Abstract
ABCA4 is an ATP binding cassette (ABC) transporter that is expressed in rod and cone photoreceptor cells and implicated in the removal of retinal derivatives from outer segments following photoexcitation. Mutations in the ABCA4 gene are responsible for a number of related retinal degenerative diseases including Stargardt macular degeneration, cone-rod dystrophy, retinitis pigmentosa, and age-related macular degeneration. In order to determine the role of the C-terminus of ABCA4 in protein structure and function and understand mechanisms by which C-terminal mutations cause retinal degenerative diseases, we have expressed and purified a series of deletion and substitution mutants of ABCA4 and ABCA1 in HEK 293T cells for analysis of their cellular localization and biochemical properties. Removal of the C-terminal 30 amino acids of ABCA4 including a conserved VFVNFA motif resulted in the loss in N-retinylidene-phosphatidylethanolamine substrate binding, ATP photoaffinity labeling, and retinal stimulated ATPase activity. This mutant was also retained in the endoplasmic reticulum (ER) of cells. Replacement of the VFVNFA motif with alanine residues also resulted in loss in function and cellular mislocalization. In contrast C-terminal deletion mutants that retain the VFVNFA motif were functionally active and localized to intracellular vesicles similar to wild-type ABCA4. Our studies indicated that the VFVNFA motif is required for the proper folding of ABCA4 into a functionally active protein. This motif also contributes to the efficient folding of ABCA1 into an active protein. Our results provide a molecular based rationale for the disease phenotype displayed by individuals with mutations in the C-terminus of ABCA4.
ABCA4, also known as ABCR or the rim protein, is a member of the ABCA subfamily of ATP binding cassette (ABC) transporters expressed in vertebrate photoreceptor cells (1–4). It is localized along the rims and incisures of rod and cone outer segment discs where it has been implicated in the binding and transport of the Schiff base adduct of all-trans retinal and phosphatidylethanolamine (PE)3 known as N-retinylidene-PE or N-ret-PE across disc membranes as part of visual cycle (3,5–9).
To date over 500 different mutations in the ABCA4 gene are known to cause Stargardt macular degeneration, an early onset, recessive disease characterized by the loss in central vision, the presence of lipofuscin deposits in RPE cells, a delay in dark adaptation, and progressive degeneration of photoreceptor and RPE cells (1,10–15). Mutations in ABCA4 are also responsible for other related, but more severe retinal degenerative diseases including autosomal recessive cone-rod dystrophy and retinitis pigmentosa (16–19). Finally, individuals heterozygous for selected disease-linked mutations in ABCA4 have been suggested to be at higher risk in developing age related macular degeneration (20). Disease-associated mutations are distributed throughout the ABCA4 gene and comprise missense, splice-site, and nonsense mutations as well as small deletion and insertions resulting in a truncated protein. Biochemical studies indicate that disease-linked mutations in ABCA4 cause complete or partial loss in retinal stimulated ATPase activity (21).
ABCA4 is most similar to ABCA1, an ABC transporter implicated in the efflux of cholesterol and phospholipids from cells (16,22,23). ABCA4 and ABCA1 are over 50% identical in amino acid sequence. Both proteins have a similar topological organization consisting of two tandem halves each containing a transmembrane segment followed by a large extracellular domain (ECD), a membrane spanning domain (MSD), and a nucleotide binding domain (NBD) (24,25). In addition, both transporters contain an extended C-terminal tail of about 170 amino acids in length downstream from the NBD in the C-terminal half (NBD2). The importance of the C-terminus is underscored by the finding that a mutation in ABCA4 which causes the removal of the C-terminal 30 amino acids of ABCA4 is responsible for cone-rod dystrophy (26) and a mutation leading to the deletion of the C-terminal 46 amino acids of ABCA1 is associated with Tangiers disease, an autosomal recessive disorder characterized by a loss in circulating high density lipoprotein and accumulation of cholesterol esters in peripheral tissues (27). Fitzgerald et al. (28) have examined several C-terminal deletion mutants of ABCA1 including the Δ46 mutation associated with Tangiers disease. Their studies suggest that a conserved VFVNFA motif present in the cytoplasmic C-terminal domain of ABCA1 interacts with an unknown cytoplasmic protein to orchestrate the binding of apoA-I to ABCA1 and efflux of cholesterol from cells.
The VFVNFA motif is also present within the C-terminal 30 amino acids of ABCA4, but its role in this transporter has not been determined. In this paper, we have characterized a number of C-terminal deletion and substitution mutants of ABCA4 including mutants known to cause cone-rod dystrophy and Stargardt macular degeneration (26,29). Our studies indicate that the VFVNFA motif present within the 30 C-terminal amino acids plays a crucial role in the folding of ABCA4 into a functionally active protein.
EXPERIMENT PROCEDURES
Reagents
All-trans retinal and ATP were purchased from Sigma, CHAPS detergent was from Anatrace (Maumee, OH), and lipids including brain polar lipids, DOPE, and DOPC were from Avanti Polar Lipids (Alabaster, AL). Extracti-Gel D detergent removal gel was obtained from Thermo Scientific (Rockford, IL) and 8-azidoadenosine 5′-[α32P] triphosphate was a product of ALT Bioscience (Lexington, KY). All-trans retinal and all-trans retinol were radiolabeled and isolated as described by Garwin and Saari (30).
Solutions
The composition of buffers was as follows: hypotonic buffer: 10 mM HEPES, pH 7.5; column buffer: 50 mM HEPES, pH 7.5, 0.1 M NaCl, 10 mM CHAPS, 1 mM DTT, 3 mM MgCl2, 10% glycerol, 0.32 mg/ml DOPE, and 0.32 mg/ml DOPC; reconstitution buffer: 25 mM HEPES, pH 7.5, 0.14 M NaCl, 1mM EDTA, 1 mM DTT, and 10% glycerol; and solubilization buffer: 50 mM HEPES, pH 7.5, 0.1 M NaCl, 18 mM CHAPS, 1 mM DTT, 3 mM MgCl2, 10% glycerol, 0.32 mg/ml DOPE, and 0.32 mg/ml DOPC. All-trans retinal concentration in ethanol was determined spectrophotometrically using an extinction coefficient of 42,900 M−1cm−1.
Monoclonal Antibodies and DNA Constructs
The Rim 3F4 monoclonal antibody directed against a defined epitope near the C terminus of ABCA4 and the Rho 1D4 monoclonal antibody to the nine-amino acid C-terminal sequence of rhodopsin have been described previously (2,31,32). The purified Rho 1D4 antibody was purchased from UBC-UILO (http://www.flintbox.com/). C-terminal deletion mutants of ABCA4 containing an added 9 amino acid 1D4 tag (-T-E-T-S-Q-V-A-P-A) were generated by PCR using 1) the human ABCA4-1D4 in the pCEP4 plasmid (Invitrogen) as a template; 2) a common forward primer that anneals between nt positions 5808 and 5831 and contains an Afl II site; and 3) unique reverse primers that anneal between nt 6778 to 6795 (ABCA4-Δ8-1D4), 6751 to 6771 (ABCA4-Δ16-1D4), 6726 to 6747 (ABCA4-Δ24-1D4) and 6710 to 6729 (ABCA4-Δ30-1D4) and contain a Nhe I site and the sequence for 1D4 tag. C-terminal deletion mutants of ABCA1 were also generated by PCR using human ABCA1-1D4 in the pCEP4 plasmid as the template, a common forward primer that anneals between nt position 6221 to 6239 of ABCA1 with a BamH I site, and unique reverse primers that anneal to nt positions 6689 to 6711 (ABCA1-Δ24-1D4), 6642 to 6663 (ABCA1-Δ40-1D4), and 6625 to 6645 (ABCA1-Δ46-1D4) and contain a BamH I site and the sequence for 1D4 tag. Chimera constructs ABCA4/A1 with the C-terminus of ABCA4 replaced with the C-terminus of ABCA1 just after the VFVNFA motif and ABCA1/A4 in which the C-terminus of ABCA1 was replaced with the C terminus of ABCA4 were generated by overlapping PCR. Alanine substitution mutants (ABCA4-Ala6 and ABCA1-Ala6) in which the VFVNFA motif in ABCA4 and ABCA1 were replaced with six alanine residues (VFVNFA→AAAAAA) were generated by overlapping PCR.
