Abstract
Monomeric CRISPR-Cas9 nucleases are widely used for targeted genome editing but can induce unwanted off-target mutations with high frequencies. Here we describe dimeric RNA-guided FokI Nucleases (RFNs) that recognize extended sequences and can edit endogenous genes with high efficiencies in human cells. The cleavage activity of an RFN depends strictly on the binding of two guide RNAs (gRNAs) to DNA with a defined spacing and orientation and therefore show improved specificities relative to wild-type Cas9 monomers. Importantly, direct comparisons show that RFNs guided by a single gRNA generally induce lower levels of unwanted mutations than matched monomeric Cas9 nickases. In addition, we describe a simple method for expressing multiple gRNAs bearing any 5′ end nucleotide, which gives dimeric RFNs a broad targeting range. RFNs combine the ease of RNA-based targeting with the specificity enhancement inherent to dimerization and are likely to be useful in applications that require highly precise genome editing.
Zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered, regularly interspaced, short palindromic repeat (CRISPR)-CRISPR-associated (Cas) nucleases are broadly useful technologies for targeted genome editing1–3. Repair of nuclease-induced double-stranded breaks (DSBs) by non-homologous end-joining (NHEJ) or homology-directed repair (HDR) can stimulate efficient introduction of variable-length insertion/deletion mutations (indels) or specific sequence alterations, respectively. Dimeric ZFNs and TALENs recognize extended sequences consisting of two “half-sites”, each bound by one monomer, and cleave intervening spacer sequences using their dimerization-dependent FokI nuclease domains2, 3. By contrast, monomeric Streptococcus pyogenes Cas9 nuclease (hereafter referred to as Cas9) can be directed to cleave specific DNA sequences by an associated ~100 nt single guide RNA (gRNA)4, 5 bearing 17 to 20 nts of target site complementarity at its 5′ end. DNA sites to be cleaved by Cas9 must also lie next to a protospacer adjacent motif (PAM) sequence of the form 5′-NGG6.
The simplicity of the Cas9 system has made it an increasingly popular tool for research but an important concern with these monomeric nucleases, particularly for human therapeutic applications, has been the frequency and magnitude of unwanted off-target indel mutations7–9. Dimerization is an attractive strategy for improving the specificity of Cas9 nucleases. Although recent work has described a paired Cas9 nickase approach for improving specificity10–12, this system is not truly dimerization-dependent. Rather, paired nickases only require co-localization of two Cas9 nickases on a segment of DNA, which then induces high efficiency genome editing via an undefined mechanism10–12. As dimerization of Cas9 nickases is not a requirement for enzymatic activity, each single Cas9 nickase in a pair can independently nick DNA and induce mutations with potentially high efficiencies via an unknown mechanism,4, 10, 11, 13 thereby creating the risk of unwanted off-target mutations. The extent of these off-target alterations caused by the activities of single nickases remains unclear because no unbiased, genome-wide method for assessing off-target effects yet exists. Thus, to our knowledge, a truly dimeric Cas9-based system with improved specificities has yet to be described.
Here we describe RNA-guided FokI nucleases in which dimerization, rather than just co-localization, is required for efficient genome editing activity. These new nucleases can robustly induce genome editing events with high frequencies in human cells and can reduce known off-target mutations to undetectable levels as judged by sensitive deep sequencing methods. We also developed a system for expressing pairs of gRNAs bearing any 5′ end nucleotide, a method that confers a useful targeting range on this platform. Finally, we show that monomeric Cas9 nickases generally introduce more undesirable indels and point mutations (to our knowledge, a previously unknown side-effect) than the RFNs in the presence of a single gRNA. Our results define a robust, user-friendly next-generation CRISPR-based platform with the specificity advantages of a well-characterized dimeric architecture and an improved mutagenesis profile relative to paired Cas9 nickases, features that will be beneficial for research and therapeutic applications requiring the highest possible genome editing precision.
Results
Strategy for designing dimeric RNA-guided nucleases
To develop RNA-guided nucleases with increased specificity, we fused the well-characterized, dimerization-dependent wild-type FokI nuclease domain to a catalytically inactive Cas9 (dCas9) protein. We imagined that, like FokI-based ZFNs and TALENs, dimers of our fusions might mediate sequence-specific DNA cleavage when bound to target sites composed of two “half-sites” (each bound by one dCas9 monomer domain) with a certain length “spacer” sequence between them (Fig. 1a). We hypothesized that these fusions would have enhanced specificity compared to the standard monomeric Cas9 nucleases and the paired nickase system, because they should require two gRNAs for activity (Fig. 1a) and because a single gRNA would presumably be unable to recruit the two FokI-containing fusion proteins required for DNA cleavage.
Figure 1. RNA-guided FokI nucleases and a Csy4-based multiplex gRNA expression system.
(a) Schematic overview of RNA-guided FokI nucleases. Two FokI-dCas9 fusion proteins are recruited to adjacent target sites by two different gRNAs in order to facilitate FokI dimerization and DNA cleavage.
(b) Schematic overview of a Csy4-based multiplex gRNA expression system. Two gRNAs (with any 5′ end nucleotide) are co-expressed in a single transcript from a U6 promoter with each gRNA flanked by Csy4 recognition sites. Csy4 cleaves and releases gRNAs from the transcript. The Csy4 recognition site remains at the 3′ end of the gRNA with a Csy4 nuclease bound to that site.
(c) Comparison of the activities of Cas9 co-expressed with either Csy4-processed single gRNAs (blue bars) or single gRNAs expressed from a standard U6 promoter (red bars) in the U2OS.EGFP disruption assay. The target sequences in EGFP gene for the three different single gRNAs tested are provided in the table to the right of the graph. Error bars represent s.e.m, n= 3.
