Abstract
Chromosome breakage in germline and somatic genomes gives rise to copy number variation (CNV) responsible for genomic disorders and tumorigenesis. DNA sequence is known to play an important role in breakage at chromosome fragile sites; however, the sequences susceptible to double-strand breaks (DSBs) underlying CNV formation are largely unknown. Here we analyze 140 germline CNV breakpoints from 116 individuals to identify DNA sequences enriched at breakpoint loci compared to 2800 simulated control regions. We find that, overall, CNV breakpoints are enriched in tandem repeats and sequences predicted to form G-quadruplexes. G-rich repeats are overrepresented at terminal deletion breakpoints, which may be important for the addition of a new telomere. Interstitial deletions and duplication breakpoints are enriched in Alu repeats that in some cases mediate non-allelic homologous recombination (NAHR) between the two sides of the rearrangement. CNV breakpoints are enriched in certain classes of repeats that may play a role in DNA secondary structure, DSB susceptibility and/or DNA replication errors.
Introduction
Genomic CNV is a major cause intellectual disability, autism spectrum disorders, epilepsy, and psychiatric disorders. Large, pathogenic CNVs are located throughout the human genome and include tens to hundreds of genes [1], [2]. Though most germline CNVs have different chromosome breakpoints [3]–[6], it is possible that breakpoint regions share common DNA features that make them susceptible to double-strand breaks (DSBs). This is true of chromosome rearrangements in leukemia that vary in location, but share repetitive DNA and/or DNAse hypersensitive sites at breakpoint cluster regions (BCRs) [7]–[9].
Classic studies of chromosomal fragile sites have revealed that DNA sequence and structure can influence chromosome breakage. Fragile sites were originally identified as breaks and gaps in metaphase chromosomes, induced under conditions of DNA replication stress [10], [11]. Mapping and sequence analyses of fragile sites have uncovered repetitive classes of DNA at many loci. Trinucleotide repeats and tandemly repeated minisatellites underlie many rare fragile sites [12], [13]. The FRA7E and FRA16D common fragile sites are made up of AT-rich repeats [14], and yeast studies have shown that the FRA16D AT-rich dinucleotide repeat intrastrand pairs to form a secondary structure that stalls DNA replication [15]. Though there is no single DNA sequence responsible for fragile sites in the human genome, in general fragile sites are made up of repetitive DNA that may form secondary structures.
Some studies of germline CNV breakpoints have attempted to identify common DNA sequences that, like fragile sites, contribute to chromosome breakage [16]–[21], but the search for breakage-prone DNA at CNV boundaries is challenging for a number of reasons. First, more than half of the human genome is made up of repetitive DNA, so finding a repeat at or near a CNV breakpoint may be a circumstantial finding unrelated to breakage. Second, DNA resection after chromosome breakage can lead to a CNV breakpoint that is kilobases (kb) away from the initial DSB [22], [23]. Thus, studies that only focus on the sequence directly adjacent to the post-repair junction will miss some DSB sites. Finally, since chromosome breaks are caused by heterogeneous factors, it is necessary to analyze breakpoints from a large cohort of annotated CNVs to find DNA motifs that are significantly enriched at breakpoints.
Using our large dataset of fine-mapped and sequenced CNV breakpoints from patients with neurodevelopmental disorders, we applied several motif and repeat discovery tools to search for DNA sequences enriched at CNV breakpoints compared to control regions of the genome. We broadened the breakpoint regions to include flanking sequence and account for DNA resection. To search for a common breakage-associated motif, we analyzed patient breakpoint regions and control sequences using Multiple EM for Motif Elicitation (MEME) and nested motif independent component analysis (NestedMICA). We also searched for repetitive DNA with Tandem Repeats Finder, QuadParser, and RepeatMasker. This large-scale analysis revealed an enrichment of tandem repeats and potential G-quadruplex sequences at human CNV breakpoints, providing insight into DNA sequences susceptible to DSBs.
