Abstract
The largest subunit of the mammalian SWI/SNF-A or BAF (BRG1-associated factor) chromatin-remodelling complex is encoded by two related cDNAs hOsa1/BAF250a and hOsa2/BAF250b that are unique to the BAF complex and absent in the related PBAF (Polybromo BAF). hOsa/BAF250 has been shown to interact with transcriptional activators and bind to DNA suggesting that it acts to target the remodelling complex to chromatin. To better understand the functions of hOsa2, we established inducible stable HeLa cell lines over-expressing FLAG-hOsa2 or a derivative lacking the ARID (AT-rich interactive domain) DNA-binding domain. Immunopurification of complexes containing hOsa2 that was followed by mass spectrometry and immunoblotting demonstrated the presence of BRG1 and known BAFs, but not hOsa1 or hBRM. Deletion of the ARID did not compromise the integrity of the complex. Induction of hOsa2 expression caused impaired cell growth and accumulation of cells in the G0/G1 cell cycle phase. Elevated levels of the p53 and p21 proteins were detected in these cells while c-Myc mRNA and protein levels were found to decrease. Chromatin immunoprecipitation and reporter assays suggested that hOsa2 had a direct effect on c-myc and p21 promoter activity. Thus hOsa2 plays an important role in controlling genes regulating the cell cycle.
Keywords: AT-rich interactive domain 1 (ARID1), chromatin, SWI/SNF remodelling complex, transcription
INTRODUCTION
The eukaryotic ATP-dependent chromatin remodelling complexes alter the structure of nucleosomes, facilitating the activation or repression of gene transcription (reviewed in [1–4]). The SWI/SNF complex is one of many distinct remodelling complexes identified in yeast, Drosophila and humans, each of which has an ATPase subunit related to the yeast SWI2/SNF2 (reviewed in [5,6]). At least two classes of SWI/SNF complex have been reported: mammalian BAF [BRG1-associated factor (SWI/SNF-A)] and PBAF (Polybromo BAF) [7,8], and Drosophila BAP (Brahma-associated protein) and PBAP (Polybromo BAP) [9]. Purification and characterization of these complexes demonstrate differences in their activity and subunit composition, implying diverse regulatory roles ascribed to complex-specific subunits. Furthermore, a recent report indicates that variations in the canonical SWI/SNF complexes exist that target specific gene promoters [10]. The SWI/SNF complex has been implicated in the development of cancer as several of its subunits have been shown to function as either tumour suppressors or to interact with known tumour suppressors and oncogenes (reviewed in [11,12]).
The mechanisms by which the SWI/SNF complex is targeted to specific genes for remodelling are not fully understood. The observation of protein-protein interactions between activators and SWI/SNF subunits has led to the notion that SWI/SNF is recruited to promoters by site-specific transcriptional regulators. In addition, it is known that several SWI/SNF subunits contain DNA-binding domains. Whether these DNA-binding domains play a direct role in the recruitment of SWI/SNF to specific genes remains to be established.
We have previously reported the isolation of two cDNA clones hOsa1 and hOsa2 [13], also known as BAF250a/p270/ARID1A/OSA1 [14–17] and BAF250b/ARID1B/hELD/p250R [18–20] respectively, that encode the largest subunits of the human BAF (SWI/SNF-A) complex. hOsa/BAF250 is related distantly to yeast SWI1 and more closely to Drosophila Osa, a component of the BRM complex [21]. Osa is required for proper photoreceptor differentiation and embryonic segmentation in Drosophila and is thought to antagonize the Wingless signalling pathway [22]. Osa contains a DNA-binding domain termed ARID (AT-rich interactive domain) that is found in yeast SWI1 and is shared by several site-specific transcription factors. Although the ARID in Osa binds to DNA with no apparent sequence specificity [21], it is possible that Osa binds to DNA in the context of chromatin to recruit or stabilize the SWI/SNF complex.
The BAF complex, purified from HeLa cells, contains both hOsa1 and hOsa2 whereas the related PBAF complex lacks these subunits but contains the unique subunits BAF180 and BAF200 [8,23]. Previous studies have led to the identification of subunits both unique and common to BAF and PBAF complexes, raising the possibility that complex-specific subunits might perform gene-specific functions. In Drosophila, knockdown of selective BAP or PBAP subunits followed by expression profiling implicated each complex to function in distinct biological processes in vivo [24]. Although highly similar in their sequences, the ATPase subunits BRG1 and hBRM appear to serve distinct functions in mammalian cells. BRG1 null mice are embryonic lethal, whereas hBRM knockout mice show little phenotypic abnormality [25,26]. Deficiency of BAF250a (hOsa1) or BAF250b (hOsa2) has been reported to compromise pluripotency in mouse embryonic stem cells [27,28]. Therefore it is thought that each SWI/SNF complex is composed of a distinct set of subunits and is targeted to a specific set of genes to regulate their transcription. BAF250 was also found to associate with an E3 ubiquitin ligase that targets histone H2B, implicating SWI/SNF in the cross-talk of chromatin remodelling and modifying activities [29].
To investigate the role of the hOsa2 subunit in the BAF complex, we generated stable HeLa cell lines expressing FLAG epitope-tagged hOsa2 and purified and characterized hOsa2-containing complexes. We found that cells induced to overexpress hOsa2 exhibited specific growth and cell cycle defects. Analysis of these cells led to the identification of potential target genes of hOsa2 that include c-myc and p21.
EXPERIMENTAL
Plasmids
The pUHD-hOsa2 and pUHD-hOsa2ΔARID plasmids were constructed by subcloning hOsa2 (16e) and hOsa2ΔARID sequences [13] into the tetracycline-responsive mammalian expression vector pUHD-10-3 [30] with a FLAG tag sequence added to the 5′ end. The T7-hOsa2 plasmid was made in a CMV (cytomegalovirus) enhancer/promoter-based mammalian expression construct (pCGT) as described previously [13]. The T7-p53 plasmid [31] was a gift of LaiYee Wong and Angus Wilson (NYU School of Medicine, 550 First Avenue, New York, NY 10016-6481, U.S.A.). The p21-luciferase and pGL3-c-myc promoter (𢈒 1616/+16) [32] constructs were gifts of Xiao-Hong Sun (Oklahoma Medical Research Foundation, 825 Northeast 13th Street, Oklahoma City, OK 73104-5097, U.S.A.) and Mitsuyasu Kato (University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305–8575 Japan) respectively.
Cell lines
HeLa, C33A and Saos-2 cells were maintained in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10 % fetal bovine serum (Gemini Bio-Products), L-glutamine and penicillin/streptomycin (Invitrogen) and then cultured in a humidified incubator with 5 % CO2. HeLa Tet-off cells (Clontech) were grown in medium that also contained 200 µg/ml Geneticin (Invitrogen) and 100 ng/ml Dox (doxycycline, Sigma). To establish cell lines that stably expressed hOsa2, HeLa Tet-off cells were plated in a 35 mm plate and transfected by the calcium phosphate method with 2.5 µg of pUHD-hOsa2, pUHD-hOsa2AΔRID or pUHD-10-3 (used as a control vector), and 50 ng of pDH, a plasmid encoding resistance to hygromycin B. Each plate was divided into two 100 mm plates the following day and hygromycin (Invitrogen) was added to 200 µg/ml 24 h after plating. The culture medium was changed every 48 h and drug-resistant colonies were selected after 14 days. Clones expressing FLAG-tagged proteins in the absence of Dox were selected by immunoblotting of the cell lysates. For induction, Dox was first reduced from 10–30 ng/ml (the concentration used for maintenance) to 1 ng/ml for 1 day and then removed entirely from the medium the following day. The cells were washed with PBS between medium changes to facilitate Dox removal.
For the cell cycle analysis, at 24, 48 and 72 h after hOsa2 induction, cells were fixed overnight with 80 % ethanol and 0.1% BSA in PBS at 4°C. After washing with PBS, the DNA was stained by incubating the cells for 1 h in PBS containing 50 µg/ml propidium iodide and 100 µg/ml RNase A at 37 °C. The stained cells were analysed by flow cytometry.
To label cells with BrdU (bromodeoxyuridine), cells that had been induced for 8 h were incubated with BrdU for 2 h and then fixed in 4 % paraformaldehyde overnight. hOsa2 was detected with a rabbit polyclonal antibody against hOsa2 (pre-absorbed, [13]) and using anti-rabbit Texas Red (Vector Labs) as the secondary antibody. BrdU was detected with a monoclonal antibody against BrdU (Amersham) followed by anti-mouse FITC (Vector Labs).
Nuclear extract preparation
Small-scale nuclear extract preparations were carried out as described previously [33]. The cells were washed with PBS and harvested by centrifugation (1 000 g for 5 min). The cell pellet was resuspended in 50 µl of buffer A [10 mM Hepes/KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1.5 mM DTT, (dithiothreitol), 0.2 mM PMSF and 0.2 mM sodium metabisulfite] and allowed to stand on ice for 10 min. The cells were lysed with seven strokes of a 1 ml syringe fitted with a 25 gauge needle. The homogenate was centrifuged for 1 min (12 000 g) at 4°C, the PNS (post-nuclear supernatant) was carefully removed and then the pellet was mixed with 30 µl of buffer C (20 mM Hepes/KOH, pH 7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.7 mM PMSF, 1.5 mM DTT and 0.2 mM sodium metabisulfite). The suspension was kept on ice for 30 min, and the nuclear extract was collected by centrifugation for 10 min at 12 000 g in an Eppendorf microcentrifuge.