(d) Validation of the multiplex, Csy4-based system. Two gRNAs targeted to adjacent sites in EGFP were expressed in a single RNA transcript using the Csy4-based system in human U2OS.EGFP cells together with Csy4 and wild-type Cas9 nuclease. Sequences of indel mutations induced in these cells are shown. The wild-type sequence is shown at the top with both target sites highlighted in yellow and PAM sequences shown as red, underlined text. Deletions are indicated by red dashes against gray background and insertions by lowercase letters against a light blue background. To the right of each sequence, the sizes of insertions (+) or deletions (Δ) are specified.
Multiplex expression of gRNAs without 5′-end nucleotide limitations
In designing a dimeric RNA-guided nuclease platform, we realized that the targeting range of such a system would be very low if we used existing gRNA expression methods. Two sequence requirements typically restrict the targeting range of a dCas9 monomer: the requirement for a PAM sequence of 5′-NGG that is specified by dCas914 and a requirement for a G nucleotide at the 5′ end of the gRNA imposed by the use of a U6 promoter in most expression vectors. If the 5′ G requirement could be relieved, then the targeting range would be improved by 16-fold.
To develop a system that permits multiplex expression of gRNAs bearing any 5′ nucleotide, we designed a plasmid from which two or more gRNAs, each flanked by cleavage sites for the Csy4 ribonuclease of Pseudomonas aeruginosa 15 could be expressed within a single RNA transcribed from a single U6 promoter (Fig. 1b). Similarly to what occurs when endogenous Csy4 processes CRISPR-derived RNAs in bacteria15, co-expressed Csy4 would be expected to cleave this transcript thereby releasing the gRNAs. Based on the known mechanism of Csy4-mediated cleavage15, 16, each processed gRNA should retain a Csy4 recognition site on its 3′ end with a Csy4 protein bound to that site (Fig. 1b). Because gRNAs are precisely cleaved out from the larger transcript, this system should, in principle, permit the expression of gRNAs bearing any 5′ nucleotide.
We initially tested whether gRNAs expressed in such a Csy4-based system would be as active in human cells as those expressed from a standard U6 promoter plasmid. To do this, we directly compared the abilities of single gRNAs expressed in each system to direct Cas9-mediated indels in human cells (note that the single gRNAs expressed in the Csy4-based system in this experiment are flanked by Csy4 cleavage sites). We performed this comparison with three gRNAs targeted to sites in the EGFP reporter gene (Fig. 1c) using a well-established human cell-based EGFP disruption assay7, 13, 17 (Online Methods), which can be used to rapidly and easily quantify the induction of Cas9-mediated indels. Our results demonstrate that single gRNAs expressed in our Csy4-based system have Cas9-mediated activities in the EGFP disruption assay that are ~90% of that observed with matched gRNAs made from a standard U6 promoter vector (Fig. 1c). These results also suggest that the presence of the Csy4 recognition site at the 3′ end of the gRNA and an associated Csy4 molecule after ribonuclease processing from the larger transcript do not appear to substantially impair the ability of these gRNAs to direct Cas9-mediated cleavage in human cells.
We next sought to determine whether our Csy4-based system could be successfully used to express more than one gRNA from a single transcript. In this experiment, we used two of the three EGFP-targeted gRNAs described above that recognize sites with an edge-to-edge distance of 112 bps (Fig. 1d). Co-expression of these two gRNAs (each flanked by Csy4 recognition sites) together with Csy4 and wild-type Cas9 nuclease in human cells led to the introduction of indel mutations at both EGFP target sites as well as deletion of the sequence between these sites (Fig. 1d). These results suggest that both gRNAs can be processed from the single parental RNA transcript and are capable of directing Cas9 nuclease activities in human cells. Additionally, the identification of alleles bearing mutations at both sites or deletion of the sequence in between the two sites demonstrates that both gRNAs are being expressed within the same cells.
Construction and optimization of dimeric RNA-guided nucleases
We constructed two different hybrid proteins: one in which the FokI nuclease domain is fused to the carboxy-terminus of dCas9 (dCas9-FokI) and the other in which it is fused to its amino-terminus (FokI-dCas9) (Fig. 2a). The dCas9-FokI protein is analogous in architecture to ZFNs and TALENs (Fig. 2a). For both fusions, we used a five amino acid linker with the sequence GGGGS to connect the two domains and the dCas9 domain was codon optimized for expression in zebrafish and human cells. We used the EGFP disruption assay to ascertain whether either or both of these fusions could mediate site-specific cleavage of DNA in human cells. Because we did not know the specific half-site orientations or spacings required for efficient cleavage, we designed 60 pairs of gRNAs targeted to various sites in EGFP. The two half-sites targeted by each of these gRNA pairs were oriented such that both of their PAM sequences are either directly adjacent to the spacer sequence (the “PAM-in” orientation) or positioned at the outer boundaries of the full-length target site (the “PAM-out” orientation) (Fig. 2b). The spacer sequence was also varied in length from 0 to 31 bps for both the PAM-in and PAM-out orientations (Fig. 2b and Supplementary Table 1).
Figure 2. Design and optimization of RNA-guided FokI nucleases.
(a) Schematic illustrations of a ZFN, TALEN, FokI-dCas9 fusion and dCas9-FokI fusion.
