. Colon) | 0.59 | 0.25–1.38 | .22 |
| MSI (Positive vs. Negative) | 1.44 | 0.52–4.00 | .49 |
| KRAS/BRAF Mutation (Positive vs. Negative) | 1.51 | 0.68–3.37 | .32 |
Abbreviations: CI = confidence interval; HR = hazard ratio; MSI = microsatellite instability.
Footnotes
This work was presented in part in abstract form at the 2009 Association for Molecular Pathology Meeting; November 18–22, 2009; Kissimmee, FL and at the 2011 American Society of Clinical Oncology Gastrointestinal Cancers Symposium; Jan 20–22, 2011; San Francisco, CA.
Disclosure
The authors have stated that they have no conflicts of interest.
References
- 1.http://www.cancer.gov/cancertopics/types/colon-and-rectal.
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–7. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 5.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Span M, Moerkerk PT, De Goeij AF, Arends JW. A detailed analysis of K-ras point mutations in relation to tumor progression and survival in colorectal cancer patients. Int J Cancer. 1996;69:241–5. doi: 10.1002/(SICI)1097-0215(19960621)69:3<241::AID-IJC15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Bazan V, Migliavacca M, Zanna I, et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13:1438–46. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 9.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 10.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Cunningham JM, Winters JL, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–12. [PubMed] [Google Scholar]
- 12.Deng G, Bell I, Crawley S, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10(1 Pt 1):191–5. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 13.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 14.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for pan-itumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 16.Lang AH, Drexel H, Geller-Rhomberg S, et al. Optimized allele-specific real-time PCR assays for the detection of common mutations in KRAS and BRAF. J Mol Diagn. 2011;13:23–8. doi: 10.1016/j.jmoldx.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundstrom M, Edlund K, Lindell M, et al. KRAS analysis in colorectal carcinoma: analytical aspects of pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2010;10:660. doi: 10.1186/1471-2407-10-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 19.Sinicrope FA, Sargent DJ. Clinical implications of microsatellite instability in sporadic colon cancers. Curr Opin Oncol. 2009;21:369–73. doi: 10.1097/CCO.0b013e32832c94bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelmann W, Yang K, Umar A, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–77. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 23.Lecomte T, Berger A, Zinzindohoue F, et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542–8. doi: 10.1002/ijc.10526. [DOI] [PubMed] [Google Scholar]
- 24.Takeda S, Ichii S, Nakamura Y. Detection of K-ras mutation in sputum by mutant-allele-specific amplification (MASA) Hum Mutat. 1993;2:112–7. doi: 10.1002/humu.1380020209. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 26.Finkelstein SD, Sayegh R, Bakker A, Swalsky P. Determination of tumor aggressiveness in colorectal cancer by K-ras-2 analysis. Arch Surg. 1993;128:526–31. doi: 10.1001/archsurg.1993.01420170056008. discussion 531–2. [DOI] [PubMed] [Google Scholar]
- 27.Finkelstein SD, Sayegh R, Christensen S, Swalsky PA. Genotypic classification of colorectal adenocarcinoma. Biologic behavior correlates with K-ras-2 mutation type. Cancer. 1993;71:3827–38. doi: 10.1002/1097-0142(19930615)71:12<3827::aid-cncr2820711207>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 28.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Miyashita K, Ando T, et al. Exclusive KRAS mutation in microsatellite-unstable human colorectal carcinomas with sequence alterations in the DNA mismatch repair gene, MLH1. Gene. 2008;423:188–93. doi: 10.1016/j.gene.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira C, Westra JL, Arango D, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–11. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- 31.Zlobec I, Kovac M, Erzberger P, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. 2010;127:2569–75. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- 32.Benhattar J, Losi L, Chaubert P, Givel JC, Costa J. Prognostic significance of K-ras mutations in colorectal carcinoma. Gastroenterology. 1993;104:1044–8. doi: 10.1016/0016-5085(93)90272-e. [DOI] [PubMed] [Google Scholar]
- 33.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 34.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 35.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–21. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 38.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–9. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poehlmann A, Kuester D, Meyer F, Lippert H, Roessner A, Schneider-Stock R. K-ras mutation detection in colorectal cancer using the pyrosequencing technique. Pathol Res Pract. 2007;203:489–97. doi: 10.1016/j.prp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Miyaki M, Iijima T, Yamaguchi T, Kadofuku T, Funata N, Mori T. Both BRAF and KRAS mutations are rare in colorectal carcinomas from patients with hereditary nonpolyposis colorectal cancer. Cancer Lett. 2004;211:105–9. doi: 10.1016/j.canlet.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 42.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986;83:5057–61. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouzourene H, Gervaz P, Cerottini JP, et al. p53 and Ki-ras as prognostic factors for Dukes’ stage B colorectal cancer. Eur J Cancer. 2000;36:1008–15. doi: 10.1016/s0959-8049(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 44.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 45.Moerkerk P, Arends JW, van Driel M, de Bruine A, de Goeij A, ten Kate J. Type and number of Ki-ras point mutations relate to stage of human colorectal cancer. Cancer Res. 1994;54:3376–8. [PubMed] [Google Scholar]
- 46.Peeters M, Douillard JY, Van Cutsem E, et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–65. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 47.He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15:6956–62. doi: 10.1158/1078-0432.CCR-09-1165. [DOI] [PubMed] [Google Scholar]
- 48.Samowitz WS, Curtin K, Wolff RK, Tripp SR, Caan BJ, Slattery ML. Microsatellite instability and survival in rectal cancer. Cancer Causes Control. 2009;20:1763–8. doi: 10.1007/s10552-009-9410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalady MF, Sanchez JA, Manilich E, Hammel J, Casey G, Church JM. Divergent oncogenic changes influence survival differences between colon and rectal adenocarcinomas. Dis Colon Rectum. 2009;52:1039–45. doi: 10.1007/DCR.0b013e31819edbd4. [DOI] [PubMed] [Google Scholar]
- 50.Gaedcke J, Grade M, Jung K, et al. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol. 2010;94:76–81. doi: 10.1016/j.radonc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenson JK, Huang SC, Herron C, et al. Pathologic predictors of microsatellite instability in colorectal cancer. Am J Surg Pathol. 2009;33:126–33. doi: 10.1097/PAS.0b013e31817ec2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chou CL, Lin JK, Wang HS, Yang SH, Li AF, Chang SC. Microsatellite instability screening should be done for right-sided colon cancer patients less than 60 years of age. Int J Colorectal Dis. 2010;25:47–52. doi: 10.1007/s00384-009-0815-y. [DOI] [PubMed] [Google Scholar]
- 53.Hoogerbrugge N, Willems R, Van Krieken HJ, et al. Very low incidence of microsatellite instability in rectal cancers from families at risk for HNPCC. Clin Genet. 2003;63:64–70. doi: 10.1034/j.1399-0004.2003.630110.x. [DOI] [PubMed] [Google Scholar]