Purification of protein complexes
To purify proteins associated with hOsa2 or hOsa2ΔARID, cells were seeded on 5–10 (150 mm) plates and induced for 24 h. Nuclear extracts were prepared as described above (adjusted for increased starting material) and 4.5 mg was loaded on to a 5 ml gradient of 12–32 % glycerol in HEMG buffer and centrifuged at 200 000 g in a Beckman SW50.1 rotor for 13 h. Next, 20 fractions of 250 µl were collected then either precipitated with TCA (trichloroacetic acid) for immunoblotting, or pooled (fractions 13–17) for purification with anti-FLAG antibody. The pooled fractions (1.2 ml) were adjusted to 0.15 M KCl and incubated with 40 µl (50 % slurry) of anti-FLAG M2-agarose beads (Sigma) for 6 h at 4°C. The beads were washed in the same buffer and the proteins eluted in 30 µl of buffer containing 250 µg/ml of FLAG peptide (Sigma). The eluted proteins were separated by SDS/PAGE (10% gel), stained with silver and analysed by Q-TOF (quadrupole-time-of-flight) mass spectrometry.
Transient transfections
Saos-2, HeLa and Tet-off HeLa (16eC3, 16eC8, control pUHD-10-3) cells were seeded at a density of 3×105 to 1×106 cells per 35 mm plate and then transfected the next day either by the calcium phosphate precipitation method or with Lipofectamine™ Plus reagent (Invitrogen). Briefly, 0.1, 0.3 or 0.5 µg of p21-luciferase reporter or 0.5 µg of c-myc-luc reporter, and where appropriate, 1 µg of T7-hOsa2 or T7-p53 were transfected along with 0.25 µg of RSV (Rous sarcoma virus)-LacZ or CMV-LacZ. The Tet-off cells were transfected in a medium containing 1 µg/ml Dox, which was removed after 24 h. Cells were harvested 1–2 days post-transfection and luciferase and β-galactosidase assays were performed as described previously [13]. For siRNA (small interfering RNA) knockdown experiments, 1 × 106 HeLa cells were transfected with 2.5 µg of siRNA (Dharmacon) against luciferase (5′-FlCUUACGCUGAGUACUUCGA-3′ where Fl is fluorescein) or hOsa2 (5′-FlAACCGCUCGGAAUUCCUUU-3′ where Fl is fluorescein) using Gene Pulser II (Bio-Rad). Cells were harvested 48 h after electroporation and the RNA prepared for RT-PCR (reverse transcription PCR) analysis. The knockdown experiments in 16eC3 cells were carried out as follows: 4×105 cells/6 cm plate were transfected with siRNA (400 pmol/plate) against luciferase, p53 (SMARTpool L-003329, Thermo Scientific) or Miz1 (SMARTpool, L-012659, Thermo Scientific) with Lipofectamine™ 2000 (Invitrogen). Each plate was split into two 35 mm plates the following day, and 2 days later, one plate was induced for hOsa2 expression for 7 h. Cells were harvested in 0.3 ml of RIPA buffer and proteins analysed by SDS/PAGE (10 % gel) and immunoblotting.
Immunoblotting
Nuclear and PNS (or cytoplasmic) cell fractions were separated by SDS/PAGE (10 % gel) and transferred to nitrocellulose. Samples loaded on to the gel were adjusted for protein concentration using the Bio-Rad protein assay reagent. The nitrocellulose was incubated in a blocking solution for 1 h (5 % non-fat milk, 20 mM Tris-HCl, pH 8, 150 mM NaCl, 0.05 % Tween 20 and 0.5 mM EDTA) and then with a primary antibody for 1 h, followed by an HRP (horseradish peroxidase)-coupled secondary antibody. The signal was detected with a chemiluminescence detection kit (Amersham or Bio-Rad). The following primary antibodies were used: anti-FLAG M2 (F3165, Sigma), anti-BRG1 (sc-10768, Santa Cruz), anti-Sp1 (sc-59), anti-hOsa2 [13], anti-p53 (sc-6243), anti-p21 (sc-397), anti-c-Myc (9E10), anti-T7 (Novagen), anti-Miz1 (AF3760, R&D Systems), anti-p53 phospho-Ser15 (9284, Cell Signaling), anti-p53 phospho-Ser37 (9289, Cell Signaling).
RT-PCR
1 × 106 16e cells were plated in 100 mm plates and harvested at 0h, 12 h and 24 h after removal of Dox. Cells were lysed in 1 ml TRI Reagent (Sigma) and total RNA was prepared according to the manufacturer’s recommendations. To detect expression of c-myc mRNA, 200 ng of total RNA was used as a template in a reaction mix containing eAMV RT and SYBR Green mix (Sigma). RT-PCR reactions were carried out in a LightCycler (Roche) using primers 5′-TCTCCACACATCAGCACAACTACGC-3′ and 5′-CGCCTCTTGACATTCTCCTCGGTG-3′. 5 ng of RNA was used as a template in RT-PCR reactions carried out with primers 5′-AAACTCTGGTGGAGGTCCGT-3′ and 5′-CTTAC-CAAAAGTGGCCCACTA-3′ to 28S rRNA, which served as a control for normalization. Primers used to detect hOsa2 mRNA were 5′-CTGTCGCCTCAGAGAC-3′ and 5′-GTTCG-ATAAAAGCGCCAT-3′. Primers for B2M (β-2-microglobulin) were also used as a control (5′-AAAGATGAGTATGCCTGCCG-3′ and 5′-CCTCCATGATGCTGCTTACA-3′).
ChIP (chromatin immunoprecipitation) assay
The preparation of chromatin and procedure for immunoprecipitation was adapted from published protocols [34–36]. To prepare chromatin, 16e cells were plated at a density of 1.75 ×106 cells/10 cm plate and induced for 24 h. Ten plates of un-induced cells and ten plates of induced cells were cross-linked with formaldehyde, lysed and then their nuclei resuspended in a total volume of 3 ml of ice-cold ChIP buffer 3 (10 mM Tris, pH 8, 1 mM EDTA, 0.5 mM EGTA and protease inhibitors) and placed on ice. Cells were sonicated using the 1/8” microtip on a Branson 250 digital sonicator, 50 % maximum amplitude for 10 min continuously. A dry ice/ethanol bath was utilized to cool the sonication mixture. The sonicated chromatin was then cleared of cell debris by centrifugation in a microcentrifuge at 10 000 g for 30 min at 4°C. Ethidium-bromide staining of sonicated chromatin run on an agarose gel showed fragment sizes 400–1000 bp.
50 µg of sonicated chromatin was diluted in ChIP buffer 3 (300 µl) and pre-cleared by adding 30 µl of blocked Protein A/G sepharose beads and agitating at 4 °C for 3 h. The supernatant was incubated overnight with 2 µg of antibody followed by 20 µl of 50 % slurry Protein A/G Sepharose (Amersham), or 20 µl of 50 % slurry FLAG EZview or FLAG M2 agarose beads (Sigma). All antibodies, except hOsa2 and pre-immune serum, were purchased, from Santa Cruz Biotechnology (anti-Pol II, sc-899; anti-p53, sc-6243; anti-p21, sc-397; anti-c-Myc, sc-764; anti-Miz-1, sc-5984). The immunoprecipitated complexes were collected, washed, and cross-link reversed as described previously [37]. RT-PCR was carried out in a LightCycler (Roche) using the following primers: c-myc core: 5′-AAGATCCTCTCTCGCTAATCTC-3′ and 5′-CTGCCTCTCGCTGGAATTACT-3′ p21 upstream primer pair 1: 5′-CCAGCCCTTTGGATGGTTTG-3′ and 5′-GCCTCCT-TTCTGTGCCTGA-3′ p21 upstream primer pair 2: 5′-AGAG-GAGAAAGAAGCCTGTCCT-3′ and 5′-TGAAACATTTGCA-GTTTTGCTT-3′ p21 core: 5′-TCTTTTCAGCTGCATTGG-GTAA-3′ and 5′-GCCCCCTTTCTGGCTCA-3′ p21 exon2: 5′-TTGGCAGAGCAGGGTTACCCTACTTGG-3′ and 5′-AGGAC-CAGGGTCCTGTTTGCCACCAGG-3′. To detect the p21 upstream region, standard PCR reaction was carried out with primer pair 1 for 15 cycles and a fraction of the product was used for a template in a RT-PCR using primer pair 2.