Expression of WT and Mutant ABCA4 and ABCA1 in HEK 293T Cells and Extraction of Membrane Proteins
HEK 293-T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine and 1.25 μg/ml fungizone. Typically, each 10-cm dish of cells at 30% confluency was transfected with 20 μg of plasmid DNA using calcium phosphate (33). After 48 h, cells from two or three plates were harvested in 1.5 ml hypotonic buffer. Membranes from homogenates of transfected cells were prepared as described previously (24), but in a smaller scale. Briefly, the cell homogenate from two or three 10-cm dishes was passed 5 times through a 26-gauge needle and subsequently applied to a step gradient consisting 5% and 60% (w/v) sucrose. Centrifugation was carried out for 30 min at 77,000 × g in a TLS 55 swing bucket (Optima TL Ultracentrifuge, Beckman, Palo Alto, CA). Membranes at the interface between the 5% and 60% layers were collected, pelleted at 100,000 × g and resuspended at a concentration of 5 mg protein/ml in buffer consisting of 10 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 0.1 mM EDTA and 10% glycerol. To compare extractability of the expressed 1D4 tagged ABCA4 mutants, 20 μl of membranes was added to 180 μl CHAPS solubilization buffer at 4 °C. The mixture was stirred for 20 min and subsequently centrifuged at 100,000 × g for 10 min in a TLA55 rotor (Beckman). The supernatant fraction (20 μl) was mixed with 4 X SDS loading buffer and resolved by SDS-PAGE for analysis by western blotting. To determine the total expression level of each mutant, 2 μl of membranes was mixed with 18 μl water, 10 μl 4 X SDS loading buffer and resolved by SDS-PAGE gel for quantification by western blotting.
Analysis of Retinoid Binding
Solid phase binding assay using [3H]-labeled all-trans retinal was carried out as described previously (7). All incubations were carried out at 4 °C. Briefly, transfected HEK 293T cells harvested from one dish were centrifuged at 2800 × g for 3 min and resuspended in 0.5 ml of solubilization buffer. The supernatant fraction obtained after centrifugation (100,000g for 10 min) was incubated with 12.5 μl Rho 1D4-Sepharose 2B immunoaffinity matrix pre-equilibrated in column buffer. After 1 h, the matrix containing the bound 1D4-tagged protein was washed several times with 0.4 ml column buffer by low speed centrifugation and mixed with 0.25 ml of 20 μM [3H]-labeled all-trans retinal (specific activity of 500 dpm/pmol, 2.5 × 106 dpm total) in column buffer for 30 min. The matrix was washed several times with 0.4 ml column buffer to remove unbound [3H]-labeled all-trans retinal and then incubated in the presence or absence of 0.5 mM ATP for 15 min. The matrix was washed 2 more times with 0.4 ml column buffer before being transferred to Ultrafree-MC (0.45 μm filter) spin column (Millipore). The samples containing the immobilized 1D4 tagged ABCA4 proteins with bound labeled retinoid was washed 3 more times before extracting the [3H]-labeled all-trans retinal from the matrix with 0.5 ml of ice-cold ethanol for 15 min at room temperature by centrifugation. Radiolabeled all-trans retinal in the ethanol extractions was determined by liquid scintillation counting. Assays were carried out in triplicate for each point determination. In general, the procedures were carried out under dim light to prevent any photoreaction of the retinoids. 1D4 tagged Na/K ATPase was treated in the same way and used as a control to monitor for background retinoid binding. The counts in the controls, typically about 20% of the test sample not treated of ATP, were subtracted from the test samples to determine specific retinoid binding. A duplicate column eluted with 60 μl 1% SDS was used to determine the amount of 1D4-tagged protein immobilized on Rho 1D4 Sepharose affinity matrix.
Reconstitution in Lipid Vesicles
For ATPase assays, 2–3 dishes of transfected HEK 293T cells were solubilized as described for retinoid binding studies. The solubilized protein was incubated with 25 μl Rim 3F4 or Rho 1D4 Sepharose for 1 h at 4 °C in an Ultrafree-MC spin column. The matrix was washed 6 times with 0.4 ml column buffer to remove unbound protein. The matrix was then incubated with 37.5 μl of 0.2 mg/ml competing peptide (3F4 (YDLPLHPRTA) or 1D4 (TETSQVAPA) peptide) in column buffer at 7 °C for 20 min. The addition of peptide and elution was repeated and two eluates were combined. The eluted ABCA4 proteins were reconstituted into brain polar lipids using the procedure of Sun et al with minor modifications (6). Briefly, 9 μl of 25 mg/ml sonicated brain polar lipid extract (33% PE, 18% phosphatidylserine, 13% PC, 4.1% phosphatidylinositol, 31% other lipids) was mixed with 6 μl of 15% n-octylglucoside (w/v) in 25 mM HEPES, pH 7.4, 140 mM NaCl, 10% glycerol. Purified ABCA4 protein (60 μl) was added, and the mixture was incubated on ice for 30 min. Reconstitution buffer (180 μl) was added rapidly, and the sample was then passed through 400 μl of Extracti-gel resin pre-equilibrated with reconstitution buffer in a Mobicol mini-column fitted with a 10 μm pore size filter (MoBiTec, Gottingen, Germany). The flow-through containing reconstituted ABCA4 or ABCA1 protein was collected at 0.8 ml/min by applying gentle pressure with a syringe, and 5 mM MgCl2 was added prior to carrying out ATPase activity measurements.
ATPase Assay
ATP assays were carried out using [α-32P]ATP (Perkin Elmer) and thin layer chromatography as previously described (34). ATPase assays were carried out in a 10-μl total reaction volume consisting of 8 μl (20–40 ng) of reconstituted protein and 1 μl of 10X retinoid or buffer. The reaction was initiated by the addition of 1 μl of a 10X ATP solution (0.2 μCi) to achieve a final concentration of 50 μM. After 30 min at 37 °C, 4 μl of 10% SDS were added. One μl of the reaction mixture was spotted onto a polyethyleneimine cellulose plate (Sigma-Aldrich) and chromatographed in 0.5 M LiCl/1 M formic acid. The plate was exposed to a storage phosphor screen for 3 h and scanned in a Typhoon Variable Mode Imager (GE Healthcare). Spots corresponding to ATP and ADP were quantified using ImageQuant TL software (Amersham Biosciences). The ratio of the amount of ADP produced to the initial amount of ATP present in the reaction mixture was calculated. Each sample was assayed in triplicate. Buffer blanks were included to determine nonenzymatic ATP hydrolysis, which was subtracted from the total ATP hydrolyzed.
Protein Determination
The amount of protein in detergent extracts and reconstituted vesicles was determined by comparing the intensity of Coomassie Brilliant Blue staining of ABCA4 with that of standard amounts of bovine serum albumin after SDS-polyacrylamide gel electrophoresis.
Western Blot Analysis
Proteins were separated by SDS gel electrophoresis on 8% polyacrylamide gels and transferred to Immobilon FL membranes (Millipore) at 13V for 30 min in a semidry transfer apparatus (Bio-Rad) using a buffer consisting of 25 mM Tris, 192 mM glycine, 10% methanol, pH 8.3. The membranes were blocked with 0.5% skim milk in PBS (140 mM NaCl, 3 mM KCl, 10 mM phosphate, pH 7.4) for 30 min, rinsed and incubated with Rho 1D4 monoclonal antibody diluted in 0.5% milk, PBS for 1 h. After extensive washing in PBS containing 0.05% Tween 20 (PBST), the membranes were treated for 30 min with secondary antibody (goat anti-mouse Ig conjugated with IRDye 680 (LI-COR, Lincoln, NE), diluted 1:10,000 in 0.5% milk, 0.02 % SDS, PBST) and washed in PBST prior to analysis with a LI-COR infrared imaging system.