(b) Screening the EGFP disruption activities of FokI-dCas9 fusion with gRNA pairs targeted to half-sites in one of two orientations: PAM-in (left panel) and PAM-out (right panel). Half-sites were separated by spacer sequences of variable lengths ranging from 0 to 31 bps. EGFP disruption was quantified by flow cytometry, n = 1. Corresponding data for the dCas9-FokI fusion and the same gRNA pairs is shown in Supplementary Fig. 1. Note the background level of EGFP-negative cells that can be observed with the control cell sample. Control shown is the same experimental sample in both of these bar graphs and in those of Supplementary Fig. 1 and is presented redundantly in each of these graphs for ease of comparison.
(c) Additional assessment of FokI-dCas9-mediated EGFP disruption activities on target sites with half-sites in the PAM-out orientation and with spacer lengths ranging from 10 to 20 bp. EGFP disruption was quantified by flow cytometry. Values shown represent the mean of duplicate measurements. Raw values of the EGFP disruption experiments can be found in Supplementary Table 2.
(d) Mean EGFP disruption values of the data from (c) grouped according to spacer length. Error bars represent s.e.m.
The dCas9-FokI protein did not show detectable EGFP disruption activity above the background level of the assay when co-expressed with any of the 60 gRNA pairs in human U2OS.EGFP cells (Supplementary Fig. 1). However, screening of the FokI-dCas9 protein with the same 60 gRNA pairs did reveal detectable EGFP disruption activity on target sites composed of half-sites in the PAM-out orientation and with spacer lengths of 13 to 17 bps, with some lower level activity also visible at the 26 bp spacer length (Fig. 2b). Further testing of FokI-dCas9 on an additional 25 target DNA sites with spacer lengths ranging from 10 to 20 bps and with half-sites in the PAM-out orientation demonstrated efficient cleavage on targets with spacer lengths of 13 to 18 bps (Figs. 2c and 2d; Supplementary Table 2). In these experiments, we were only able to test one site each for spacer lengths of 17 or 18 bps and not all sites with a 13 bp spacer length showed activity. Analysis of a subset of these successfully targeted sites using T7 Endonuclease I (T7EI) assay (Online Methods) and Sanger sequencing (Online Methods) confirmed the presence of indels at the intended location (data not shown). We conclude that FokI-dCas9 can be directed by two appropriately positioned gRNAs to efficiently cleave a full-length target site of interest. For simplicity, we refer to the complex of two FokI-dCas9 fusions and two gRNAs as RNA-guided FokI Nucleases (RFNs).
To extend our initial findings with the EGFP reporter gene and to ascertain whether RFNs could be used to perform routine genome editing of endogenous human genes, we designed gRNA pairs for 12 different target sites in nine different human genes (Table 1). For these experiments, we used a FokI-dCas9 fusion that we had further optimized for activity by adding nuclear localization signal (NLS) sequences to its amino- and carboxy-terminal ends and by codon-optimizing the FokI domain for expression in human cells (data not shown and Online Methods). Eleven of the 12 RFNs we tested introduced indels with high efficiencies (range of 3 to 40%) at their intended target sites in human U2OS.EGFP cells as judged by T7EI assay (Table 1). Similar results were obtained with these same 12 RFN pairs in HEK293 cells (Table 1). Sanger sequencing of successfully targeted alleles from U2OS.EGFP cells revealed the introduction of a range of indels (primarily deletions with only a small number of insertions) at the expected cleavage site (Supplementary Fig. 2). Direct comparison experiments performed at three of the endogenous gene sites show that the indel frequencies induced by RFNs are only somewhat lower than those observed with wild-type Cas9 nuclease and single gRNAs from those pairs (average of ~82%; Supplementary Fig. 3). Taken together, our experiments performed in two different human cell lines demonstrate the robustness of RFNs for modifying endogenous human genes.
Table 1.
Mutation frequencies induced by RFNs at 12 endogenous human gene sites in human U2OS and HEK293 cells.
| Target name | Target site sequence (half-sites in CAPS, spacer in lowercase, PAM underlined) | Spacer Length | U2OS Mutation Frequency (%)± SEM | HEK293 Mutation Frequency (%)± SEM |
|---|---|---|---|---|
| APC | CCGGGCGGCGCTCGTACTTCTGG ccactgggcgagcgtc TGGCAGGTGAGTGAGGCTGCAGG | 16 | 34.6 ± 2.5 | 15.8 ± 0.5 |
| BRCA1 | CCGAAGCTGACAGATGGGTATTC tttgacggggggtagg GGCGGAACCTGAGAGGCGTAAGG | 16 | 14.1 ± 3 | 20.7 ± 2.1 |
| EMX1 | CCACTCCCTGGCCAGGCTTTGGG gaggcctggagtcatg GCCCCACAGGGCTTGAAGCCCGG | 16 | 3 ± 0.6 | 2.9 ± 0.1 |
| FANCF - site 1 | CCAAGGTGAAAGCGGAAGTAGGG ccttcgcgcacctcat GGAATCCCTTCTGCAGCACCTGG | 16 | 35.7 ± 0.8 | 17.8 ± 0.8 |
| FANCF - site 2 | CCCATTCGCACGGCTCTGGAGCG gcggctgcacaaccag TGGAGGCAAGAGGGCGGCTTTGG | 16 | 27 ± 1.3 | 12 ± 0.3 |
| GLI1 | CCCACCACCAATCAGTAGCTATG gcgagccctgctgtct CCGGCCCCTCCCCAGTCAGGGGG | 16 | 7.4 ± 1.6 | 6.2 ± 0.8 |
| MLH1 | CCGTTGAGCATCTAGACGTTTCC ttggctcttctggcgc CAAAATGTCGTTCGTGGCAGGGG | 16 | 27.9 ± 1.6 | 15.7 ± 0.9 |
| RARA | CCGCTTGGCATGGCCAGCAACAG cagctcctgcccgaca CCTGGGGGCGGGCACCTCAATGG | 16 | 3.8 ± 1.1 | 6.1 ± 0.2 |
| SS18 | CCGGGCCCGGAGTCGACCGGGCC gaggcggaggcgggcc TGCTGGGAATCAGCAGTGTTTGG | 16 | not detected | not detected |
| VEGFA - site 1 | CCAGGAGCAAACTCCCCCCACCC cctttccaaagcccat TCCCTCTTTAGCCAGAGCCGGGG | 16 | 25.3 ± 3.5 | 11.9 ± 0.4 |
| VEGFA - site 2 | CCACCTCCTCCCCGGCCGGCGGC ggacagtggacgcggc GGCGAGCCGCGGGCAGGGGCCGG | 16 | 5 ± 1.7 | 6.2 ± 0.1 |
| VEGFA - site 3 | CCAGAGCCGGGGTGTGCAGACGG cagtcactagggggcg CTCGGCCACCACAGGGAAGCTGG | 16 | 40 ± 1.5 | 19.9 ± 0.5 |
Mutation frequencies were measured by T7EI assay with means of triplicate measurements shown (n = 3). SEM = Standard errors of the mean.