Materials and Methods
CNV breakpoint and control sequences
We previously fine-mapped and/or sequenced CNV breakpoints from 116 individuals with abnormal clinical cytogenetic testing results [5], [6]. Patients have unique deletions and duplications that alter the copy number of different genes, so they do not share a common phenotype. In general, individuals with large pathogenic CNVs exhibit developmental delay, intellectual disability, autism spectrum disorders, and/or congenital anomalies. We analyzed 48 terminal deletions, 41 inverted duplications adjacent to terminal deletions, 11 translocations, 10 interstitial deletions, four interstitial duplications and two terminal duplications. Terminal deletions and terminal duplications have one breakpoint per rearrangement. Translocations, interstitial deletions and interstitial duplications have two breakpoints per rearrangement. For 18q-71c's translocation, we only identified the chromosome 18 breakpoint; the other breakpoint on chromosome 4 is cryptic [5]. Thus, there are a total of 140 breakpoints in 116 individuals.
We calculated 4-kb windows surrounding 140 CNV breakpoints and downloaded the corresponding DNA sequence from the NCBI 36.1/hg18 build of the human genome assembly using the Table Browser from the UCSC Genome Browser (http://genome.ucsc.edu/). Four-kb CNV breakpoint regions were unique and did not overlap with one another. We also randomly selected 4-kb control sequences from the same genome build. We concatenated the coordinates of chromosomes 1–22, X and Y that make up the 3,080,419,480 bp in the haploid human genome. Next we used a random number generator to select 10,000 numbers between one and 3,080,419,480. We added four kb to each number to produce start and stop coordinates for 10,000 regions and downloaded the associated DNA sequences from the UCSC Genome Browser. We excluded control regions with “N” bases that correspond to sequencing gaps, resulting in 9243 ungapped 4-kb sequences. From these, we randomly selected 140 sequences 20 times to make up 20 datasets of 140 control sequences. We saved 140 CNV breakpoint sequences and 140 control sequences per dataset in FASTA format for analysis.
MEME and NestedMICA
We searched for common motifs in the CNV breakpoint and 20 control datasets using MEME [24] and NestedMICA [25] with default parameters to find a single ungapped 50-basepair (bp) motif. We ran NestedMICA motif inference tool (NMinfer) and NestedMICA motif scanner module (NMscan) with a cutoff of −15 to determine the number of motifs per sequence. MEME and NestedMICA programs were executed on the Emory Human Genetics Computing Cluster (HGCC). We used NMinfer to identify the 50-bp NestedMICA motif in all 21 datasets and MochiView [26] to align the motifs to the sequences in the CNV breakpoint and control datasets.
Repeat searches
To identify Alu repeats in the CNV breakpoint and control datasets, we ran RepeatMasker using default settings [27]. We identified tandem repeats and G-quadruplex sequences using Tandem Repeats Finder (TRF) [28] and QuadParser [29], respectively. All three programs were executed with default parameters. We used custom scripts to calculate the lengths of non-overlapping tandem repeats. We calculated the GC content of sequences using the geecee program (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::geecee) within the European Molecular Biology Open Software Suite (EMBOSS) [30].
We used chi-squared goodness-of-fit tests to test whether each type of repeat was proportionally distributed across CNV types (i.e., independent of CNV type). We used two-sided binomial tests to test each type of CNV breakpoint for enrichment with each type of repeat. Since we performed 18 of these enrichment tests (testing six CNV types for enrichment for three repeat types), we performed Bonferroni adjustment for the 18 tests and used a p-value cutoff of .00278 = .05/18 to assess significance.
Results
Human CNV breakpoints
Our goal was to identify DNA sequence motifs that are overrepresented in human CNV breakpoint regions compared to control regions of the genome. We analyzed the breakpoint sequences of pathogenic CNVs ascertained from 116 children with phenotypes including intellectual disability, developmental delay, congenital abnormalities, and autism spectrum disorders. We excluded recurrent CNVs mediated by NAHR between segmental duplications. The 140 breakpoints from 116 CNVs have been fine-mapped by high-resolution array comparative genome hybridization (CGH) [5], [6]. Thirty-two out of 116 CNV junctions have been sequenced, resolving the breakpoints to the bp. Most of the sequenced junctions were simple with little or no microhomology at the breakpoint junctions; three had more complex junctions with short insertions 10–16 bp long [5], [6] (Table S1). The other 84 CNV junctions were fine-mapped with custom microarrays that had, on average, one oligonucleotide probe per 200 bp. Oligonucleotide spacing is not uniform throughout the genome due to repetitive sequences that confound unique probe design. Thus, the mean and median resolutions of breakpoints are 468 bp and 101 bp, respectively (Table S1).