Photoaffinity Labeling of ABCA4
Five μl membrane protein (25 μg protein) in resuspension buffer (25 mM HEPES, pH 7.5, 0.15 M NaCl, 5 mM MgCl2) was mixed with 15 μl of 8-azido [α-32P]ATP (4.5 μM, 0.05 mCi/ml) in the same buffer. The sample was immediately exposed to 254 nm UV at a distance of 10 cm for 10 min. SDS sample buffer (10 μl) was mixed with each sample without heating, and the proteins were resolved by SDS-PAGE and analyzed by autoradiography. Triplicate cell expression samples were carried out for each mutant. The same amount of unlabeled membranes was resolved by SDS-PAGE and subjected to western blotting analysis for protein quantification.
Immunofluorescence Labeling of Cells
HEK293T cells were grown on glass coverslips and transfected with the plasmid using calcium phosphate. After 24 h, the cells were washed in 0.1M phosphate buffer, and fixed for 20 min in 4% paraformaldehyde in 0.1M phosphate buffer at 22°C. Cells were then blocked and permeabilized for 30 min at 22 °C in the same buffer containing 10% (v/v) normal goat serum and 0.1% (v/v) Triton X-100. For single labeling experiments, the cells were treated with the Rho 1D4 antibody for 1 h at 22 °C, washed in PBS buffer and treated with goat anti-mouse Ig coupled Cy3 diluted 1:1000 in phosphate buffer and counterstained with the nuclear dye DAPI. For double labeling studies, ABCA4 protein was labeled with Rho 1D4 followed by goat antimouse Ig conjugated to Alexa 488 and calnexin was labeled with a rabbit anti-calnexin polyclonal antibody followed by goat anti-rabbit Ig conjugated to Alexa 594. Immunofluorescence labeling was visualized using a Meta 510 Zeiss Confocal microscope or a Zeiss Axioplan 2 fluorescence microscope.
RESULTS
C-terminal 1D4 Tag Does Not Affect the ATPase Activity of ABCA4
In previous studies the Rim 3F4 monoclonal antibody coupled to Sepharose was used to purify ABCA4 from bovine rod outer segments and recombinant ABCA4 from transfected cells for structure-function studies (2,6,7,21,34). Since the 3F4 epitope is located close to the C-terminus (amino acids 2252-2262) of ABCA4, the Rim 3F4 antibody can not be used to purify ABCA4 C-terminal deletion mutants for biochemical studies. To circumvent this problem, we have added a 9 amino acid 1D4 tag to the C-terminus of WT and mutant ABCA4. This tag recognized by the Rho 1D4 monoclonal antibody (31) has been shown previously to be extremely effective for the immunoaffinity purification of membrane proteins for structure-function studies (35,36).
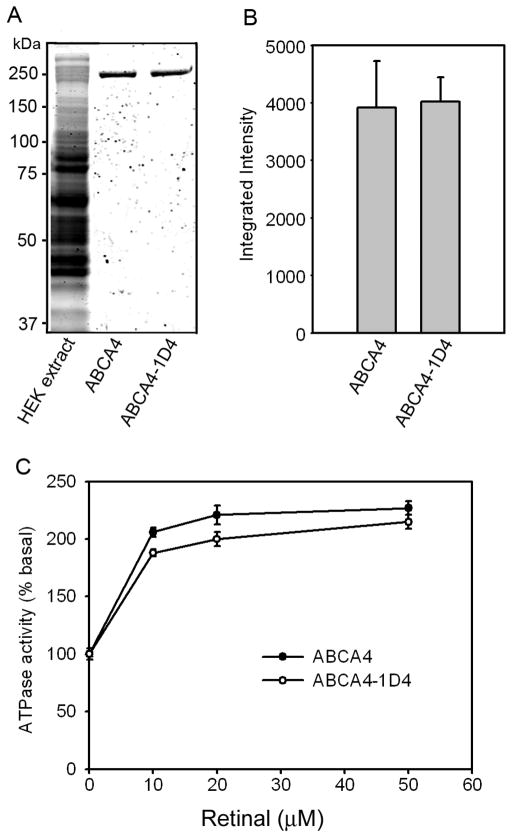
First, it was necessary to determine if addition of the 1D4 tag has any effect on the biochemical properties of ABCA4. This was determined by expressing ABCA4 and ABCA4-1D4 in HEK 293T cells and purifying the proteins from CHAPS solubilized membranes on a Rim 3F4 - Sepharose immunoaffinity matrix (Figure 1A,B). Both proteins expressed at similar levels and were solubilized to the same extent in CHAPS detergent. Elution of the proteins bound to the immunoaffinity matrix with the 3F4 peptide yielded similar amounts of highly pure 250 kDa ABCA4 as quantified on SDS gels stained with Coomassie blue. After reconstitution into brain lipid vesicles, both ABCA4 and ABCA4-1D4 displayed similar basal and all-trans-retinal stimulated ATPase activities (Figure 1C). The basal activities for ABCA4 and ABCA4-1D4 were 54.3 ± 0.9 and 48.9 ± 2.5 nmoles ATP hydrolyzed per min per mg protein, respectively. HEK 293T cells transfected with vector alone were used in control studies. No contaminating proteins or ATPase activity was observed when the detergent solubilized extract from these cells was passed through either a Rim 3F4 or Rho 1D4 immunoaffinity column (data not shown). These results indicate that the C-terminal 1D4 tag does not affect the expression, purification, or enzymatic activity of ABCA4, and the basal and retinal stimulated ATPase activities are solely due to expressed ABCA4.
Figure 1. Expression and ATPase activity of ABCA4 and ABCA4-1D4.
ABCA4 and ABCA4-1D4 containing a 9 amino acid 1D4 epitope were expressed in HEK 293T cells, solubilized in CHAPS detergent, purified on a Rim 3F4 – Sepharose immunoaffinity matrix and reconstituted into liposomes for determination of basal and all-trans retinal stimulated ATPase activity measurements. A. SDS gel stained with Coomassie blue showing a typical membrane extract from HEK 293T cells expressing ABCA4 (lane 1), immunoaffinity purified ABCA4 (lane 2), and immunoaffinity purified ABCA4-1D4 (lane 3). B. Relative expression levels of ABCA4 and ABCA4-1D4 quantified on Western blots of membranes from HEK293 cells expressing these proteins. C. ATPase activity of purified and reconstituted ABCA4 and ABCA4-1D4 as a function of added all-trans retinal. For each assay 20–40 ng of protein was used. Data show the average value for 3 experiments ± SD.
1D4-tagged ABCA4 and ABCA1 Mutants
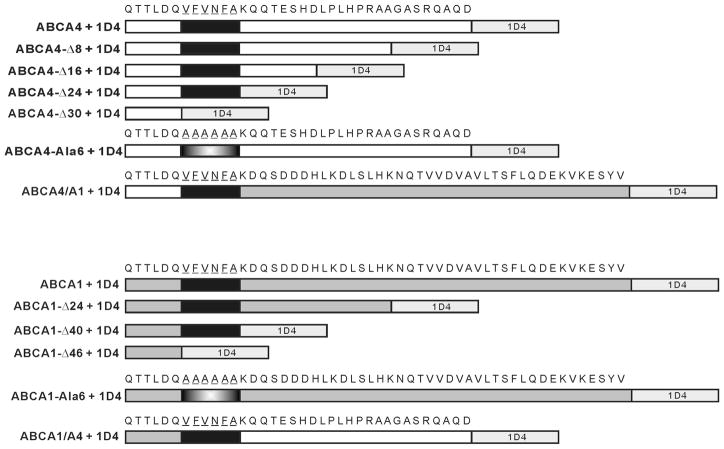
To investigate the importance of the C-terminus on the biochemical properties of ABCA4, we generated four C-terminal deletion mutants, one C-terminal chimera mutant (ABCA4/A1) in which the C-terminal 24 amino acid segment of ABCA4 was replaced with the C-terminal 40 amino acid segment of ABCA1, and one C-terminal mutant in which a conserved VFVNFA motif was replaced with six alanine residues (ABCA4-Ala6) (Figure 2). Several related ABCA1 mutants were also constructed for comparative studies. The WT and mutant proteins contained a 1D4 C-terminal tag for detection and purification. For simplicity we have omitted 1D4 in the designation of the various ABCA4 proteins used in this study.