RFNs possess specificities for extended cleavage sites
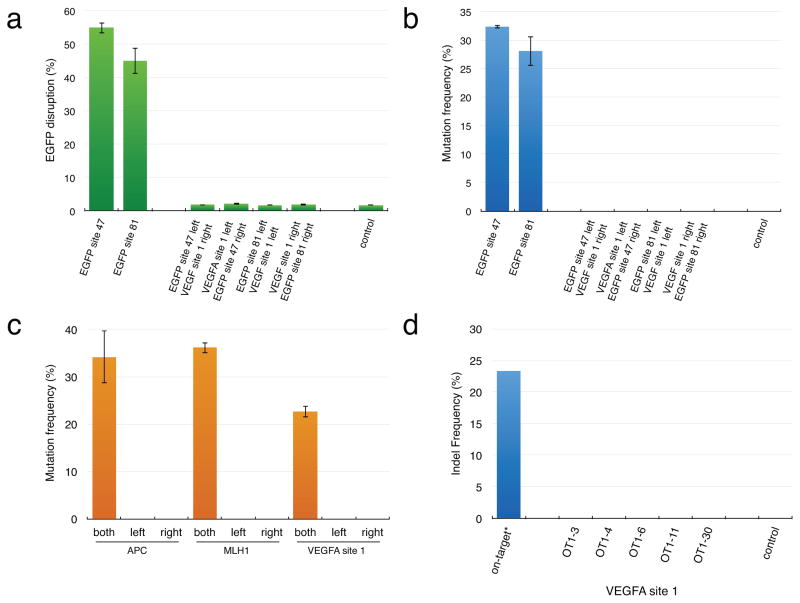
To test whether RFNs possess enhanced recognition specificities expected with dimerization, we examined whether these nucleases strictly depend upon the presence of both gRNAs in a pair. For this we used two pairs of gRNAs that we had shown could efficiently direct FokI-dCas9-induced indels to their target sites in the EGFP reporter gene (EGFP sites 47 and 81) in human U2OS.EGFP cells (Fig. 2c above). Replacement of one or the other gRNA in each of these two pairs with another targeted to an unrelated site in the human VEGFA gene resulted in reduction of EGFP disruption activity to a level observed with a negative control (Fig. 3a) and reduction of targeted mutations to undetectable levels as judged by T7EI assays (Fig. 3b). We performed a similar experiment using gRNA pairs we had shown could efficiently introduce FokI-dCas9-mediated indels in the human APC, MLH1 and VEGFA genes (Table 1) and again observed loss of detectable RFN-induced indels by T7EI assay when only one of the two gRNAs in a pair was present (Fig. 3c). These results demonstrate that efficient induction of genome editing by an RFN requires two appropriately targeted gRNAs.
Figure 3. Dimerization of RFNs is required for efficient genome editing activity.
(a) EGFP disruption activities of two RFN pairs assessed in the presence of correctly targeted gRNA pairs (to EGFP sites 47 and 81) and pairs in which one or the other of the gRNAs has been replaced with another gRNA targeted to a non-EGFP sequence (in the VEGFA gene). EGFP disruption was quantified by flow cytometry. EGFP, Enhanced Green Fluorescent Protein; VEGFA, Vascular Endothelial Growth Factor A. Error bars represent standard errors of the mean (s.e.m.), n = 3.
(b) Quantification of mutagenesis frequencies by T7EI assay performed with genomic DNA from the same cells used in the EGFP disruption assay of (a). Error bars represent s.e.m., n = 3.
(c) Activities of RFNs targeted to sites in the APC, MLH1 and VEGFA genes. For each target, we co-expressed FokI-dCas9 with a pair of cognate gRNAs, only one gRNA for the “left” half-site, or only one gRNA for the “right” half-site. Rates of mutagenesis were measured by T7E1 assay. APC, Adenomatous polyposis coli; MLH1, mutL homolog 1; VEGFA, Vascular Endothelial Growth Factor A. Error bars represent s.e.m., n = 3.
(d) Mutagenesis frequencies of RFNs targeted to VEGFA site 1 at the on-target site and at five previously known off-target (OT) sites for one of the gRNAs used to target VEGFA site 1. Frequencies of mutation were determined by deep sequencing and these data are also shown in Supplementary Table 3. Each value reported was determined from a single deep sequencing library prepared from genomic DNA pooled from three independent transfection experiments. The value shown for the on-target VEGFA site 1 (marked with an asterisk) is the same as the one shown in Fig. 4a below and is only shown here for ease of comparison with the values presented in this figure.