To characterize a diverse collection of CNV breakpoints, we included breakpoints from 48 terminal deletions, 41 inverted duplications adjacent to terminal deletions, 11 translocations, 10 interstitial deletions, four interstitial duplications and two terminal duplications. Interstitial deletions and duplications have two breakpoints in the same chromosome arm, and translocations have two breakpoints in different chromosomes. Terminal deletions and duplications have a single breakpoint. Inverted duplications adjacent to terminal deletions are a specific type of CNV where the deletion and duplication form as part of one chromosome rearrangement. In this case, the terminal deletion is the site of the initial DSB [5], [6], so we only included that breakpoint in the analysis. Since our CNV dataset is enriched in terminal deletions and duplications, breakpoints are overrepresented towards chromosome ends. The mean and median distances from the CNV breakpoint to the end of the chromosome are 4.8 Mb and 2.3 Mb, respectively (Table S2). It is important to recognize that all CNVs in this study extend beyond the terminal segmental duplications that make up human subtelomeres. Thus, none of the CNV breakpoints lie in subtelomeric segmental duplications.
Array CGH and junction sequencing can resolve chromosome breakpoints to a relatively small region; however, that may not correspond to the exact DSB site. After the initial DSB, 5′ to 3′ DNA resection can lead to a CNV breakpoint that is up to 1.3 kb away [22], [23]. We included additional sequence around each of the 140 breakpoints to account for DNA resection and array resolution. Breakpoint regions are based on the normal locus in the reference genome (before breakage), not the patient's CNV (after breakage). In the case of sequenced breakpoint junctions, we added sequence two kb proximal and two kb distal of the breakpoint. For breakpoints that were resolved by array but not sequencing, we added two kb proximal and distal to the midpoint between the abnormal and normal probes that defined the CNV breakpoint. Thus, each breakpoint region is four kb and centered around the post-repair chromosome breakpoint. We downloaded the 140 4-kb breakpoint regions from the reference genome assembly (NCBI Build 36.1/hg18).
As a comparison, we analyzed 4-kb control regions from the human genome. We compiled 140 control sequences 20 times to make up 20 control datasets (see Methods). In the following experiments, we compared the motifs in the CNV breakpoint dataset to those in the 20 control datasets. We applied motif-finding tools to search for DNA motifs with the potential to form secondary structures susceptible to DSBs. Since most of our breakpoints are fine-mapped, but not sequenced, we focused on long repeats that span much of the breakpoint region. We did not analyze short motifs reported at other chromosome breakpoints (e.g., 6-8-bp translin target sites) due to the imprecision of most CNV junctions in our study.
Common motif search
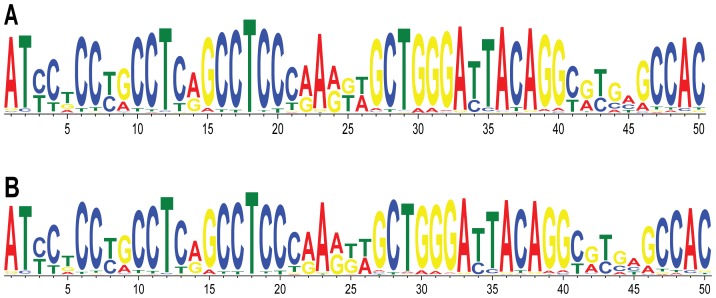
It is possible that CNVs are caused by breakage in a common DNA sequence motif present in many or all CNV breakpoints. To look for common motifs among the 140 CNV breakpoints, we performed MEME [24] and NestedMICA [25] searches. We queried the top 50-bp motif in the CNV breakpoint dataset using default parameters for both programs. MEME and NestedMICA output almost identical motifs; only one bp is different between the two consensus sequences, at position 25 (Figure 1). We performed NestedMICA searches with the same parameters in the 20 control datasets and found motifs that were very similar to each other, and very similar to the motif present in the CNV breakpoint dataset (Figure S1). Further review of this common motif revealed that it is part of the Alu repeat sequence. This 50-bp motif is present twice in the ∼300-bp Alu consensus (Figure 2) and includes a 26-bp core sequence that is a hotspot within Alus involved in gene rearrangements [31].