RESULTS
Purification of protein complex associated with hOsa2
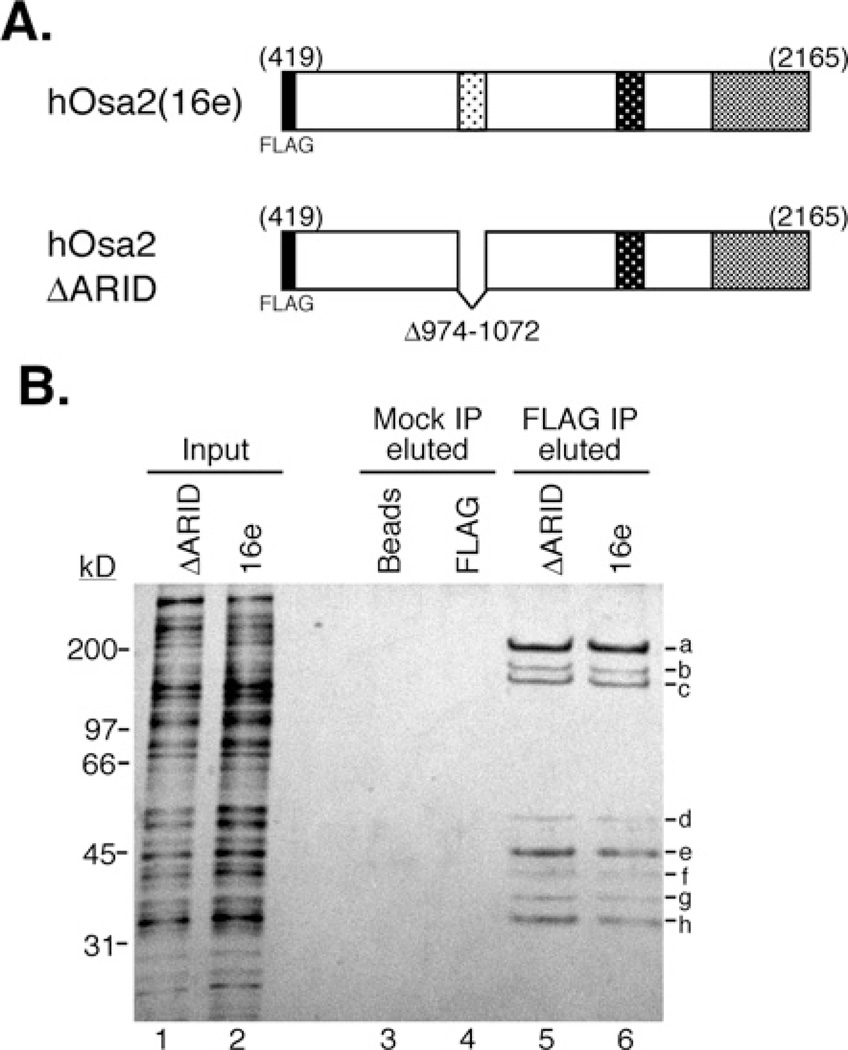
We have previously reported [13] molecular cloning of the two largest subunits of the human SWI/SNF complex, hOsa1 and hOsa2, also known as BAF250a/p270/ARID1A/OSA1 [14–16] and BAF250b/ARID1B/hELD/p250R [18–20]. Immunoblotting and mass spectrometry confirmed the presence of both hOsa proteins in SWI/SNF-A purified from HeLa cell nuclear extracts by conventional chromatography [13]. To biochemically determine components of the hOsa2-containing protein complex, we made stable HeLa Tet-off cell lines inducibly overexpressing the C-terminal 1746 amino acids of the hOsa2 protein (termed 16e, [13]) tagged with FLAG at the N-terminus (Figure 1A). Larger forms of hOsa2 did not express stably in transfected cells. hOsa2 with different N-terminal sequences have been reported in the literature including a paper indicating a polypeptide of 1740 amino acids as full-length hOsa2 [19]. There appears to be isoforms that differ in the N-terminal sequence; however, this is a region of hOsa2 that is most divergent from its homologue hOsa1 and orthologues in other species. Because the 16e construct used in the present study has all the conserved domains and can form a complex with endogenous SWI/SNF components (see below) and the size of the 16e polypeptide approximately corresponds to the lower form of the doublet detected for the endogenous hOsa2 protein [13], we chose to use cells expressing the stable 16e protein in the present study. Additionally, we made cell lines expressing FLAG-hOsa2 that lack the 99 residues that correspond to the DNA-binding domain ARID (hOsa2ΔARID).
Figure 1. Purification of protein complexes associated with hOsa2 and hOsa2ΔARID.
(A) The domain structure of FLAG-tagged hOsa2 (16e) and derivative hOsa2ΔARID lacking the DNA-binding domain ARID. The three shaded regions are conserved between human and Drosophila Osa protein as described previously [13]. (B) HeLa Tet-off cells induced to express FLAG–hOsa2 (cell line 16eC3) or FLAG-hOsa2ΔARID were used to purify hOsa2-associated proteins. Nuclear extracts were loaded on to a glycerol gradient and fractions containing hOsa2 (lower third of the gradient) were pooled and subjected to immunopurification with anti-FLAG antibody and elution with FLAG peptide. The input represents 0.25% of the pooled gradient fractions (lanes 1 and 2). Mock IP (immunoprecipitation) was performed with Protein G beads (lanes 3 and 4) and eluted using the same conditions as with the anti-FLAG antibody (lanes 5 and 6). Protein bands labelled a-h were excised and subjected to analysis by mass spectrometry. Peptides for the following SWI/SNF subunits were identified: hOsa2 (a), BRG1 (a), BAF170 (b), BAF155 (b and c) and BAF60 (e). The recovery of peptides from bands d and f–h was insufficient for positive identification. The molecular mass in kDa is indicated on the left-hand side.
Nuclear extracts were prepared from cells induced for 24 h by Dox removal and proteins were fractionated by glycerol gradient sedimentation. The gradient fractions containing hOsa2 were pooled and subjected to immunopurification with anti-FLAG antibody-coupled beads. The proteins eluted with the FLAG peptide were separated by SDS/PAGE and stained with silver (Figure 1B). The pattern of the protein bands that co-purified with hOsa2 and hOsa2ΔARID appeared to be identical, indicating that the deletion of the ARID did not affect the integrity of the complex. Mass spectrometric analysis of the excised bands identified peptides for the following subunits of SWI/SNF-A: hOsa2 (band a), BRG1 (band a), BAF170 (band b), BAF155 (bands b and c) and BAF60 (band e). We also identified a peptide for PRMT5 (protein arginine N-methyltransferase 5) in band d, confirming the previous report of the association of PRMT5 with SWI/SNF [38]. Immunoblotting confirmed the presence of hOsa2, BRG1, BAF170 and BAF155 in the purified complex (results not shown). No peptide unique to hOsa1 or hBRM was recovered from the purified complex, suggesting that hOsa1 and hOsa2 are present in distinct SWI/SNF-A complexes and that hOsa2 is associated predominantly with BRG1 in HeLa cells.