Figure 2. Schematic showing the sequence of the various C-terminal deletion and chimera mutants of 1D4 tagged ABCA4 and ABCA1 used in this study.
Only the C-terminal 36 and 52 amino acids are shown for full-length ABCA4 and ABCA1, respectively. The 9 amino acid 1D4 epitope (TETSQVAPA) is shown in light gray; ABCA4 in white; ABCA1 in darker gray; the VFVNFA motif in black, and the Ala substitution in blend.
The Effect of C-terminal Deletions on the Expression and Solubilization of ABCA4
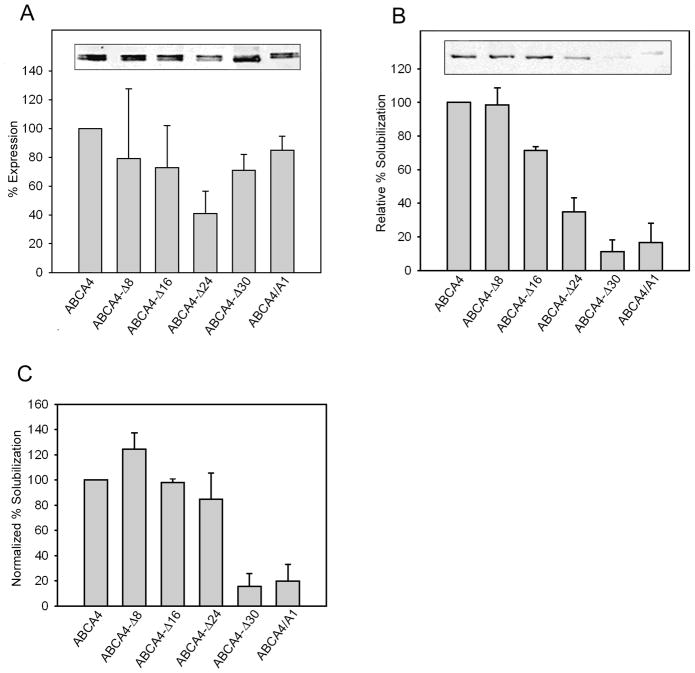
The level of expression of WT ABCA4 and the various mutants was determined by directly solubilizing membranes from HEK 293T cells in SDS for analysis by SDS-PAGE and western blotting (Figure 3A). The WT and mutants migrated as tightly spaced doublets, presumably arising from differential posttranslational processing of the proteins. The level of expression of the mutants was typically within 75% that of WT ABCA4 with the exception of ABCA4-Δ24 which expressed at only 40% of WT ABCA4. The ability of CHAPS to effectively solubilize the ABCA4 mutants from HEK 293T cell membranes was also investigated. Figure 3B shows that all CHAPS-solubilized mutants migrated as a sharp single band, but at variable levels. The single bands suggest that CHAPS solubilized only a single class of ABCA4 proteins, presumably nonaggregated proteins having a native-like structure. ABCA4-Δ8 was solubilized in CHAPS at a level comparable to WT ABCA4, whereas the ABCA4-Δ16 and ABCA4-Δ24 mutants on average solubilized at about 75% and 35% of WT ABCA4. The ABCA4-Δ30 and the ABCA4/A1 mutants solubilized at only 10–20% that of WT ABCA4. When CHAPS solubilization of the mutants was normalized to the level of protein expression as determined by SDS solubilization, only the ABCA4-Δ30 and ABCA4/A1 mutants showed significantly reduced levels compared to WT ABCA4 (Figure 3C) suggesting that a major fraction of these mutants (~80–90%) were highly misfolded.
Figure 3. Expression and solubilization of ABCA4 C-terminal mutants.
Membranes from HEK 293T cells expressing WT and mutant ABCA4 and containing the 1D4 tag were treated with detergent and the amount of ABCA4 was quantified on Western blots labeled with the Rho 1D4 antibody. (A) Relative expression of ABCA4 mutants was determined from membranes solubilized with SDS; (B) Relative solubilization was determined by solubilizing membranes in 18 mM CHAPS and removing unsolubilized material by centrifugation prior to SDS gel electrophoresis and Western blotting. (C) Normalized solubilization was determined from the ratio of CHAPS solubilization to the level of expression determined from SDS solubilized membranes and expressed as a % of WT ABCA4. Inserts show representative examples of western blots of the ABCA4 mutants. A doublet band was present in SDS solubilized samples and a single sharp band was observed for CHAPS solubilized samples suggesting that poorly processed ABCA4 was not solubilized by CHAPS. Quantitative data are an average from 3 or more independent experiments ± SD.
8-Azido-ATP Photoaffinity Labeling of ABCA4 Mutants
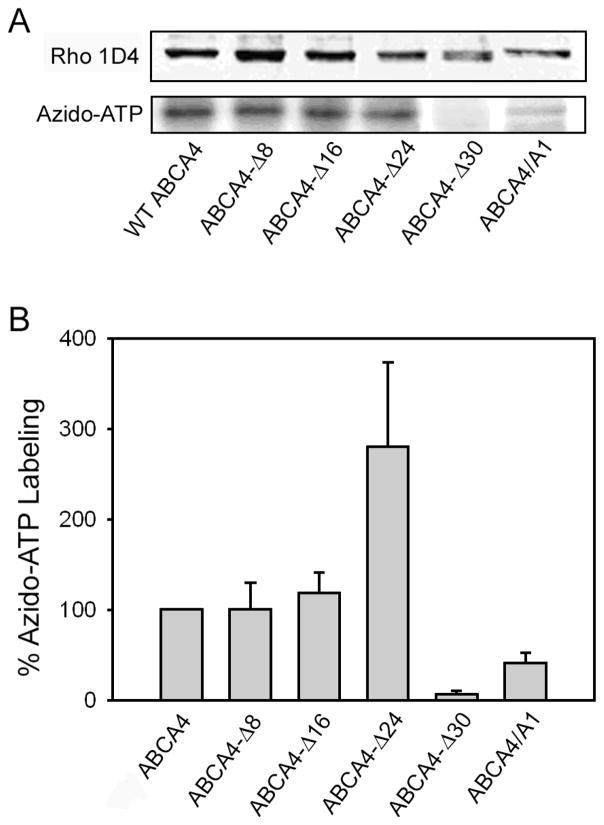
The capacity of the C-terminal ABCA4 deletion mutants to bind ATP was investigated by photoaffinity labeling. Membranes from transfected HEK 293T cells were treated with 8-azido-[32P]ATP, irradiated with ultraviolet light, and resolved by SDS-PAGE for quantitative analysis of bound radiolabeled azido-ATP on a phosphor imager. As shown in Figure 4A and 4B, WT ABCA4, ABCA4-Δ8, and ABCA4-Δ16 were labeled to a similar extent. The ABCA4-Δ24 mutant displayed a 3-fold higher level of 8-azido-ATP labeling when normalized for the amount of protein present, while the ABCA4/A1 chimera mutant showed significantly reduced labeling and the ABCA4-Δ30 lacking a conserved VFVNFA motif did not label.
Figure 4. Azido-ATP photoaffinity labeling of ABCA4 mutants.