Figure 1. Common 50-bp motif in CNV breakpoint dataset.
Logo plots for (A) MEME and (B) NestedMICA motifs show nearly identical consensus sequences.
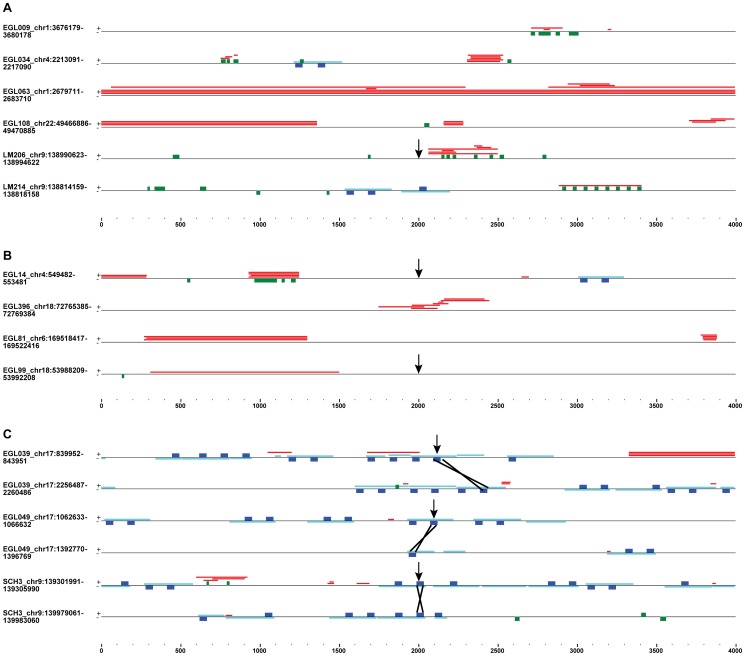
Figure 2. Repeats in 4-kb CNV breakpoint regions.
The location of tandem repeats (red), G-quadruplexes (green), Alus (light blue) and 50-bp motif (dark blue) sequences are shown for a subset of terminal deletion (A), inverted duplication adjacent to terminal deletion (B) and interstitial deletion (C) breakpoint regions. Repeats on the positive (+) and negative (−) strands are shown on the top and bottom of the black line, respectively. Arrows point to sequenced breakpoint junctions. The breakpoint region from EGL108 underlies terminal deletions and one interstitial duplication. Black Xs show Alu-Alu recombination sites for sequenced junctions (C).
It is possible that although the 50-bp motif is present in both breakpoint and control datasets, it is enriched in CNV breakpoints. To explore this, we examined the number of motifs in CNV breakpoints and control regions. Using the 50-bp NestedMICA motif detected in the CNV breakpoint dataset, we visualized the motif alignments with MochiView [26] and counted the number of motifs in all 21 datasets. The number of 4-kb sequences with at least one motif is not enriched in the CNV breakpoint dataset (68) versus controls (56–85), and the total number of motifs in the CNV breakpoint dataset (161) is not greater than the number of motifs in the control datasets (137–209) (Figures S2 and S3, Table S3). Since Alus are the most abundant mobile element in the human genome [32], it is not surprising that we find a substring of the Alu sequence as the most common 50-bp sequence in the CNV breakpoint dataset and the 20 control datasets; however, this motif is not overrepresented in breakpoint regions compared to control regions of the genome.
Repeats enriched at CNV breakpoints
Diverse types of repetitive sequences may be involved in DSBs that give rise to CNVs. This is the case for fragile sites, which are made up of various classes of satellite DNA, dinucleotide and trinucleotide repeats. In addition, different types of repetitive DNA predicted to form secondary structures underlie many BCRs in tumor genomes [7]–[9], [16], [33], [34]. To investigate repetitive DNA sequences involved in germline CNVs, we searched for tandem repeats and predicted G-quadruplex DNA in the CNV breakpoint and control datasets.