Induced overexpression of hOsa2 leads to reduced growth rate and accumulation of cells in the G0/G1 phase
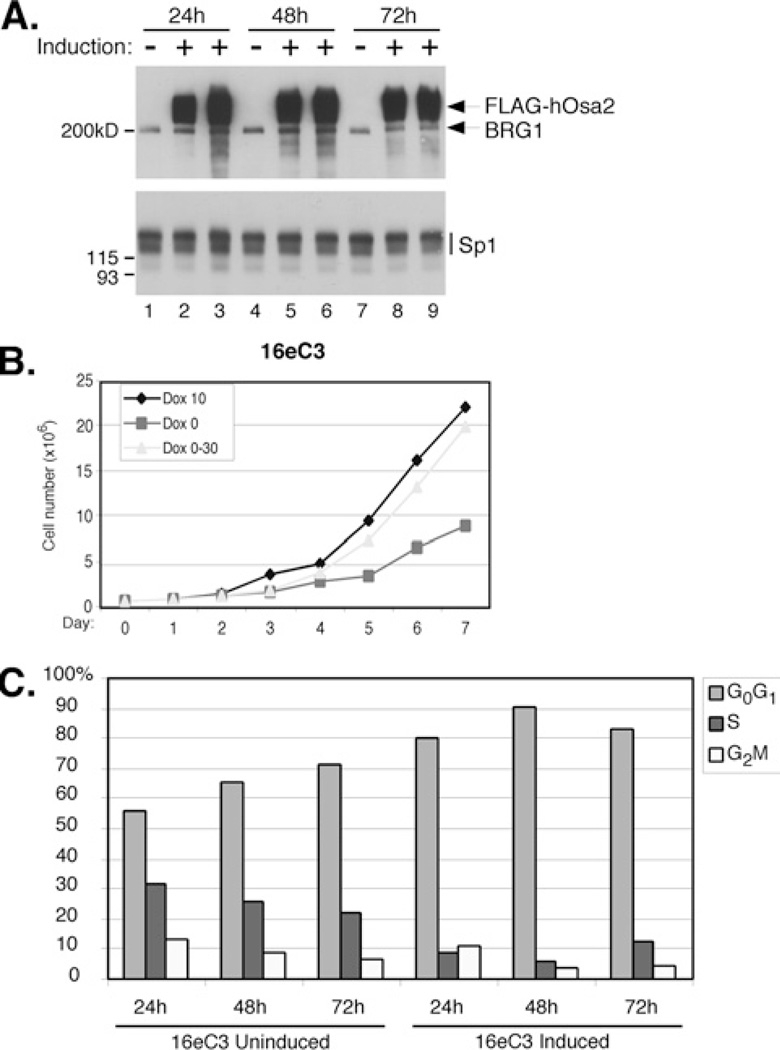
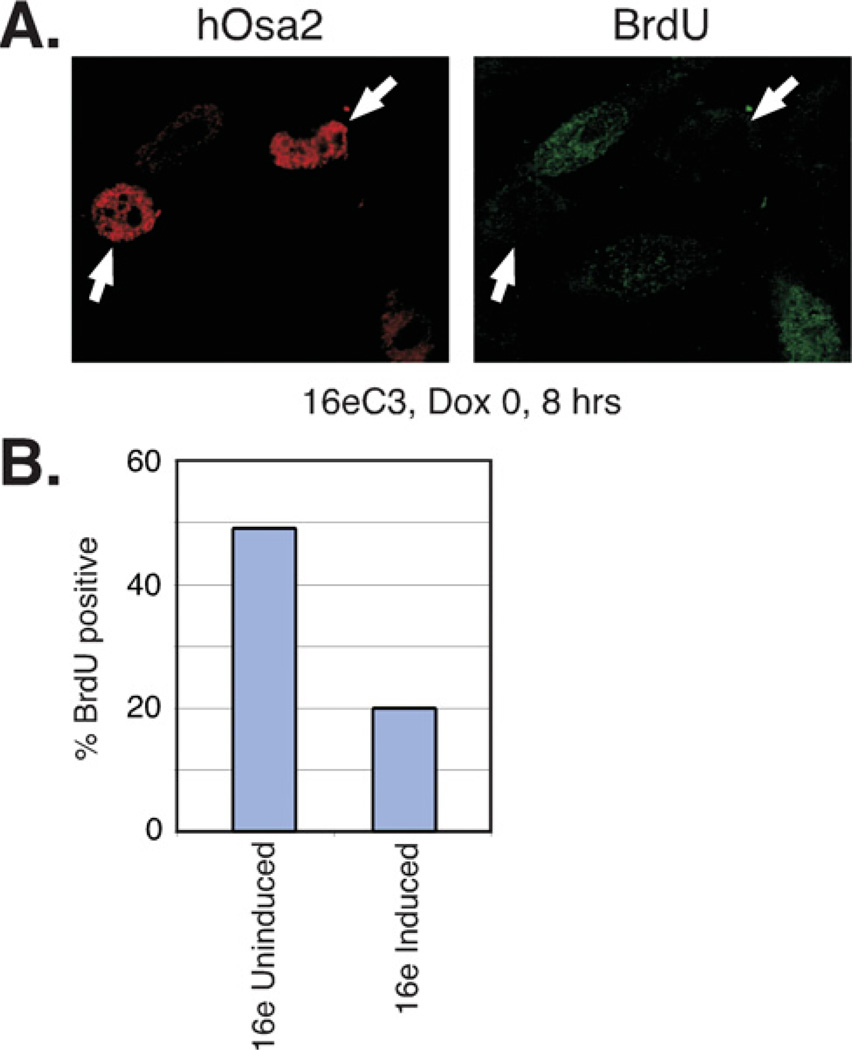
As the subunits of SWI/SNF have been implicated in cancer development [39] and expression of the largest subunit was reported to be altered in human tumour cells [40,41], we examined the effect of hOsa2 expression on cell cycle regulation. We conducted analyses of two independent clonal cell lines 16eC3 and 16eC8 that express FLAG–hOsa2, as well as control cells generated with the pUHD-10-3 vector. The induction of FLAG– hOsa2 was regulated tightly as determined by immunoblotting (Figure 2A). The protein levels of BRG1 and Sp1 remained the same after hOsa2 induction indicating that hOsa2 did not affect gene expression globally. However, the 16eC3 cell line showed a substantial reduction in cell number after a few days after the induction of hOsa2 (Dox 0) compared with uninduced cells (Dox 10) (Figure 2B). 16eC8 cells were consistently more impaired for growth than 16eC3 cells, and the presence of apoptotic cells was confirmed by the TUNEL (terminal deoxynucleotidetransferase-mediated dUTP nick end labelling) assay in 16eC8 cells induced from 24 to 72 h (results not shown). The induced and uninduced cells were also subjected to cell cycle analysis. We detected accumulation of cells in the G0/G1 phase soon after hOsa2 induction in 16eC3 cells (Figure 2C), 16eC8 cells and cells expressing hOsa2ΔARID (results not shown). Control cells with stably integrated empty vector DNA showed no change in their cell cycle profile after induction (results not shown). To test whether induced cells would recover after hOsa2 expression was abrogated, Dox was re-added to separate plates of cells 2 days after it was first removed (Dox 0–30 in Figure 2B). Both cell lines showed partial recovery of growth (Figure 2B) and restoration of normal cell cycle progression (results not shown), indicating that the effects of hOsa2 overexpression were reversible at least up to day 2. To confirm the growth arrest of 16e cells after hOsa2 expression, we measured the incorporation of BrdU in 16eC3 cells induced for 8 h (Figure 3). Consistent with the impaired growth properties observed with hOsa2-expressing cells, the incorporation of BrdU was substantially reduced in 16e cells expressing high levels of hOsa2.
Figure 2. Induced expression of hOsa2 results in impaired cell growth and accumulation of cells in the G1 phase.
(A) Nuclear extracts were prepared from 16eC3 cells uninduced (lanes 1, 4 and 7) or induced to express FLAG–hOsa2 (lanes 2, 5 and 8: 0.01 ng/ml Dox; lanes 3, 6 and 9: 0 ng/ml Dox). Shown is an immunoblot probed sequentially with antibodies against FLAG, BRG1 and Sp1. The molecular mass in kDa is indicated on the left-hand side. (B) 16eC3 cells were plated at 5×105 cells per plate in 10 ng/ml Dox and induced the next day (day 0, Dox 0, 0 ng/ml) or left uninduced (day 0, Dox 10, 10 ng/ml). The total cell number was determined each day for a period of 7 days. Cells induced to overexpress hOsa2 showed a substantial and increased reduction in cell number compared with uninduced cells. The cell number did not change in the control cells that were treated similarly with Dox (results not shown). Cells that were induced on day 0, but were refed in a medium containing 30 ng/ml Dox on day 2 (Dox 0–30) to shut down hOsa2 expression, showed a partial recovery in cell number. Similar results were obtained with an independent cell line 16eC8. (C) Uninduced and induced 16eC3 cells were subjected to DNA staining and flow cytometry. Upon removal of Dox, 16eC3 cells and 16eC8 cells (results not shown) accumulated in the G0/G1 phase of the cell cycle. The control cells showed no change in cell cycle distribution in the same experiment (results not shown).
Figure 3. Induced expression of hOsa2 decreases BrdU incorporation.
(A) 16eC3 cells induced for 8h were labelled with BrdU for 2h then fixed and examined by indirect immunofluorescence with an antibody to hOsa2 or BrdU. These cells showed heterogeneous expression of hOsa2. The arrows point to those expressing hOsa2 at high levels.
(B) Quantification of the percentage of BrdU positive cells in uninduced and induced 16eC3 cell cultures.
Induced expression of hOsa2 increases p53 and p21 protein levels
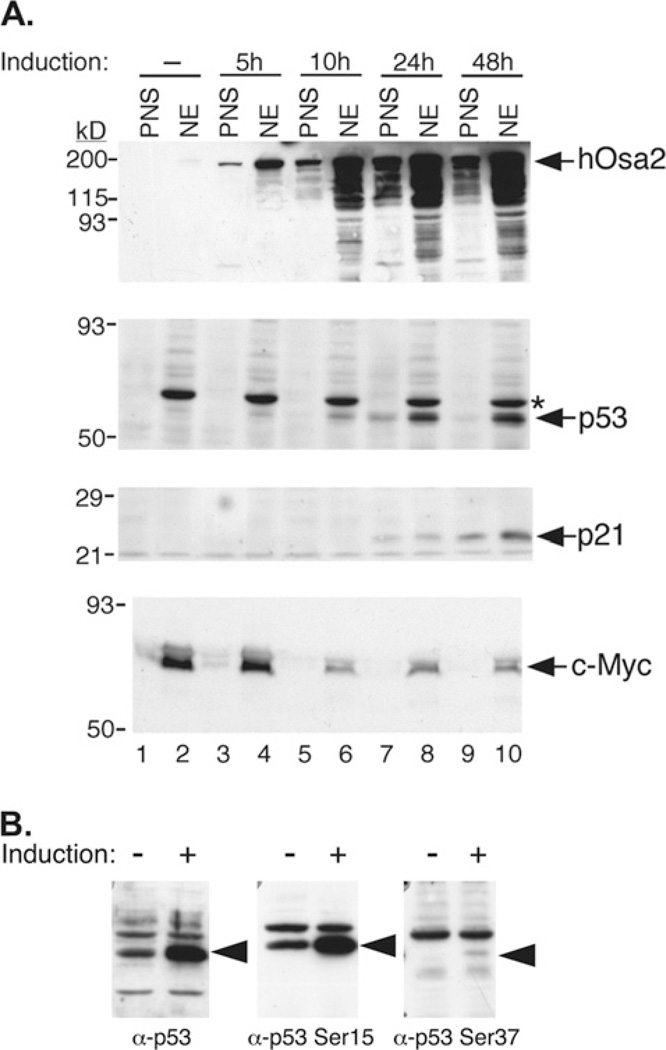
To examine the changes in specific cell cycle regulator levels that accompany overexpression of hOsa2 in 16eC3 cells, we prepared immunoblots of nuclear and PNS fractions from cells induced over a period of 48 h (Figure 4A). Most notably, the amount of p53 protein began to increase 5–10 h after induction. An early time course experiment indicated that FLAG–hOsa2 mRNA and protein were detected at 4 h and an increase in p53 protein was detected as early as 6 h post-induction (results not shown). Northern blotting, RT-PCR and gene array analyses showed no change in the amount of p53 mRNA, indicating that the p53 protein level increased through a post-translational mechanism that led to reduced p53 turnover. Indeed, we detected increased phosphorylation of p53 at Ser15 and Ser37, after hOsa2 induction (Figure 4B), which is probably mediated by ATR (S. Giannakopoulos and N. Tanese, unpublished work) and contributed to the stabilization of p53. The p21 protein level was found to increase concomitantly, consistent with the known role of p53 in activating the p21 gene.