Membranes from HEK 293T cells expressing the ABCA4 mutants tagged with the 1D4 epitope were photoaffinity labeled with 8-azido-adenosine 5′-[α 32P] triphosphate prior to analysis on SDS gels. A. Representative western blots of 1D4 tagged ABCA4 proteins labeled with the Rho 1D4 antibody and azido-ATP labeling analyzed on a phosphor imager. B. Quantitative analysis of azido-ATP labeling of mutants relative to WT ABCA4 after normalization for the amount of ABCA4. Data represent an average of 3 experiments ± SD.
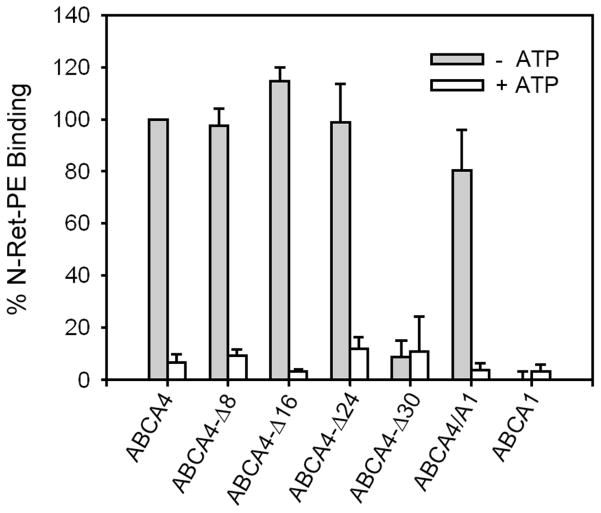
Binding of N-retinylidene-PE (N-ret-PE) to C-terminal Mutants
In previous studies, we have shown that purified ABCA4 from bovine ROS immobilized on an immunoaffinity matrix specifically bound N-ret-PE in the absence of ATP (7). The addition of ATP caused N-ret-PE to dissociate from ABCA4. We have used this assay to measure N-ret-PE binding to the C-terminal mutants (Figure 5). All mutants except ABCA4-Δ30 bound N-ret-PE in the absence of ATP at levels comparable to WT ABCA4. N-ret-PE substrate binding was lost upon the addition of ATP. As part of this study, we also determined if purified ABCA1 was capable of binding N-ret-PE. As shown in Figure 5, no binding of N-ret-PE to ABCA1 was observed.
Figure 5. Binding of N-ret-PE to ABCA4 mutants in the absence and presence of ATP.
WT and mutant ABCA4 tagged with the 1D4 epitope were immobilized on a Rho 1D4-Sepharose matrix and incubated with [3H]-labeled all-trans retinal in the presence of PE. The matrix was washed to remove unbound substrate and incubated in the absence or presence of 0.5 mM ATP. The bound N-ret-PE was eluted with ethanol and quantified by scintillation counting. Data represent the average of 3 or more experiments ± SD.
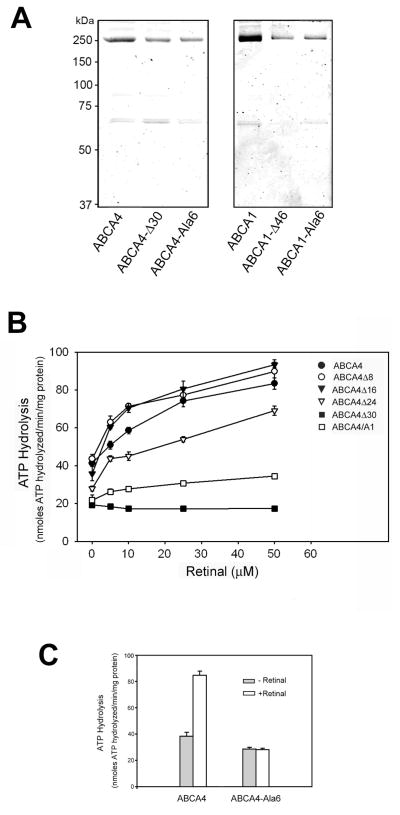
Immunoaffinity purification of ABCA1 and ABCA4
For analysis of ATPase activities, 1D4-tagged WT and mutant ABCA4 and ABCA1 were purified from CHAPS solubilized extracts of HEK293 cells on a Rho 1D4 immunoaffinity matrix. Figure 6A shows representative Coomassie blue stained SDS gels of the protein preparations eluted from the matrix with the competing 1D4 peptide. The ABCA4 and ABCA1 transporters were present as intensely stained bands having an apparent molecular mass of 250 kDa. Another faint 65 kDa protein band was often observed in both WT and mutant ABCA4 and ABCA1 preparations. The identity of this protein remains to be determined, but it most likely represents a chaperone protein that is tightly bound to a portion of these heterologously expressed ABCA transporters.
Figure 6. Purification of WT and mutant 1D4-tagged ABCA4 and ABCA1 and analysis of retinal stimulated ATPase activities of ABCA4 mutants.
ABCA4 and ABCA1 proteins containing the 1D4 tag were expressed in HEK 293T cells, purified on Rho1D4-Sepharose matrix, and reconstituted into brain lipid vesicles. The ATPase activity of ABCA4 mutants was measured as a function of increasing all-trans retinal concentrations. A. Coomassie Blue stained SDS gels showing the intense 250 kDa ABCA4 or ABCA1 transporters and an additional faint 65 kDa protein. B. Retinal stimulated ATPase activity of purified and reconstituted ABCA4 proteins. Data represent an average of 3 experiments ± SD. C. ATPase activity of wild-type ABCA4 and the ABCA4-Ala6 mutant in which the VFVNFA motif was replaced with six alanine residues. Data represent an average of 3 experiments ± SD.
The Effect of C-terminal Mutations on Basal and Retinal Activated ATPase Activity
Basal and all-trans retinal stimulated ATPase activity was measured for the immunoaffinity purified C-terminal ABCA4 mutants reconstituted into brain polar lipid vesicles containing PE. All mutants retained varying levels of basal ATPase activity (Figure 6B). The ABCA4-Δ8 and ABCA4-Δ16 mutants exhibit basal ATPase activity that was stimulated by the addition of all-trans retinal at levels similar to WT ABCA4. The ABCA4-Δ24 mutant had reduced basal and all-trans retinal stimulated ATPase activities, whereas the ABCA4-Δ30 mutant lacking the VFVNFA motif displayed low basal activity and no stimulation by all-trans retinal. The ABCA4/A1 chimera mutant exhibited low basal activity and only limited stimulation by all-trans retinal.
To further investigate the importance of the VFVNFA motif in retinal stimulated ATPase activity, we constructed and expressed an ABCA4 mutant in which the 6 amino acids of this motif were replaced with alanine residues (ABCA4-Ala6). As seen in Figure 6C, this mutant, like the ABCA4Δ30 mutant, was devoid of retinal stimulated ATPase activity.