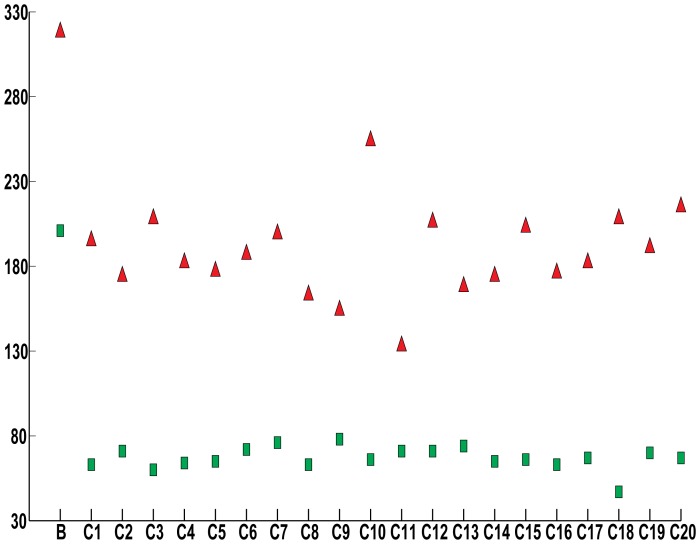
We used Tandem Repeats Finder with default settings to identify tandem repeats in the 21 datasets. Tandem repeats are not based on a consensus sequence, rather they are defined by two or more duplicated sequences arrayed head-to-tail. Since tandem repeats may overlap one another (Figure 2), we counted the total number of tandem repeats as well as the total non-overlapping bp occupied by at least one tandem repeat in each 4-kb sequence. In the CNV breakpoint dataset, 104 out of 140 breakpoint sequences had at least one tandem repeat (Table S4). For the 104 sequences with tandem repeats, 25–4000 bp were occupied by tandem repeats, and the mean and median amounts of sequence including at least one tandem repeat were 330 bp and 133 bp, respectively. The 20 control datasets had 71–95 out of 140 sequences with at least one tandem repeat (mean = 82; median = 83). For those control sequences with at least one tandem repeat, the mean and median numbers of bp occupied by a tandem repeat per 4-kb sequence were 156 bp and 63 bp, respectively. Since all 21 datasets had the same number of bp analyzed (4 kb * 140 sequences = 560 kb), we can compare the number of tandem repeats without adjusting for the size of the dataset. The CNV breakpoint dataset had 318 tandem repeats, whereas the control datasets had 133–254 tandem repeats (Figure 3). Thus, the CNV breakpoint regions are enriched in the number and density of tandem repeats compared to the control regions.
Figure 3. Number of repeats per dataset.
The total number of tandem repeats (red triangles) and G-quadruplex (green rectangles), sequences per CNV breakpoint (B) and control (C1-C20) datasets are plotted.
Tandem repeats include other classes of repetitive sequences, including triplet repeats and satellite DNA. It is possible that the enrichment in tandem repeats in the CNV breakpoint dataset is due largely to one particular type of duplicated sequence. Instead, we found that the tandem repeats in the breakpoint regions vary in repeat unit size, repeat array size, AT- and GC-content. We concatenated the tandem repeats in 104 CNV breakpoints to assemble the non-overlapping tandem repeat sequences and avoid counting segments of breakpoint regions more than once. Non-overlapping tandem repeat regions have mean and median GC percentages of 44% and 49%, respectively. Thus, both AT-rich and GC-rich tandem repeats are present at CNV breakpoints.
We also investigated sequences predicted to form G-quadruplexes in CNV breakpoint and control datasets. Sequences that contain four tracts of at least three guanines separated by other bases can form G-quadruplexes by intrastrand pairing between the four G-rich tracts. Such G-rich sequences can assemble G-quadruplex structures in vitro [35] and cause chromosome breakage and genomic instability in vivo [36], [37]. We searched for the G-quadruplex consensus sequence, G3+N1–7G3+N1–7G3+N1–7G3+, using the QuadParser program [29]. Sixty-eight out of 140 CNV breakpoint regions have at least one G-quadruplex, whereas 38 to 52 control regions in the 20 control datasets have at least one G-quadruplex (mean and median = 42). There are 201 G-quadruplexes in the CNV breakpoint dataset and 47–78 G-quadruplexes in the control datasets (Figure 3; Table S5). Thus, CNV breakpoints are enriched in the G-quadruplex consensus sequence.