Figure 4. Changes in p53, p21 and c-Myc protein levels after induction of hOsa2.
(A) 16eC3 cells were treated with Dox for the indicated hours and then were harvested and fractionated into NE (nuclear) and PNS fractions. Proteins were separated by SDS/PAGE and immunoblotted with antibodies against hOsa2, p53, p21, or c-Myc. Molecular size markers are indicated at left. (B) Lysates of 16eC3 cells induced for 24 h (+) were separated by SDS/PAGE and probed for total p53, p53 phospho-Serine 15, and p53 phospho-Serine 37. *Non-specific cross-reacting protein.
hOsa2 represses c-myc gene expression and promoter activity
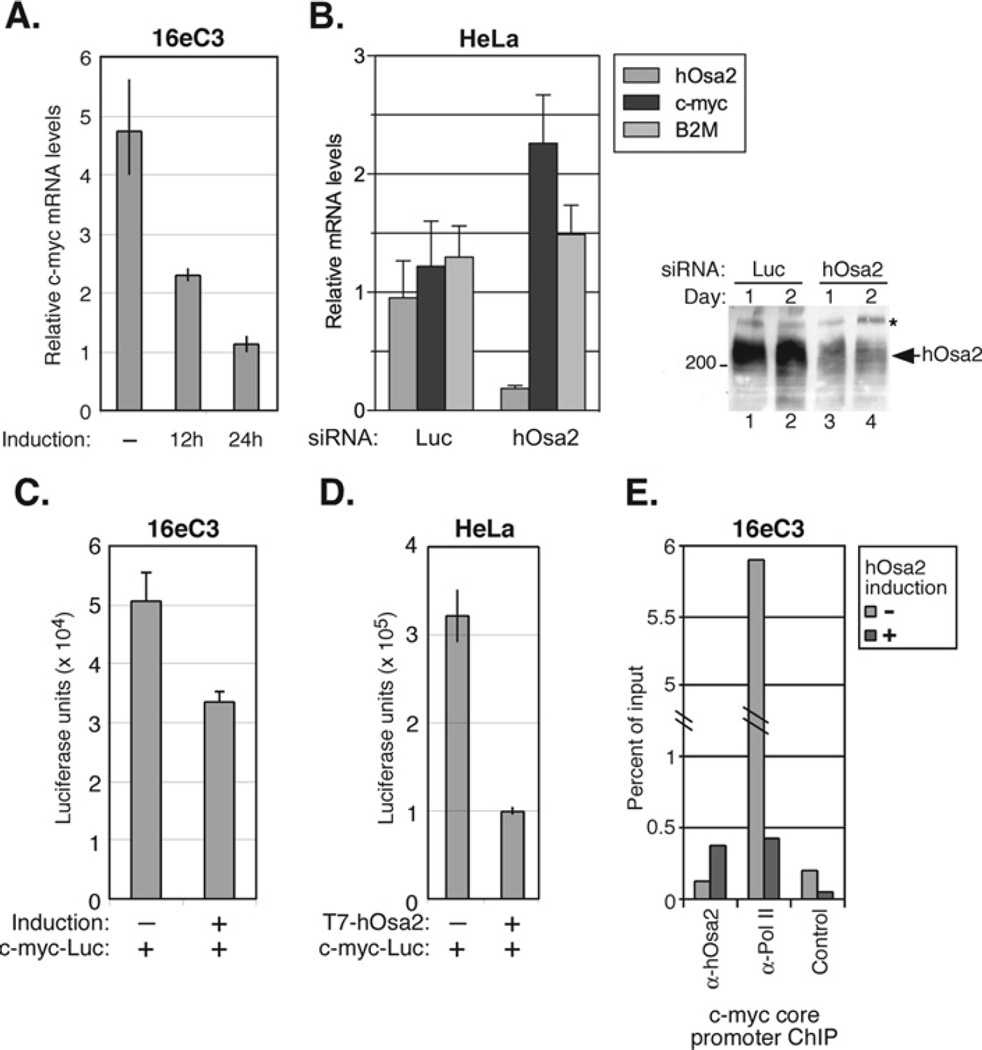
Among the regulators of the cell cycle that we examined, we found the levels of c-Myc protein decreased as early as 10 h after induction of hOsa2 (Figure 4A). This observation was verified by RT-PCR in 16e cells after hOsa2 induction during which we detected an approx. 4-fold decrease in c-myc mRNA after 24 h (Figure 5A). We next measured c-myc mRNA levels after knockdown of endogenous hOsa2 by RNAi (RNA interference) in HeLa cells (Figure 5B). Transfection of siRNA against hOsa2 reduced its expression by 5-fold compared with siRNA against the control luciferase gene. By contrast, the c-myc mRNA levels increased by approx. 2-fold when hOsa2 was knocked down by siRNA. We also examined the activity of a reporter plasmid containing the c-myc gene promoter sequence (− 1616/+16, [32]) in 16eC3 cells (Figure 5C) as well as in HeLa cells co-transfected with a hOsa2-expressing plasmid (Figure 5D). In both cases, the expression of hOsa2 reduced c-myc promoter activity, indicating that hOsa2 might serve a repressive role in the transcription of the c-myc gene. Lastly, ChIP assays performed with chromatin from 16eC3 cells either uninduced or induced for 24 h demonstrated a reproducible enrichment of hOsa2 at the core promoter region of the c-myc gene upon induction of hOsa2 and reduced occupancy by RNA polymerase II (Figure 5E). Together, the results suggest that hOsa2 represses c-myc mRNA expression.
Figure 5. c-myc mRNA levels are reduced after the expression of hOsa2, but increased after the knockdown of endogenous hOsa2.
(A) The total RNA prepared from16eC3 cells uninduced or induced for 0, 12 or 24 h was used in RT-PCR with primers corresponding to c-myc exon1. The value obtained for each reaction was normalized to that of 28S rRNA. The experiment was performed in duplicate and the spread of the mean is shown for each sample. (B) HeLa cells were transfected with siRNA against luciferase or hOsa2 by electroporation and the total RNA was prepared 48 h later. RT-PCRs were carried out using primers to hOsa2, c-myc and B2M. The experiment was performed in triplicate. The immunoblot on the right shows reduced levels of endogenous hOsa2 protein, 1 and 2 days after transfection of HeLa cells with siRNA. (C) 16eC3 cells were transiently transfected with a c-myc promoter–luciferase reporter plasmid (0.5 µg) using Lipofectamine™ Plus, and hOsa2 expression was induced 8 h later. The cells were harvested after 24 h and luciferase activity was measured and normalized to β-galactosidase activity from the co-transfected LacZ plasmid (0.25 µg). The assay was performed in triplicate. (D) HeLa cells were transiently transfected by the calcium phosphate precipitation method with a c-myc promoter–luciferase reporter plasmid (0.5 µg) and a plasmid expressing T7-hOsa2(16e) (1 µg). The cells were harvested after 48 h and the luciferase activity was measured and normalized to β-galactosidase activity from the co-transfected LacZ plasmid (0.25 µg). The experiment was performed in duplicate and the spread of the mean is shown for each sample. (E) 16eC3 cells uninduced ( − ) or induced for 24 h (+) were cross-linked with formaldehyde and a ChIP assay was performed using the indicated antibodies. The products were detected by real time PCR with primers specific for the c-myc core promoter region and plotted as percentage of input. The control antibody was anti-p21 antibody.