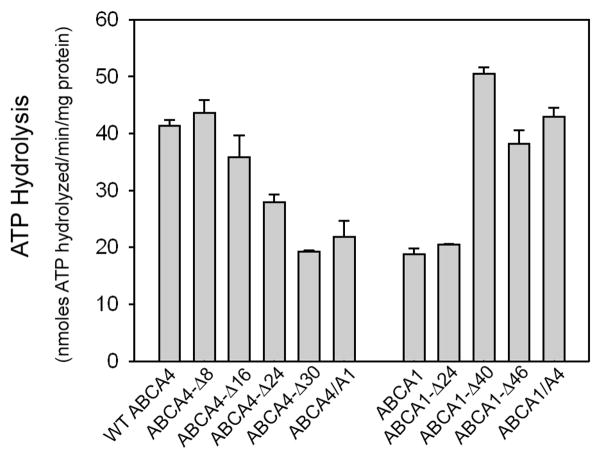
Differential Effect of C-terminal Deletion and Chimera Mutations on the Basal ATPase Activities of ABCA1 and ABCA4
Although the effect of ABCA1 C-terminal deletion mutations on the efflux cholesterol in the presence and absence of apoA-1 has been reported (28), their effect on the ATPase activity of the purified transporter was not investigated. As part of our study, we compared the basal ATPase activity of C-terminal deletion mutants of ABCA4 with several C-terminal ABCA1 deletion mutants (Figure 7). A gradual decrease in basal ATPase activity of ABCA4 was observed with increasing deletion of the C-terminus. An opposite trend was observed for ABCA1 deletion mutants. A higher ATPase activity was observed for the deletion mutants missing the C-terminal 40 and 46 amino acids. In addition two chimera mutants ABCA4/A1 and ABCA1/A4 were examined (Figure 7). Whereas the ABCA4/A1 mutant showed significantly reduced basal ATPase activity relative to WT ABCA4, the ABCA1/A4 mutant showed increased activity relative to WT ABCA1. These results suggest that segments of ABCA4 downstream of the VFVNFA motif enhance the basal ATPase activity of ABCA4 and ABCA1, whereas segments of ABCA1 downstream of this motif reduce the basal ATPase activity of these transporters. The basal activities, however, do not appear to reflect the true function of these transporters since the ABCA1-Δ46 mutant has been reported to be defective in cholesterol efflux (28).
Figure 7. Basal ATPase activity of ABCA4 and ABCA1 mutants.
ABCA4 and ABCA1 mutants were expressed in HEK 293T cells, immunoaffinity purified on a Rho 1D4-Sepharose matrix and reconstituted into brain lipid vesicles for analysis of basal ATPase activity.
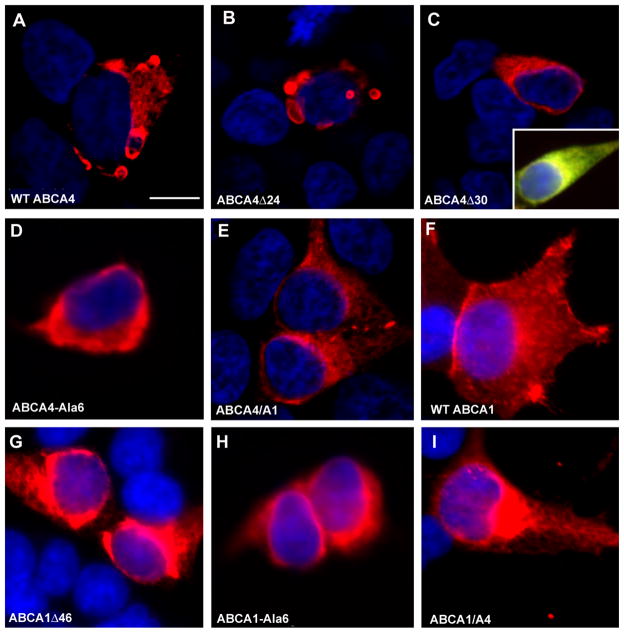
Immunofluorescence Localization of ABCA4 Mutants
Previous studies in our laboratory have shown that WT ABCA4 expressed in COS-1 cells preferentially localize to intracellular vesicles of varying sizes consistent with the intracellular localization of ABCA4 in photoreceptor cells (2,37). In the present study, we have compared the subcellular localization of the 1D4 tagged WT and mutant ABCA4 expressed in HEK 293T cells. WT ABCA4 and deletion mutants ABCA4-Δ8, ABC4-Δ16 and ABCA4-Δ24 all exhibited a similar subcellular distribution with the majority of the protein being localized to intracellular vesicular structures (Figure 8 A–B). In contrast essentially all the ABCA4-Δ30 and ABCA4-Ala6 mutants deficient in the VFVNFA motif and most of the ABCA4/A1 mutant showed a perinuclear, reticular distribution characteristic of ER retention (Fig 8 C–E). Double labeling using the ER marker calnexin confirmed the localization of ABCA4-Δ30 within the ER of cells (Fig 8C).
Figure 8. Immunofluorescence microscopy of HEK 293T cells expressing 1D4-tagged ABCA4 mutants.
HEK 293T cells transfected with WT or mutant ABCA4 plasmids were fixed in paraformaldehyde, permeabilized in 0.1% Triton X-100, labeled with the Rho 1D4 antibody and Cy3-tagged goat anti-mouse Ig, and counterstained with DAPI nuclear stain. A – B. WT and ABCA4-Δ24 mutants are localized to large intracellular vesicular structures. A similar labeling pattern was observed for the ABCA4-Δ8 and ABCA4-Δ16 mutants (data not shown). C–D ABCA4-Δ30 and ABCA4-Ala6 mutants labeling shows a perinuclear pattern characteristic of ER localization. Inset in C shows a merged image of a cell double labeled for ABCA4-Δ30 (red) and the ER marker calnexin (green). Colocalization is indicated by a yellow color. E. ABCA4/A1 mutants show primarily a perinuclear pattern characteristic of ER although some intravesicular labeling is observed. F. WT ABCA1 mutant shows a pattern of labeling characteristic of plasma membrane localization. G–H. ABCA1-Δ46 and ABCA1-Ala6 mutants show a perinuclear pattern of labeling characteristic of ER localization. I. ABCA1/A4 mutant shows strong perinuclear labeling along with some plasma membrane labeling. Bar = 5 μ;m..
As part of this study, we also investigated the distribution of the 1D4 tagged WT and mutant ABCA1 proteins expressed in HEK 293 cells. WT-ABCA1 localized to the plasma membrane of HEK293 cells as previously reported (42), whereas essentially all of the ABCA1-Δ46 and most of the ABCA1-Ala6 mutant displayed a perinuclear distribution characteristic of retention in the ER (Fig 8F–H). The ABCA1/A4 mutant localized to both the plasma membrane and ER (Fig 8I).
These studies suggest that the major fraction of ABCA4-Δ8, ABCA-Δ16 and ABCA4-Δ24 mutants are folded in a native-like conformation that allows the protein to exit the ER and accumulate as intracellular vesicles in HEK 293T cells. In contrast, essentially all of the ABCA4-Δ30 and ABCA4-Ala6 mutants are highly misfolded such that they are retained in the ER by the quality control system of cells. WT ABCA1 goes to the plasma membrane, while most of the ABCA1-Δ46 is retained in the ER.
DISCUSSION
In this study we have expressed, purified and characterized a series of C-terminal deletion and substitution mutants of ABCA4 in order to define the role of the C-terminus of ABCA4 in protein structure and function and to gain insight into the molecular basis for retinal degenerative diseases associated with mutations in this segment of the ABCA4 transporter. Our studies indicate that a conserved VFVNFA motif is required for the production of a functional protein. The ABCA4-Δ30 mutant lacking the VFVNFA motif failed to bind the substrate N-ret-PE, was not labeled with 8-azido ATP, and showed limited basal and no retinal stimulated ATPase activity. Similarly, the ABCA4-Ala6 mutant in which the VFVNFA motif was replaced with six alanine residues was not stimulated by all-trans retinal. In contrast the ABCA4-Δ8, ABCA4-Δ16, and ABCA4-Δ24 mutants which have the VFVNFA motif are all functionally active.