It is possible that the enrichment in G-quadruplex sequences in the CNV breakpoint datasets stems from an increase in overall GC-richness in CNV breakpoints. The human genome is organized into GC-rich and AT-rich isochores [38], and the genome-wide average of GC content is 41% [39]. The mean GC content of the 20 control datasets ranged from 39.8–41.6%, whereas the mean GC content of the CNV breakpoint regions was 47.2% (Table S6). CNV breakpoints are 133 kb to 75 Mb from the nearest telomere; the mean and median distances are 4.8 Mb and 2.3 Mb, respectively (Table S2). Thus, many breakpoints lie within the terminal chromosome band that is known to be elevated in GC content [40], [41]. Since terminal deletion and duplication CNV breakpoints lie closer to chromosome ends than other CNV breakpoints, we would expect them to be GC-rich. However, breakpoint regions from all six types of CNV had a higher GC percentage than the genome average (Table 1). Therefore, the GC enrichment in CNV breakpoints is not due to only a subset of terminal chromosome rearrangements.
Table 1. Repeats enriched and depleted in CNV breakpoints. The GC content and number of breakpoints are listed for the six CNV types.
| CNV type | %GC | Breakpoints | Tandem repeats | G-quads | Alus |
| Terminal deletion | 50.1 | 48 | 122 | 99** | 62 |
| Inverted duplication with terminal deletion | 43.6 | 41 | 70 | 39* | 36* |
| Translocation | 46.1 | 21 | 40 | 24 | 42 |
| Interstitial deletion | 50.0 | 20 | 60 | 30 | 72** |
| Interstitial duplication | 45.4 | 8 | 25 | 9 | 28** |
| Terminal duplication | 41.0 | 2 | 1 | 0 | 2 |
| Total | 140 | 318 | 201 | 242 |
The number of tandem repeats, G-quadruplexes, and Alus per type of CNV are shown. Significant depletion (*) and enrichment (**) for repeats were determined by binomial test p-values <.00278 (.05/18, based on Bonferroni adjustment for 18 tests).
Repeats enriched in different types of CNV
We analyzed the enrichment of tandem repeats, G-quadruplexes and Alus in the breakpoints from the six types of CNVs (Table 1). Interstitial and terminal duplications are underrepresented in the 140 breakpoints (n = 10), whereas terminal deletions and inverted duplications adjacent to terminal deletions make up more than half of the 140 breakpoints. Tandem repeats, G-quadruplexes and Alus were not distributed proportionally across CNV types according to chi-square goodness-of-fit tests (1e-15<p<.0024). Terminal deletion breakpoint regions have an average GC content of 50.1% and are enriched in G-quadruplexes (p = 9.2e-6). Inverted duplication terminal deletion breakpoints were slightly depleted for G-quadruplexes (p = .0019) and Alus (p = 2.4e-7) (Table 1). Breakpoints from interstitial duplications and deletions were enriched in Alus (p = 3.9e-10 and 3.0e-4, respectively). The enrichment of motifs at certain types of CNV breakpoints was striking and points to specific repetitive DNA being involved in various types of chromosome rearrangements.
Discussion
Our analysis of 140 CNV breakpoints revealed an enrichment in tandem repeats and G-quadruplexes. It is possible that some of these sequences assemble secondary structures that are susceptible to DNA replication errors or DSBs. Tandem repeats have been described at other CNV breakpoints and are predicted to form a range of secondary structures [17]–[19]. In our CNV breakpoints, we find both AT-rich and GC-rich tandem repeats. Additional studies of DNA secondary structure and chromosome fragility are necessary to pinpoint the factors required for DSBs in particular classes of tandem repeats.