hOsa2 stimulates transcription of the p21 gene independent of p53
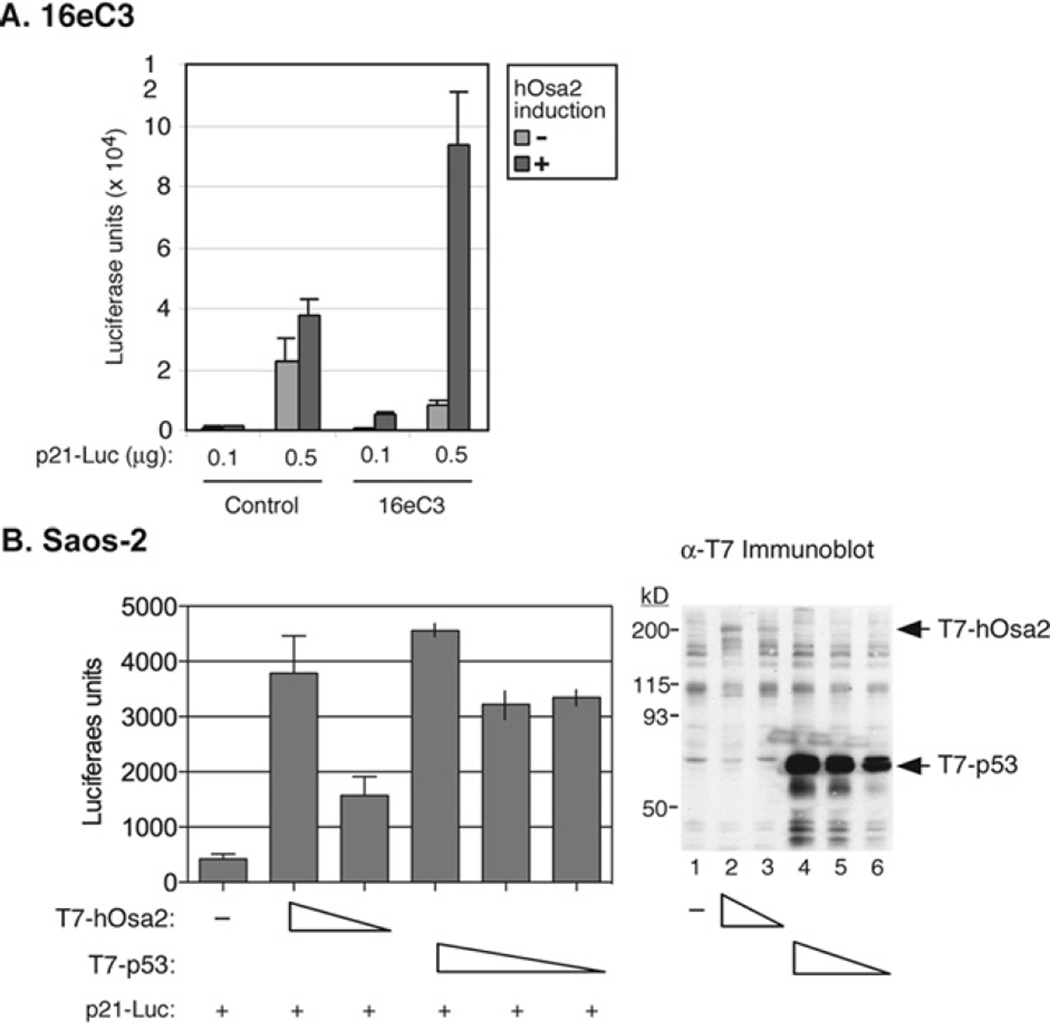
We next focused on the activation of the p21 gene in 16eC3 cells. Transient transfection of the p21 promoter–luciferase reporter plasmid in 16e cells resulted in as much as 10-fold activation of the reporter after induction of hOsa2 (Figure 6A). By contrast, the control cells showed little change in the reporter activity after Dox removal. The p21 gene is a target of p53; therefore, we asked if the presence of functional p53 was necessary for hOsa2-mediated induction of p21. Transient transfection of a plasmid expressing hOsa2 into cells lacking functional p53 (Saos-2 cells, Figure 6B, and C33A cells, results not shown) also activated the p21-Luc reporter plasmid. Thus hOsa2 appears to stimulate p21 promoter activity through an additional mechanism independent of p53.
Figure 6. Activation of the p21 promoter upon co-expression of hOsa2.
(A) 16eC3 and control cells were transiently transfected by the calcium phosphate precipitation method with a p21 promoter-driven luciferase reporter plasmid with the indicated amounts of DNA. The expression of hOsa2 was induced (+) the following day by removal of Dox. The cells were harvested and luciferase activity measured after 24 h. (B) Saos-2 cells were transiently transfected with p21 promoter-reporter plasmid (0.1 µg), a plasmid expressing T7-hOsa2(16e) (1.5 and 1 µg) or a plasmid expressing T7-p53 (1, 0.5 and 0.1 µg) using the Lipofectamine™ Plus reagent. The cells were then harvested and luciferase activity measured after 24 h. The experiment was performed in triplicate (lanes 1–3) and in duplicate (lanes 4–6) and the average or the spread of the mean is shown. An immunoblot of the transfected cell lysates with anti-T7 antibody is shown in the right-hand panel.
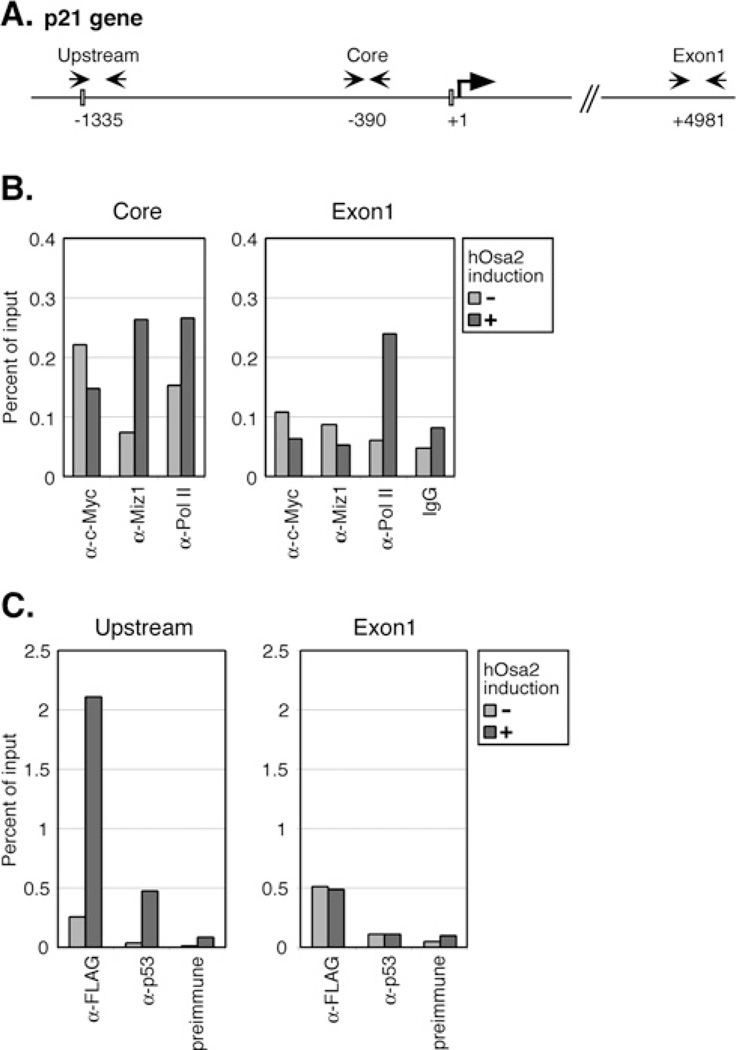
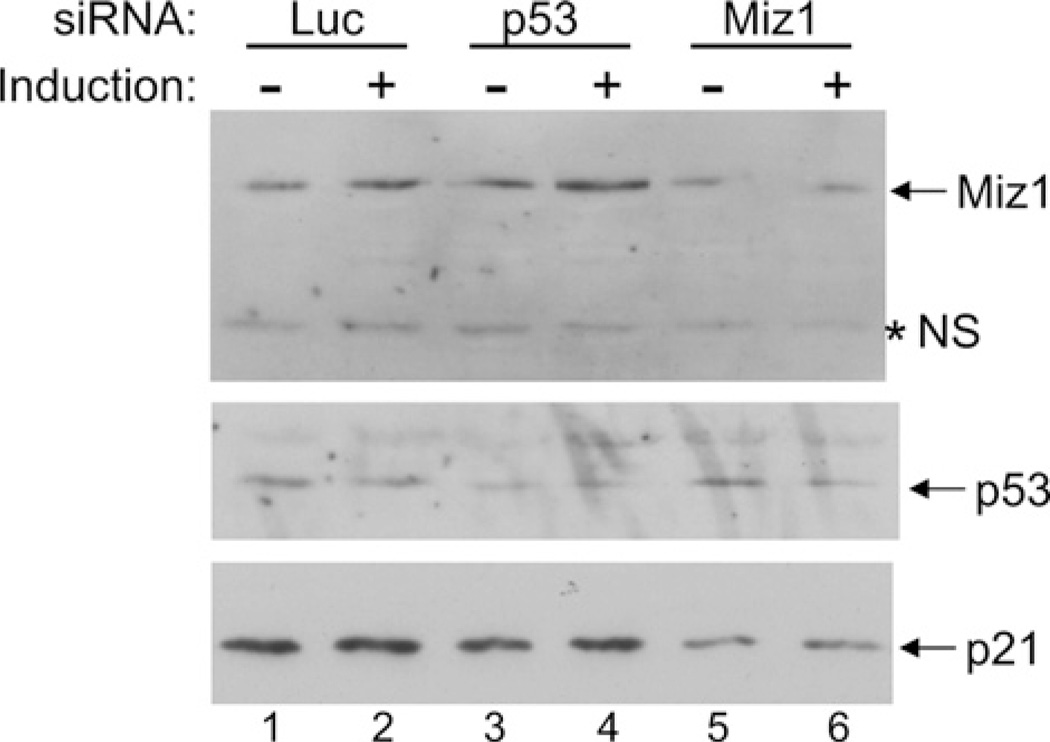
Studies have shown that c-Myc associates with the repressor protein Miz-1 to prevent transcription of the p21 gene [42]. Because the levels of c-Myc protein and mRNA were found to decrease upon induction of hOsa2 in 16e cells (Figures 4A and 5A), we hypothesized that reduced expression of c-Myc could contribute further to the activation of the p21 gene promoter. To address this, we carried out ChIP assays on the p21 core promoter region that binds Miz-1 (Figures 7A and 7B), as well as an upstream region containing a p53 binding site (Figure 7C). Consistent with our hypothesis, we detected reduced association of c-Myc and increased occupancy of Miz-1 at the core promoter region of p21 after hOsa2 induction. As expected, the upstream region was found to associate with p53, but we also detected substantial increase in occupancy by FLAG–hOsa2 after induction. The downstream exon 1 of the p21 gene was used as a control, which showed no change with either anti-FLAG or anti-p53 antibody. Taken together, the results suggest that hOsa2 plays a role in p21 gene activation by binding to the upstream regulatory region as well as by reducing the levels of the c-Myc protein. In support of these findings, knockdown of Miz1, but not p53 reduced the expression of p21 in 16e cells (Figure 8).