Given the dependence of RFNs on the expression of two gRNAs, we hypothesized that their mutagenic effects on known off-target sites of single gRNAs in the pair should be negligible. However, performing these direct comparisons requires knowledge of the Cas9-induced off-target sites of single gRNAs in the pair. We had previously identified five off-target sites for one of the gRNAs we used to target the dimeric RFN site in the human VEGFA gene7, 13. Using deep sequencing, we found that the frequency of indel mutations induced by the dimeric RFNs was indistinguishable from background at all five off-target sites (Fig. 3d and Supplementary Table 3; these are the same cells we used for the T7EI assay shown in Fig. 3c). These results demonstrate that the use of RFNs can essentially eliminate Cas9-induced off-target effects of a single gRNA and are consistent with our observations that a single gRNA co-expressed with FokI-dCas9 does not efficiently induce indels. At present, it is not possible to perform these direct comparisons for additional sites – such experiments will have to await the identification of off-target sites for more single gRNA sites that can also target the half-site of a dimeric RFN. Nonetheless, we conclude that dimeric RFNs have enhanced specificities relative to standard monomeric Cas9 nucleases.
Monomeric Cas9 nickases mutagenize more efficiently than single gRNA/FokI-dCas9 complexes
As noted above, an important weakness of the paired Cas9 nickase approach is that single monomeric nickases can introduce indel mutations with high frequencies at certain target sites4, 10, 11, 13. This lack of dimerization-dependence in the paired Cas9 nickase system is a potential source of off-target effects, with each of the two monomeric nickases having the ability to create unwanted indel mutations elsewhere in the genome. We hypothesized that because FokI nuclease activity is dimerization-dependent, our FokI-dCas9 fusions should generally show less undesirable indel activity in the presence of only one gRNA when compared directly with monomeric Cas9 nickases.
To test this hypothesis, we compared the activities of FokI-dCas9 and Cas9 nickase in the presence of a single gRNA at six dimeric human gene target sites (a total of 12 gRNAs for 12 half-sites; Supplementary Table 4). We chose these six particular sites because we had previously determined that monomeric Cas9 nickases directed by just one and/or the other gRNA in these pairs could induce indel mutations at these targets (data not shown). Using deep sequencing, we assessed the genome editing activities of FokI-dCas9 or Cas9 nickase in the presence of both or only one or the other gRNA. We found that FokI-dCas9 and Cas9 nickase induced indels at all six target sites with high efficiencies in the presence of two gRNAs (Supplementary Table 4). As expected, monomeric Cas9 nickases directed by the 12 single gRNAs induced indels with frequencies ranging from 0.0048% to 3.04% (mean of 1.02%) (Fig. 4a and Supplementary Table 4). By contrast, FokI-dCas9 directed by the same 12 single gRNAs induced indels at lower frequencies ranging from 0.0045% to 0.473% (mean of 0.15%) (Fig. 4a and Supplementary Table 4). Comparing these data directly, FokI-dCas9 induced indels with lower frequencies than Cas9 nickase for 10 of the 12 single gRNAs (Fig. 4a and Supplementary Table 4). In addition, FokI-dCas9 showed greater fold-reductions in indel frequencies than Cas9 nickase at 11 of the 12 half-sites when comparing single gRNA rates to the paired gRNA rates (Fig. 4b).
Figure 4. Mutagenic activities of a Cas9 nickase or FokI-dCas9 co-expressed with a single gRNA.
(a) Indel mutation frequencies induced by FokI-dCas9 (blue bars) or Cas9 nickase (red bars) in the presence of one or two gRNAs targeted to six different human gene sites. For each gene target, we assessed indel frequencies with both gRNAs, only one gRNA for the “left” half-site, or only the other gRNA for the “right” half-site. Mutation frequencies were determined by deep sequencing and this data is presented in Supplementary Table 4. Each indel frequency value reported was determined from a single deep sequencing library prepared from genomic DNA pooled from three independent transfection experiments. VEGFA, Vascular Endothelial Growth Factor A; DDB2, Damage-Specific DNA Binding Protein 2; FANCF, Fanconi Anemia, Complementation Group F; FES, Feline Sarcoma Oncogene; RUNX 1, Runt-Related Transcription Factor 1.
(b) Data from (a) presented as a fold-reduction in the indel frequency comparing values obtained for each target site with a gRNA pair to each of the single gRNA experiments or to the control experiment (no gRNA and no Cas9 nickase or FokI-dCas9). This fold-reduction was calculated for both FokI-dCas9 (blue bars) and Cas9 nickase (red bars).
Our deep sequencing experiments also uncovered a previously undescribed side-effect of certain monomeric Cas9 nickases: the introduction of point mutations at particular positions within their target sites. Cas9 nickase co-expressed with a single gRNA for the “right” half-site of the VEGFA target induced base substitutions at position 15 of the recognition site at a frequency of 10.5% (Fig. 5a). Similar results were observed with Cas9 nickase and single gRNAs directed to the “right” half-site of FANCF target site 1 (mutation frequency of 16.3% at position 16) (Fig. 5b) and to the “right” half-site of the RUNX1 target site (mutation frequency of 2% at position 17) (Fig. 5c). We also detected these mutations at the VEGFA and FANCF1 target sites using Sanger sequencing, demonstrating that these base substitutions are not artifacts caused by the use of a deep sequencing method (Supplementary Fig. 4). In addition, mutations at these positions are not seen above background levels in control samples in which no Cas9 nickase or gRNA are expressed in the cell (Figs. 5a–5c). These point mutations are observed at much lower frequencies (five to 100-fold lower) in cells that express FokI-dCas9 protein and the same gRNAs (Fig. 5a–c), suggesting that these mutations are more efficiently induced by Cas9 nickases. Overall, we conclude that single Cas9 nickases can, at certain sites, induce mutagenic indel and point mutations with high frequencies, an effect that is not observed with FokI-dCas9 nucleases directed by matched single gRNAs.