Our results showing that the VFVNFA motif is required for the production of a functionally active ABCA4 protein is in general agreement with the studies of Fitzgerald et al (28) showing that this conserved motif is critical for the function of ABCA1 in apoA-1 binding and cholesterol efflux from cells (28). However, the VFVNFA motif may play distinct mechanistic roles for each transporter. In the case of ABCA1, the VFVNFA motif has been suggested to interact with an unidentified cytoplasmic protein to induce apoA-1 binding to an extracellular domain of ABCA1 and promote cholesterol efflux from cells (28). Our studies on ABCA4 indicate that the VFVNFA motif plays a crucial role in proper folding of the protein into a functionally active conformation. First, no candidate proteins that bind to WT ABCA4, but not the ABCA4-Δ30 or ABCA4-Ala6 mutants, were observed in purified protein preparations as analyzed on Coomassie blue stained SDS gels. Second, the ABCA4-Δ30 mutant lacking the VFVNFA motif solubilized poorly in CHAPS detergent relative to WT and other ABCA4 deletion mutants suggesting that most of the expressed protein was misfolded. Third, even the mutant protein lacking the VFVNFA motif that was solubilized was functionally inactive except for residual basal ATPase activity. Fourth, the subcellular distribution of ABCA4-Δ30 and ABCA4-Ala6 differed significantly from that of WT ABCA4 and the other functionally active deletion mutants as revealed by immunofluorescence labeling of cells. In particular, WT ABCA4, ABCA4-Δ8, ABCA4-Δ16 and ABCA4-Δ24 mutants, all preferentially localized to large intracellular vesicular structures in HEK 293T cells, a distribution previously observed for heterologously expressed peripherin/rds and rom-1, two photoreceptor proteins that co-localize with ABCA4 along the rim region of rod and cone photoreceptors (37–39). In contrast, the ABCA4-Δ30 and ABCA4-Ala6 mutants localized in a diffuse perinuclear pattern characteristic of retention of these proteins in the ER. Together, these studies indicate that VFVNFA motif of ABCA4 is required for the proper folding of ABCA4 into its native, functional state.
Our studies suggest that the VFVNFA motif contributes to the proper folding of ABCA1. The ABCA1-Δ46 and ABCA1-Ala6 mutants lacking the VFVNFA motif, and previously reported to be devoid of cholesterol efflux activity (28), were largely retained in the ER suggesting that the loss of function of these mutant is due protein misfolding as the possible loss in protein-protein interactions as previously suggested (28).
Some differences in the role of VFVNFA in ABCA4 and ABCA1, however, are evident in our analysis of the basal ATPase activity of the deletion mutants. The ABCA1 C-terminal deletion mutant, ABCA1-Δ46 and ABCA1-Δ40 displayed a significantly higher ATPase activity than WT ABCA1, whereas the corresponding ABCA4 mutants, ABCA4-Δ30 and ABCA4-Δ24 mutants, showed a marked reduction in ATPase activity compared to WT ABCA4 (Figure 7). While the VFVNFA motif is critical for functional activity of ABCA4 through it effect on protein folding, segments just downstream of this motif appear to play a role in modulating the functional properties of ABCA4. ABCA4-Δ24 exhibited 3-fold higher 8-azido ATP labeling, but lower basal and retinal stimulated ATPase activity suggesting that amino acids just downstream of the VFVNFA motif may directly or indirectly interact with the NBDs to modify ATP binding and hydrolysis in NBD2 (37). The importance of downstream sequences is further supported in the expression and functional analysis of the ABCA4/A1 chimera mutant in which the C-terminal 24 amino acids of ABCA4 is replaced with the C-terminal segment of ABCA1. This mutant retained its ability to bind N-ret-PE, but showed a marked reduction in both azido-ATP labeling and retinal stimulated ATPase activity. Furthermore, this mutant solubilized poorly with CHAPS and exhibited a subcellular distribution consistent with a major fraction of the protein being retained in the ER. These results suggest that the ABCA1 C-terminal segment interferes with efficient folding of ABCA4 and has a negative effect on nucleotide binding and hydrolysis activities. Interestingly, an opposite effect was observed for the reverse chimera mutant ABCA1/A4 in which the C-terminal segment of ABCA1 was replaced with that of ABCA4. In our study, the basal ATPase activity of this mutant was twice the activity of WT ABCA1 (Figure 7) and in an earlier published study (28), a higher cholesterol efflux activity in the presence or absence of apoA-1 was observed for this mutant relative to WT ABCA1. Hence, the C-terminus of ABCA4 appears to contain residues that enhance the functional activity of ABCA1, possibly through interactions with the NBDs, whereas the C-terminus of ABCA1 has residues that inhibit the folding of ABCA4 and diminish its functional activity.
Alignment of C-terminal ABCA4 sequences from various species show a conserved KQQ(T/N)E region just downstream of the VFVNFA motif which may contribute to enhanced ATPase activity of the ABCA1/A4 chimera mutant (Figure 9). ABCA1 orthologs also contain a conserved region (KDQSDD) just downstream of the VFVNFA motif. Additional mutational studies should further define the importance of these conserved regions in modulating the nucleotide dependent activities of these ABCA transporters.
Figure 9. Sequence alignments of the C-terminal segments from several ABCA4 and ABCA1 orthologues.

The VFVNFA motif shown in bold type is underlined. Sequences just downstream of the VFVNFA are conserved for ABCA4 and ABCA1 and may contribute in modulating the functional activities of these transporters.
Mutations in the ABCA4 gene have been shown to cause a number of retinal degenerative diseases of varying severity. A model has been proposed in which the severity of the disease phenotype is inversely proportional to the level of residual functional activity displayed by the mutant ABCA4 proteins (40,41). Retinitis pigmentosa, the most severe disease phenotype with complete loss in vision, would arise when mutations in both alleles cause the complete loss in function such as for the case of frameshift mutations causing severely truncated proteins. At the other end of the spectrum, age-related macular degeneration, the mildest phenotype can occur in individuals who are heterozygous for selected disease-linked mutations in ABCA4. These individuals retain a significant proportion of ABCA4 functional activity from the wild-type allele. Cone rod dystrophy and Stargardt macular degeneration are intermediate in severity with the former resulting from mutations that cause a significant, but not complete, loss in ABCA4 function and the latter resulting from mutations which result in only partial loss in activity. Our studies on C-terminal deletion mutants are consistent with this model and provide mechanistic insight into the different phenotypes displayed by two patients with deletion mutations in the C-terminus of ABCA4. A patient with cone-rod dystrophy was reported to be compound heterozygous for a 6730-16del44 mutation which causes the loss in the C-terminal 29 amino acids of ABCA4 including the VFVNFA motif and a R212C missense mutation (26). The relatively severe phenotype of this patient is consistent with the complete loss in functional activity observed for the ABCA4-Δ30 mutant. The R212C missense mutation appears to retain some of its activity at least with respect to azido-ATP photoaffinity labeling (21). Another patient diagnosed with a mild form of Stargardt macular degeneration was found to be compound heterozygous for a 6748delA mutation in one allele and a S1066P missense mutation in the other allele (29). The 6748delA mutant protein is comparable to the ABCA4-Δ24 mutant studied here in that the protein is truncated after the VFVNFA motif. As shown in our study, the ABCA4-Δ24 mutant retains significant functional activity. The functional consequence of the S1066P mutation in ABCA4 has not been examined. However, based on analysis of other disease-linked missense mutation in the ECD1 region of ABCA4 (21), it is likely that this mutant also retains at least some functional activity. Hence, together these mutations are likely to provide sufficient functional activity to generate the milder phenotype of Stargardt macular degeneration.
In summary, we have shown that the VFVNFA motif is required for proper folding of ABCA4 into a functionally active protein. Segments downstream of this motif appear to modulate the activity of ABCA4 possibly through direct or indirect interaction with the NBD domains of ABCA4. Functional analysis of the C-terminal deletion mutants provides a molecular based rationale for the phenotype displayed by patients with disease-associated mutations in this region of ABCA4.
Acknowledgments
We thank Frank Dyka for advice on the construction of various ABCA4 and ABCA1 mutants.
Footnotes
This work was supported by Canadian Institutes for Health Research (CIHR) grant MT 5822.