Figure 7. ChIP analysis of the p21 core promoter and the upstream region.
(A) The p21 gene indicating the transcriptional start site (+1) and upstream, core and exon1 regions amplified by PCR in ChIP assays. A p53-binding site is shown as a box in the upstream region; a box near the start site represents a reported binding site for Miz-1. (B) 16eC3 cells uninduced (−) or induced for 24 h (+) were cross-linked with formaldehyde and ChIP assays of the p21 core promoter region and exon1 were performed using the indicated antibodies. The products were amplified with primers specific to the core promoter region of the p21 gene, as well as p21 exon1 as a control, and plotted as a percentage of input. IgG, normal mouse IgG. (C) ChIP assays of the p21 upstream region containing p53-binding sites and exon1 as a control. anti-FLAG ChIP was performed with antibody-conjugated agarose beads (EZview beads, Sigma). Preimmune, rabbit serum.
Figure 8. Knockdown of Miz-1 but not p53 affects p21 gene expression in 16e cells.
16eC3 cells transfected with the indicated siRNA were induced for 7 h and then the cell lysates were analysed by immunoblotting with antibodies against proteins indicated to the right of the gel. *NS, Non-specific cross-reacting protein.
DISCUSSION
HeLa cells stably expressing FLAG-tagged hOsa2, or a version lacking the ARID, permitted affinity-purification of specific subunits associated with hOsa2 using an anti-FLAG antibody. We found that the deletion of ARID did not affect the overall integrity of the complex although subtle differences in the composition of complexes associated with wild-type and ARID-deleted hOsa2 might exist. The ARID is a DNA-binding domain, and in the context of certain transcription factors recognizes a specific target sequence. In the case of hOsa proteins, among others, it is thought that the domain binds to DNA with little or nosequence specificity [21,43]. It is possible that the ARID in hOsa2 serves to stabilize the binding of SWI/SNF to target gene promoters. In chromatin-binding assays we have found hOsa2 to associate with chromatin through several of its domains (T. Matsumura and N. Tanese, unpublished work).
The mass spectrometric analysis of co-purified proteins of hOsa2 yielded known SWI/SNF subunits such as BRG1, BAF170, BAF155 and BAF60, as well as a previously identified SWI/SNF-associated protein PRMT5. Significantly, no peptide corresponding to hOsa1 was recovered, which is consistent with the studies reporting that the two largest subunits are present in distinct complexes [18,44]. Although we previously reported that the conserved C-terminus of hOsa1 and hOsa2 associates with both BRG1 and hBRM [13], we did not find any peptide unique to hBRM in the purified hOsa2 complex. This suggests that in HeLa cells hOsa2 associates predominantly with BRG1. A similar finding was reported for the purification of BAF250b–containing complex from a retinoblastoma cell line Y79 [18]. However, another study reported co-precipitation of hBRM with ARID1B (hOsa2) from TSU-Pr1 cells that lack BRG1 as well as from HeLa cells [44]. It is possible that hBRM is present at a lower concentration than BRG1, which does not permit efficient recovery by immunopurification and identification by mass spectrometry. A recent study, describing the purification of SWI/SNF complexes under low stringency conditions, found multiple variations in the composition of SWI/SNF including co-precipitation of BAF250 (hOsa) and a PBAF subunit BAF180 [10]. Thus multiple distinct SWI/SNF complexes may exist under physiological conditions.
We found that induced overexpression of hOsa2 in HeLa cells resulted in impaired growth and accumulation of cells in the G0/G1 phase. This was accompanied by increased p53 protein levels, decreased c-Myc protein after 10 h and increased p21 protein after 24 h. The impaired cell growth properties are consistent with the increased levels of p53 and p21 protein as well as phosphorylated p53. We were interested in determining whether hOsa2 had a direct effect on transcription of select target genes. We found that c-Myc protein expression decreased as early as 10 h in 16eC3 cells. Furthermore, reduced expression of endogenous hOsa2 in HeLa cells by RNAi resulted in increased expression of c-myc mRNA, suggesting that hOsa2 plays a direct role in the repression of c-myc gene expression. A previous study in Drosophila reported that Osa might function to repress target genes of the Wingless pathway [45]. We speculate that hOsa2 has a similar role in repressing target genes of the mammalian Wnt signalling pathway, one target of which is c-myc [46]. The decreased levels of c-Myc could contribute to the activation of the p21 gene promoter by relieving its inhibitory effect on Miz-1, an activator of the p21 gene [42,47] (Figure 9).
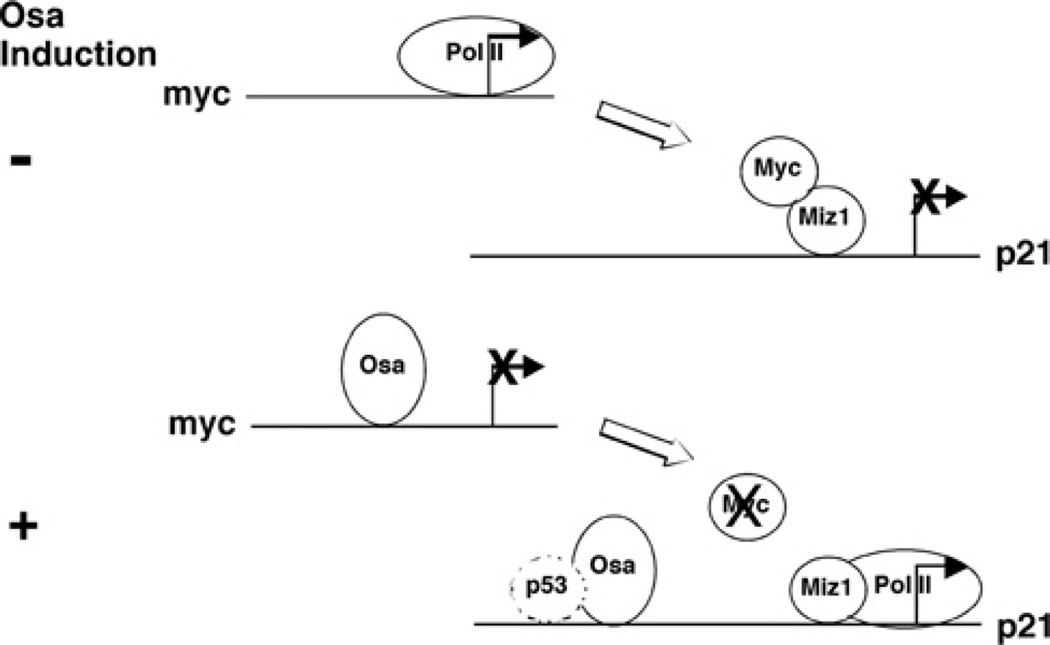
Figure 9. hOsa2 represses c-myc and activates p21 gene transcription.
The transcriptional regulatory region of the c-myc and the p21 gene before and after the induction of hOsa2. In uninduced cells, the p21 promoter region is bound by c-Myc, which represses activation by Miz-1. Increased expression of hOsa2 represses c-myc gene expression, permitting Miz-1 to activate the p21 gene. hOsa2 also binds to the upstream regulatory region of the p21 gene and increases promoter activity through a p53-independent mechanism.
In 16eC3 cells, the up-regulation of p21 could be explained in part by the increase in p53 protein; however, we also found hOsa2-activated p21 promoter activity in cells that did not express functional p53. In ChIP assays we detected enrichment of hOsa2 in the upstream region of the p21 gene after induction, but its association with the core promoter region was not detected. Previous studies have found a direct role for BRG1 in the activation of p21 through a p53-independent mechanism [48,49]. Additionally, occupancy of BRG1 by ChIP in the region upstream of p21 that is dependent on STAT3 (signal transducer and activator of transcription 3) [50] or p53 [51] has been reported. p53 was also demonstrated to interact with BAF60a [52]. We did not detect an interaction between p53 with hOsa2; thus targeting of hOsa2 to the p21 upstream region may be mediated by other SWI/SNF subunits in a p53-dependent manner, or through other transcription factors that regulate the p21 gene in a p53-independent manner. The p21 gene has also been shown to be regulated by BAF180 [53].