Figure 5. Single Cas9 nickases can introduce point mutations with high efficiencies into their target sites.
Frequencies of different point mutations found at each position in half-sites targeted by single gRNAs for (a) VEGFA, (b) FANCF and (c) RUNX1 gene targets in the presence of FokI-dCas9, Cas9 nickase or a tdTomato control. Mutation frequencies were determined by deep sequencing. Each point mutation value reported was determined from a single deep sequencing library prepared from genomic DNA pooled from three independent transfection experiments. Note that the genomic DNA used for these experiments was isolated from the same cells analyzed for indel mutations in Fig. 4. VEGFA, Vascular Endothelial Growth Factor A; FANCF, Fanconi Anemia, Complementation Group F; RUNX 1, Runt-Related Transcription Factor 1.
Discussion
In this report, we have described customizable dimeric RNA-guided nucleases for performing robust and highly specific genome editing in human cells. A key advantage of our RFNs is that, in contrast to standard monomeric Cas9 nucleases, they function as dimers and require two gRNAs for enzymatic activity. The engineering of an active RFN required fusion of the FokI nuclease domain to the amino-terminal end of the dCas9 protein, an architecture different from ZFNs and TALENs in which the FokI domain is fused to the carboxy-terminal end of engineered zinc finger or transcription activator-like effector repeat arrays. RFNs also require that the half-sites bound by each Fok-dCas9/gRNA complex have a particular relative orientation (PAMs-out) with a restricted intervening spacer length of 14 to 17 bps (although activity may be possible at additional spacings but with less consistent success). In retrospect, it is now possible to rationalize why the FokI-dCas9 architecture worked in light of recent structural information about the dCas9/gRNA/DNA complex18, 19, though we did not have these insights at the time we designed and optimized the activity of our RFNs.
A key component of our RFN platform is the Csy4-based methodology for expressing pairs of gRNAs processed from a single transcript. This method enables multiple gRNAs bearing any nucleotide at their 5′ ends to be expressed, in contrast to standard vectors that utilize a U6 promoter and therefore require a 5′ G for efficient expression of a single gRNA. The relief of this 5′-end sequence restriction is critically important because it enables a more reasonable RFN targeting range of at least one site in every 64 bps of random DNA sequence (assuming that the two half-sites are spaced by 14–17 bps) rather than once in every 1024 bps of random DNA sequence (Supplementary Disucssion). Beyond improving RFN targeting range, our multiplex gRNA system might also be used to enable the expression of gRNAs from RNA polymerase II promoters, thereby providing potential opportunities for inducible and/or cell-type-specific control. Our Csy4-based system should also prove generally useful for any application requiring the expression of two or more gRNAs (e.g.—the targeting of multiple dCas9-activator proteins to achieve synergistic upregulation of gene expression20–22). Other systems, such as a recently described ribozyme-based system23, might also provide alternative methods for accomplishing similar goals.
The dimeric nature of RFNs provides important specificity advantages relative to standard monomeric Cas9 nucleases. Our data demonstrate that FokI-dCas9 directed by a single gRNA induces very little or no mutagenesis at RFN half-sites. Given that the FokI nuclease domain must dimerize to cleave DNA, we presume that any indels we observe with a single gRNA are likely due to recruitment of two FokI-dCas9 molecules to the DNA. Regardless of mechanism, given that we observed only very low level or no mutagenesis when testing FokI-dCas9 with single gRNAs at 12 on-target half-sites, it is very unlikely that any mutagenesis will be induced at partially mismatched, off-target half-sites. Consistent with this, we confirmed that a RFN targeted to VEGFA did not induce detectable mutations at known off-target sites of one of the gRNAs as judged by deep sequencing.
We also expect that RFNs will not induce appreciable off-target mutations as dimers in human cells. RFNs, directed by a pair of gRNAs to cleave a full-length sequence composed of two half-sites, would be expected to specify up to 44 bps of DNA in their target sites. A sequence of this length will, by chance, almost always be unique (except in certain circumstances where the target might lie in duplicated genome sequence). In addition, the most closely matched sites in the genome to this longer dimeric site should, in most cases, possess a large number of mismatches, which in turn would be expected to minimize or abolish cleavage activity by an RFN dimer. For example, we computationally identified all sites in the human genome that bear 0 to 16 mismatches (and that allow for spacers of length 14 to 17 bps) for the 15 full-length sequences we successfully targeted with RFNs in this study (Online Methods). This analysis found that all 15 target sequences were unique and that the most closely matched sites harbored 7 to 12 mismatches (Supplementary Table 5). As expected, deep sequencing experiments did not detect indel mutations above background at any of the eight most closely mismatched sites for three of these target sequences (Supplementary Table 6). It will be interesting in future studies to examine more potential genomic off-target sites for additional dimeric RFNs targeted to endogenous gene targets. Overall, we conclude that dimeric RFNs should possess a high degree of specificity in human cells but the ultimate characterization of specificity will await the development of unbiased methods that can comprehensively define RFN specificity across the entire genome.