The abbreviations used are: PE, phosphatidylethanolamine; N-ret-PE, N-retinylidenephosphatidylethanolamine; RPE: retinal pigment epithelial; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; PBS, phosphate-buffered saline; HEPES, (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid). ECD, exocytoplasmic domain; NBD, nucleotide binding domain; MSD, membrane spanning domain; DOPC, dioleoylphosphatidylcholine; DOPE, dioleoylphosphatidylethanolamine; Ig, immunoglobulin; DAPI, 4,6 – diamido-2-phenylindol, ER, endoplasmic reticulum, WT, wildtype
References
- 1.Allikmets R, Singh N, Sun H, Shroyer NE, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li YX, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. Nature Genetics. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 2.Illing M, Molday LL, Molday RS. J Biol Chem. 1997;272:10303–10310. doi: 10.1074/jbc.272.15.10303. [DOI] [PubMed] [Google Scholar]
- 3.Molday LL, Rabin AR, Molday RS. Nat Genet. 2000;25:257–258. doi: 10.1038/77004. [DOI] [PubMed] [Google Scholar]
- 4.Azarian SM, Travis GH. FEBS Lett. 1997;409:247–252. doi: 10.1016/s0014-5793(97)00517-6. [DOI] [PubMed] [Google Scholar]
- 5.Weng J, Mata NL, Azarian SM, Tzekov RT, Birch DG, Travis GH. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Molday RS, Nathans J. J Biol Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 7.Beharry S, Zhong M, Molday RS. J Biol Chem. 2004;279:53972–53979. doi: 10.1074/jbc.M405216200. [DOI] [PubMed] [Google Scholar]
- 8.Molday RS. J Bioenerg Biomembr. 2007;39:507–517. doi: 10.1007/s10863-007-9118-6. [DOI] [PubMed] [Google Scholar]
- 9.Papermaster DS, Schneider BG, Zorn MA, Kraehenbuhl JP. J Cell Biol. 1978;78:415–425. doi: 10.1083/jcb.78.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allikmets R. Am J Hum Genet. 2000;67:793–799. doi: 10.1086/303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozet JM, Gerber S, Souied E, Ducroq D, Perrault I, Ghazi I, Soubrane G, Coscas G, Dufier JL, Munnich A, Kaplan J. Mol Genet Metab. 1999;68:310–315. doi: 10.1006/mgme.1999.2925. [DOI] [PubMed] [Google Scholar]
- 12.Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, Weber BH. Am J Hum Genet. 2000;67:800–813. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelisken O, De Laey JJ. Int Ophthalmol. 1985;8:225–235. doi: 10.1007/BF00137651. [DOI] [PubMed] [Google Scholar]
- 14.Fishman GA, Farber M, Patel BS, Derlacki DJ. Ophthalmology. 1987;94:809–814. doi: 10.1016/s0161-6420(87)33533-x. [DOI] [PubMed] [Google Scholar]
- 15.Fishman GA, Farbman JS, Alexander KR. Ophthalmology. 1991;98:957–962. doi: 10.1016/s0161-6420(91)32196-1. [DOI] [PubMed] [Google Scholar]
- 16.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Hayden MR, et al. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 17.Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Deutman AF, Hoyng CB. Hum Mol Genet. 1998;7:355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzalez-Duarte R, Balcells S. Nat Genet. 1998;18:11–12. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- 19.Fishman GA, Stone EM, Eliason DA, Taylor CM, Lindeman M, Derlacki DJ. Arch Ophthalmol. 2003;121:851–855. doi: 10.1001/archopht.121.6.851. [DOI] [PubMed] [Google Scholar]
- 20.Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M. Science. 1997;277:1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 21.Sun H, Smallwood PM, Nathans J. Nat Genet. 2000;26:242–246. doi: 10.1038/79994. [DOI] [PubMed] [Google Scholar]
- 22.Attie AD. Trends Biochem Sci. 2007;32:172–179. doi: 10.1016/j.tibs.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Lawn RM, Wade DP, Garvin MR, Wang X, Schwartz K, Porter JG, Seilhamer JJ, Vaughan AM, Oram JF. J Clin Invest. 1999;104:R25–31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bungert S, Molday LL, Molday RS. J Biol Chem. 2001;276:23539–23546. doi: 10.1074/jbc.M101902200. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald ME, Tolley E, Frase S, Zagvazdin Y, Miller RF, Hodos W, Reiner A. Vis Neurosci. 2001;18:299–317. doi: 10.1017/s0952523801182143. [DOI] [PubMed] [Google Scholar]
- 26.Stenirri S, Battistella S, Fermo I, Manitto MP, Martina E, Brancato R, Ferrari M, Cremonesi L. Clin Chem Lab Med. 2006;44:533–537. doi: 10.1515/CCLM.2006.116. [DOI] [PubMed] [Google Scholar]
- 27.Brousseau ME, Schaefer EJ, Dupuis J, Eustace B, Van Eerdewegh P, Goldkamp AL, Thurston LM, FitzGerald MG, Yasek-McKenna D, O’Neill G, Eberhart GP, Weiffenbach B, Ordovas JM, Freeman MW, Brown RH, Jr, Gu JZ. J Lipid Res. 2000;41:433–441. [PubMed] [Google Scholar]
- 28.Fitzgerald ML, Okuhira K, Short GF, 3rd, Manning JJ, Bell SA, Freeman MW. J Biol Chem. 2004;279:48477–48485. doi: 10.1074/jbc.M409848200. [DOI] [PubMed] [Google Scholar]
- 29.Fumagalli A, Ferrari M, Soriani N, Gessi A, Foglieni B, Martina E, Manitto MP, Brancato R, Dean M, Allikmets R, Cremonesi L. Hum Genet. 2001;109:326–338. doi: 10.1007/s004390100583. [DOI] [PubMed] [Google Scholar]
- 30.Garwin GG, Saari JC. Methods Enzymol. 2000;316:313–324. doi: 10.1016/s0076-6879(00)16731-x. [DOI] [PubMed] [Google Scholar]
- 31.MacKenzie D, Arendt A, Hargrave P, McDowell JH, Molday RS. Biochemistry. 1984;23:6544–6549. doi: 10.1021/bi00321a041. [DOI] [PubMed] [Google Scholar]
- 32.Hodges RS, Heaton RJ, Parker JM, Molday L, Molday RS. J Biol Chem. 1988;263:11768–11775. [PubMed] [Google Scholar]
- 33.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn J, Wong JT, Molday RS. J Biol Chem. 2000;275:20399–20405. doi: 10.1074/jbc.M000555200. [DOI] [PubMed] [Google Scholar]
- 35.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Proc Natl Acad Sci U S A. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loewen CJ, Moritz OL, Molday RS. J Biol Chem. 2001;276:22388–22396. doi: 10.1074/jbc.M011710200. [DOI] [PubMed] [Google Scholar]
- 37.Ahn J, Beharry S, Molday LL, Molday RS. J Biol Chem. 2003;278:39600–39608. doi: 10.1074/jbc.M304236200. [DOI] [PubMed] [Google Scholar]
- 38.Moritz OL, Molday RS. Invest Ophthalmol Vis Sci. 1996;37:352–362. [PubMed] [Google Scholar]
- 39.Goldberg AF, Moritz OL, Molday RS. Biochemistry. 1995;34:14213–14219. doi: 10.1021/bi00043a028. [DOI] [PubMed] [Google Scholar]
- 40.Maugeri A, van Driel MA, van de Pol DJ, Klevering BJ, van Haren FJ, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Dahl N, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Am J Hum Genet. 1999;64:1024–1035. doi: 10.1086/302323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shroyer NF, Lewis RA, Allikmets R, Singh N, Dean M, Leppert M, Lupski JR. Vision Res. 1999;39:2537–2544. doi: 10.1016/s0042-6989(99)00037-1. [DOI] [PubMed] [Google Scholar]
- 42.Tamehiro N, Zhou S, Okuhira K, Benita Y, Brown CE, Zhuang DZ, Latz E, Hornemann T, von Eckardstein A, Xavier RJ, Freeman MW, Fitzgerald ML. Biochemistry. 2008;47:6138–6147. doi: 10.1021/bi800182t. [DOI] [PMC free article] [PubMed] [Google Scholar]