As HeLa cells express hOsa1 and hOsa2, it is possible that overexpression of hOsa2 changes the ratio of hOsa1- and hOsa2-containing SWI/SNF complexes in favour of hOsa2 and that alterations in gene expression reflect this shift in the equilibrium. However, when endogenous hOsa2 was knocked down by RNAi, we observed increased c-myc mRNA (2-fold; Figure 5B). hOsa2 induction also repressed SGK (serum/glucocorticoid regulated kinase) mRNA 6-fold, compared with knockdown of hOsa2 that increased SGK mRNA 6-fold (E. Kono and N. Tanese, unpublished work). These results are consistent with a specific role of hOsa2 in the repression of these target genes. Thus the gene-specific effects of hOsa2 knockdown suggest that hOsa1, whose protein level remains unchanged in the knockdown cells, does not fully substitute for the loss of hOsa2. Furthermore, ChIP analysis has shown that endogenous BAF250b (hOsa2), and not BAF250a (hOsa1), occupies the p21 promoter [10].
Nagl Jr et al. [54] reported knockdown of ARID1A (hOsa1) and ARID1B (hOsa2) in the MC3T3-E1 preosteoblast cell line that undergoes cell cycle arrest upon differentiation with ascorbic acid. Although both ARID1A-depleted and ARID1B–depleted cells failed to express alkaline phosphatase, an early differentiation marker after induction with ascorbic acid, only ARID1A-depleted cells failed to undergo cell cycle arrest and did not express the p21 gene. A follow-up study showed that SWI/SNF is involved directly in the repression of c-myc gene expression in differentiating cells and that induction of p21 is dependent on c-myc repression [55]. Furthermore, using the same ARID1A- and ARID1B-knockdown cell lines Nagl Jr et al. [56] showed each ARID1 subunit has a distinct role in activating or repressing target gene transcription and consequently have opposing effects on the cell cycle. They found that in serum-depleted ARID1B-knockdown cells, c-Myc did not activate upon serum re-stimulation indicating that ARID1B is required for activation of c-Myc.
Mouse embryonic stem cells deficient in ARID1B have been created; the cells display reduced proliferation, aberrant cell cycle progression and a failure to maintain an undifferentiated state [28]. The c-myc mRNA levels were the same as in wild-type ES cells 1 day after re-plating, but reduced significantly after 3 days suggesting an indirect effect of ARID1B. The hOsa2-inducible HeLa cells used in the present study were not serum-depleted or restimulated. In this system we found by overexpression, knockdown and ChIP assays that hOsa2 (ARID1B) activates the p21 gene and down-regulates c-myc. ChIP analysis previously reported [56] indicated the presence of ARID1B at the c-myc promoter both before and after c-myc induction, suggesting that it is probably involved in activation as well as repression of the c-myc gene in association with HATs (histone acetyltransferases), HDACs (histone deacetylases) and E2F family of transcription factors [56]. The SWI/SNF complex has been shown to be involved in retinoblastoma-mediated repression of target genes [39]; consistent with this, the induction of hOsa2 in the present study showed reduction in c-myc and N-myc mRNA levels both of which contain E2F–binding sites in the promoter (E. Kono and N. Tanese, unpublished work).
ACKNOWLEDGEMENTS
We thank Brian Dynlacht and his laboratory for help with ChIP assays, Jiri Zavadil for expression profiling studies, and Tom Neubert and the NYU Protein Analysis Facility for the mass spectrometric analysis. We thank LaiYee Wong, Angus Wilson, Xiao-Hong Sun and Mitsuyasu Kato for reagents and Michael Garabedian and Angus Wilson for critical reading of the manuscript prior to submission.
FUNDING
This work was supported in part by the American Cancer Society [grant number RSG-01-248-01-CCE].
Abbreviations used
- ARID
AT-rich interactive domain
- B2M
β-2-microglobulin
- BAF
BRG1-associated factor
- BAP
Brahma-associated protein
- BrdU
bromodeoxyuridine
- BRM
Brahma
- ChIP
chromatin immunoprecipitation
- CMV
cytomegalovirus
- Dox
doxycycline
- DTT
dithiothreitol
- ES
embryonic stem
- IP
immunoprecipitation
- PBAF
Polybromo BAF
- PBAP
Polybromo BAP
- PNS
post-nuclear supernatant
- PRMT5
protein arginine N-methyltransferase 5
- RNAi
RNA interference
- RT-PCR
reverse transcription PCR
- siRNA
small interfering RNA
- SGK
serum/glucocorticoid regulated kinase
Footnotes
AUTHOR CONTRIBUTION
Hiroko Inoue, Stavros Giannakopoulos and Naoko Tanese conceived the study and designed the experiments, Hiroko Inoue, Stavros Giannakopoulos, Christopher N. Parkhurst, Tatsushi Matsumara, Evelyn A. Kono, Takako Furukawa and Naoko Tanese performed the experiments and evaluated the data. Hiroko Inoue and Naoko Tanese wrote the paper.
REFERENCES
- 1.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J. Cell. Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 2.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. BioEssays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 3.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 5.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 9.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryme J, Asp P, Bohm S, Cavellan E, Farrants AK. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J. Cell. Biochem. 2009;108:565–576. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- 11.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 13.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 2002;277:41674–41685. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 14.Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human SWI/SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol. Cell. Biol. 2000;20:3137–3146. doi: 10.1128/mcb.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozmik Z, Machon O, Kralova J, Kreslova J, Paces J, Vlcek C. Characterization of mammalian orthologues of the Drosophila osa gene: cDNA cloning, expression, chromosomal localization, and direct physical interaction with Brahma chromatin-remodeling complex. Genomics. 2001;73:140–148. doi: 10.1006/geno.2001.6477. [DOI] [PubMed] [Google Scholar]
- 17.Xi Q, He W, Zhang X, Le H, Massague J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor B transcriptional program. J. Biol. Chem. 2007;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie Z, Yan Z, Chen EH, Sechi S, Ling C, Zhou S, Xue Y, Yang D, Murray D, Kanakubo E, et al. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 2003;23:2942–2952. doi: 10.1128/MCB.23.8.2942-2952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurlstone AF, Olave IA, Barker N, van Noort M, Clevers H. Cloning and characterization of hELD/OSA1, a novel BRG1 interacting protein. Biochem. J. 2002;364:255–264. doi: 10.1042/bj3640255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeuchi T, Ohtsuki Y, Sugano S, de Bruijn DR, Chait BT, Roeder RG. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J. Biol. Chem. 2002;277:5498–5505. doi: 10.1074/jbc.M108702200. [DOI] [PubMed] [Google Scholar]
- 21.Collins RT, Furukawa T, Tanese N, Treisman JE. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 1999;18:7029–7040. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treisman JE, Luk A, Rubin GM, Heberlein U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 2007;27:651–661. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Z, Wang Z, Sharova L, Sharov A, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko M. BAF250B–associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF: a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 2010;30:1673–1688. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong LY, Matchett GA, Wilson AC. Transcriptional activation by the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 2004;78:10074–10085. doi: 10.1128/JVI.78.18.10074-10085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki T, Suzuki H, Yagi K, Furuhashi M, Yao R, Susa S, Noda T, Arai Y, Miyazono K, Kato M. Lymphoid enhancer factor 1 makes cells resistant to transforming growth factor beta-induced repression of c-myc. Cancer Res. 2003;63:801–806. [PubMed] [Google Scholar]
- 33.Lee KA, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Techn. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 34.Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 36.Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol. Cell. Biol. 2003;23:7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5- containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts CW, Orkin SH. The SWI/SNF complex: chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 40.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell. Physiol. 2001;186:136–145. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Nagl NG, Jr, Flowers S, Zweitzig D, Dallas PB, Moran E. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int. J. Cancer. 2004;112:636–642. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 42.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 43.Wilsker D, Patsialou A, Zumbrun SD, Kim S, Chen Y, Dallas PB, Moran E. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 2004;32:1345–1353. doi: 10.1093/nar/gkh277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Nagl NG, Jr, Wilsker D, van Scoy M, Pacchione S, Yaciuk P, Dallas PB, Moran E. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem. J. 2004;383:319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins RT, Treisman JE. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 2000;14:3140–3152. doi: 10.1101/gad.854300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 47.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 48.Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell. Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell. Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 51.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 52.Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. BAF60a interacts with p53 to recruit the SWI/SNF complex. J. Biol. Chem. 2008;283:11924–11934. doi: 10.1074/jbc.M705401200. [DOI] [PubMed] [Google Scholar]
- 53.Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagl NG, Jr, Patsialou A, Haines DS, Dallas PB, Beck GR, Jr, Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65:9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 55.Nagl NG, Jr, Zweitzig DR, Thimmapaya B, Beck GR, Jr, Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 56.Nagl NG, Jr, Wang X, Patsialou A, van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]