Because RFNs are a true dimeric system, they possess a number of important advantages over co-localized paired Cas9 nickases, which do not possess dimerization-dependent activities. First, direct comparisons show that single Cas9 nickases generally induce unwanted indel mutations with higher rates than FokI-dCas9 fusion proteins directed by the same individual gRNAs. Second, monomeric Cas9 nickases can in some cases also induce unwanted base pair substitutions into their target sites with high efficiencies, to the best of our knowledge a previously unknown mutagenic side-effect that we uncovered in this study (see additional discussion below). Again, Cas9 nickases induce these unwanted point mutations at substantially higher rates than FokI-dCas9 fusions guided by the same matched single gRNAs. Third, paired Cas9 nickases show greater promiscuity in the orientation and spacing of target half-sites than dimeric RFNs and therefore have a greater potential range of sites at which off-target mutations might be induced. Paired nickase half-sites can be oriented in a PAM-in or PAM-out configuration and with spacer sequences ranging in length from 0 to 1000 bps10–12. By contrast, RFNs are much more stringent in their specificities --half-sites must be in a PAM-out orientation and must be spaced apart by 14 to 17 bps, a requirement presumably enforced by the need to appropriately position two FokI cleavage domains to allow efficient dimerization.
The mechanism of the induction of base substitution mutations within the recognition sequence of certain sites when using single or paired Cas9 nickases is presently unclear. For all three sites at which we observed these mutations, the base that is most efficiently altered falls within a strand-separated region of the target DNA, which has been reported to be susceptible to P1 nuclease in vitro in a dCas9/gRNA/target DNA complex19. Perhaps DNA distortions induced by binding of Cas9 might trigger the base excision repair pathway, leading to mutagenesis. However, our observation that FokI-dCas9 directed by a single gRNA appears to induce these mutations at much lower frequencies suggests that strand separation alone may not be sufficient to induce base substitutions and that perhaps the nick induced by Cas9 nickases enhances the mutagenic effect. Notably, for all three cases, the base position mutagenized within the binding site is a cytosine preceded by a thymine on the non-complementary strand, perhaps suggesting the involvement of APOBEC proteins that preferentially recognize and deaminate cytosines at TC sequences and that can be overexpressed in transformed cancer cell lines24, 25. Further studies will be needed to clarify the mechanism by which these substitutions are induced by Cas9 nickases and to determine whether this effect might also occur in non-cancer cell lines.
In the course of developing RFNs, we have created software tools that should be useful for applying the technology. To aid in identification of potential full-length RFN target sites, we have added a new module to our existing ZiFiT Targeter software26 (http://zifit.partners.org) that can search user-defined DNA sequences. The output from ZiFiT Targeter also provides the sequences of oligonucleotides that need to be synthesized and cloned for expression of specific gRNAs in our Csy4-based system. In addition, we developed a software program to identify potential off-target sites of RFN dimers in the human genome based on mismatches to the full-length target site. This program can be used to determine whether a given dimeric full-length RFN target site is unique in the human genome as well as to provide information about the orthogonality of any given site relative to the rest of the genome. We have made this program freely available for download at this website: https://bitbucket.org/vishalthapar/casper-scan.
As the mechanism of genome editing by hybrid proteins harboring FokI nuclease domains has been well-studied and extensively characterized, further improvements in specificity can potentially be added to the basic RFN architecture described here by making well-defined alterations to the FokI domain. For example, the residual mutagenic activity observed in the presence of single gRNAs might be further reduced or eliminated by constructing RFNs that harbor obligate heterodimeric FokI domains27–29 and other orthologous Cas9/gRNA pairings30. Additionally, strategies used to improve the specificity of monomeric Cas9 nucleases (e.g.—truncated gRNAs recently described by our laboratory13) might also be used to eliminate any potential residual off-target effects induced by RFNs. The improved specificities of RFNs should make them broadly useful for research and therapeutic applications requiring high precision genome editing.
Online Methods
FokI-dCas9 and gRNA expression plasmids
Csy4 and FokI-dCas9 were co-expressed from a CAG promoter as a single protein separated by a self-cleaving T2A peptide. When expressed from the optimized plasmid vector (pSQT1601), the FokI-dCas9 fusion protein is also flanked by SV40 nuclear localization signal (NLS) sequences present on both its N-and C-termini following T2A cleavage. The FokI nuclease and dCas9 domains were joined by a five amino acid linker of sequence GGGGS. The Csy4 protein expressed in this system does not have an appended NLS sequence. Schematic maps, DNA sequences and protein sequences of these various components are provided in Supplementary Figs. 5 and 6.
Plasmids encoding single or multiplex gRNAs were assembled in a single-step ligation of annealed target site oligosduplexes (Integrated DNA Technologies) (Supplementary Table 7 and Supplementary Fig. 7) and a constant region oligoduplex (for multiplex gRNAs) with BsmBI-digested Csy4-flanked gRNA backbone (pSQT1313; to be deposited with Addgene http://www.addgene.org) (Supplementary Fig. 8).
Tissue culture and transfections
All cell culture experiments were carried out in HEK 293 cells, U2OS cells or in U2OS cells harboring a stably integrated, single-copy, destabilized EGFP gene (U2OS.EGFP cells). Cell lines were cultured in Advanced DMEM (Life Technologies) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies) and penicillin/streptomycin at 37C with 5% CO2. Additionally, U2OS.EGFP cells were cultured in the presence of 400 μg/ml of G418.
U2OS cells and U2OS.EGFP cells were transfected using the DN-100 program of a Lonza 4D-Nucleofector according to the manufacturer’s instructions. In Csy4 optimization experiments, 500 ng of pCAG-Csy4-T2A-Cas9 (pSQT834) and 250 ng of gRNA encoding plasmids were transfected with 50 ng of tdTomato expression plasmid (Clontech) as a transfection control. In initial FokI-dCas9 activity screens and focused spacer length analysis experiments, 750 ng of pCAG-Csy4-FokI-dCas9-nls nuclease plasmid (pJAF1484) and 250 ng of gRNA encoding plasmids were transfected together with 50 ng tdTomato expression plasmid as a transfection control. In all other experiments in U2OS and U2OS.EGFP cells, 975 ng of human codon optimized pCAG-hCsy4-T2A-nls-hFokI-dCas9-nls (pSQT1601) or pCAG-Csy4-T2A-Cas9-D10A nickase (pNW3) were transfected along with 325 ng of gRNA vector and 10 ng of tdTomato expression plasmid and analyzed 3 days after transfection. HEK293 cells were transfected with 750 ng of pSQT1601 nuclease plasmid, 250 ng of gRNA expression plasmid and 10 ng of Td tomato, using Lipofectamine (Life Technologies) according to the manufacturer’s instructions and analyzed for NHEJ-mediated mutagenesis 3 days after transfection.
Single transfections were performed for the initial spacer activity screen, duplicate transfections for the focused spacer length analysis and quadruplicate transfections for the comparison of the activity of FokI-dCas9 with wildtype Cas9. All other transfections were performed in triplicate.
EGFP disruption assay
The EGFP disruption assay was performed as previously described17 using U2OS.EGFP reporter cells. Cells were assayed for EGFP and tdTomato expression using an BD Biosciences LSR II or Fortessa FACS analyzer.
Quantification of nuclease- or nickase-induced mutation rates by T7EI assay
T7E1 assays were performed as previously described17. Briefly, genomic DNA was isolated 72 hours post transfection using the Agencourt DNAdvance Genomic DNA Isolation kit (Beckman Coulter Genomics) according to the manufacturer’s instructions with a Sciclone G3 liquid-handling workstation (Caliper). PCR reactions to amplify genomic loci were performed using Phusion Hot-start Flex DNA polymerase (New England Biolabs). Samples were amplified using a two-step protocol (98 °C, 30 sec; (98 °C, 7 sec; 72 °C, 30 sec) × 35; 72 °C, 5 min) or a touchdown PCR protocol ((98 °C, 10 s; 72–62 °C, −1 °C/cycle, 15 s; 72 °C, 30 s) × 10 cycles, (98 °C, 10 s; 62 °C, 15 s; 72 °C, 30 s) × 25 cycles). 200 ng of purified PCR amplicons were denatured, hybridized and treated with T7 Endonuclease I (New England Biolabs). Mutation frequency was quantified using a Qiaxcel capillary electrophoresis instrument (Qiagen) as previously described17.
Sanger sequencing of mutagenized genomic DNA
The same purified PCR products used for T7EI assay were Topo-cloned (Life Technologies) and plasmid DNA of individual clones was isolated and sequenced using an M13 reverse primer (5′-GTAAAACGACGGCCAG-3′).
Illumina Library Preparation and Analysis
Short 200–350 bp PCR products were amplified using Phusion Hot-start FLEX DNA polymerase. PCR products were purified using Ampure XP beads (Beckman Coulter Genomics) according to manufacturer’s instructions. Dual-indexed TruSeq Illumina deep sequencing libraries were prepared using a high-throughput library preparation system (Kapa Biosystems) on a Sciclone G3 liquid-handling workstation. Final adapter-ligated libraries were quantified using a Qiaxcel capillary electrophoresis instrument (Qiagen). 150 bp paired end sequencing was performed on an Illumina MiSeq Sequencer by the Dana-Farber Cancer Institute Molecular Biology Core.
MiSeq paired-end reads were mapped to human genome reference GChr37 using bwa31. Reads with an average quality score >30 were analyzed for insertion or deletion mutations that overlapped the intended target or candidate off-target nuclease binding site. Mutation analyses were conducted using the Genome Analysis Toolkit (GATK) and Python.
Off-target search algorithm
We implemented a target-site matching algorithm that looks for matches with less than a specified number of mismatches in a sliding window across the human genome. This program can be used to identify potential dimeric off-target sites and is available for download at: https://bitbucket.org/vishalthapar/casper-scan.
Supplementary Material
Acknowledgments
We thank Y. Fu and M. Maeder for helpful discussions and Y. Fu, J. Angstman and B. Kleinstiver for comments on the manuscript. This work was funded by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378), NIH R01 GM088040, NIH P50 HG005550, and the Jim and Ann Orr Massachusetts General Hospital (MGH) Research Scholar Award. This material is based upon work supported by, or in part by, the U. S. Army Research Laboratory and the U. S. Army Research Office under grant number W911NF-11-2-0056.
Footnotes
J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. and S.Q.T. are inventors on patent applications describing the FokI-dCas9 technology and the multiplex gRNA expression method. Plasmids described in this work will be deposited with and made available through the non-profit plasmid distribution service Addgene (http://www.addgene.org).
Author Contributions
S.Q.T., N.W. and J.K.J. conceived of and designed experiments. S.Q.T., N.W., C.K., J.A.F. and M.J.G. performed experiments. D.R. developed the updated version of the ZiFiT Targeter software and V.T. and M.J.A. wrote the software program for identifying potential RFN off-target sites. S.Q.T., N.W. and J.K.J. wrote the paper.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech. 2014 doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013 doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotech. 2014 doi: 10.1038/nbt.2808. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotech. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014 doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed]
- 20.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2013 doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 24.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander JD, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 28.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 29.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 30.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.