Breaks in repetitive sequences may facilitate particular types of chromosome rearrangements. DSBs that give rise to terminal deletions may be repaired by synthesis of a new telomere at the deletion breakpoint [5], [42]–[44]. Breaks that occur in or resect to G-rich DNA are ideal substrates for telomerase to prime a new telomere sequence, (5′-TTAGGG-3′)n. In addition, G-rich sequences with the ability to form secondary structures are susceptible to DSBs. Thus, sequences that underlie terminal deletion breakpoints may be G-rich due to the propensity for DSBs plus the likelihood of recovering a chromosome break repaired by a new telomere. We sequenced 13 of 48 terminal deletion junctions to pinpoint the post-repair CNV junction. None of the 13 terminal deletion junctions lies in a G-quadruplex or tandem repeat; however, LM206's chromosome 9q terminal deletion junction is 65 bp proximal of a cluster of G-rich tandem repeats and G-quadruplexes (Figure 2). It is tempting to speculate that a DSB in the G-rich repeat region resected 65 bp and was the site of telomere addition. In this case, the new telomere lies directly adjacent to a G-rich sequence at the breakpoint, 5′-GGGGCGGAGGGGCCGAAGCTGGCTGGTGG-3′ [5].
Though Alu repeats were not enriched in the entire CNV breakpoint dataset compared to control datasets, they were enriched in interstitial deletion and duplication breakpoint regions (Table 1). Alus that recombine to form interstitial deletions and duplications are oriented in the same direction, share high sequence homology (typically >85% identical), and crossover at a homologous site within the Alus [5], [45]–[48]. NAHR generates a hybrid Alu at the breakpoint that merges the two sides of the CNV, which is detectable by breakpoint sequencing. We sequenced three of the ten interstitial deletion junctions and none of the four interstitial duplication junctions. Sequence analysis revealed that EGL039, EGL049 and SCH3 breakpoints are the product of recombination between two highly identical Alu repeats [5] (Figure 2). In all three cases, the sequenced Alu-Alu breakpoints lie within a 50-bp motif. Other interstitial deletion breakpoints may have Alus nearby, but are not the product of Alu-Alu NAHR. EGL094's sequenced interstitial deletion junction is the product of non-homologous end-joining (NHEJ) between two breakpoints that are not in Alus [5]. However, EGL094's proximal breakpoint region has a cluster of five Alus in four kb (Figure S2). Other studies of chromosome breakpoints have also found an enrichment of Alus at interstitial deletion junctions [16], [20].
CNV breakpoint regions in our study were significantly more GC-rich than the genome average. A previous study of germline and somatic breakpoints suggested that deletion breakpoints were AT-rich, whereas translocation breakpoints were GC-rich [16]. The deletion and translocation breakpoints in our study are both GC-rich, with GC contents of 50% and 46%, respectively (Table 1). This difference is likely due to the chromosome rearrangements selected for the two studies: Abeysinghe et al. examined a large cohort of mostly somatic chromosome rearrangements, whereas our study included only germline chromosome rearrangements. In addition, 87/140 (87%) of the CNV breakpoints in our study occur in the last 10 Mb of chromosomes, which are more GC-rich than the genome average. There are likely different biases in GC content for deletions, depending on the origin of the deletion (germline vs. somatic) and the location of the deletion breakpoints. In their large-scale analysis of 663,446 breakpoints from diverse cancer genomes, De and Michor found an enrichment of tandem repeats and Alus [34]. Thus, some classes of DNA repeats are shared between germline and somatic breakpoints.
Repeat density may also play a role in chromosome breakage. For example, both of EGL039's breakpoints are made up of several Alus and tandem repeats. EGL063's terminal deletion breakpoint is entirely covered by tandem repeats across the 4-kb region, and LM206's terminal deletion breakpoint has overlapping tandem repeats and G-quadruplexes (Figure 2). In some cases, G-quadruplexes are part of the tandem repeat structure, rather than separate sequences (see EGL99 and LM214 breakpoints). In other breakpoints, different types of repeats are dispersed across the 4-kb region (see EGL034, EGL108 and EGL81 breakpoints). Repeats at CNV breakpoints could have an additive effect, whereby more repeats lead to a greater propensity for chromosome breakage and/or recombination. On the other hand, there may be only one repeat per locus that is responsible for chromosome rearrangement.
Our analysis of CNV breakpoint regions revealed an enrichment in tandem repeats and sequences predicted to form G-quadruplexes. Furthermore, particular classes of repeats are overrepresented at breakpoints of different types of CNV. Thus, when interpreting mechanisms of CNV formation, it is important to consider the DNA at breakpoints as well as the resulting chromosome rearrangement. Functional analysis of individual DNA motifs will delineate the sequences responsible for gross chromosomal rearrangement [49]–[51]. In addition, motif mining of even larger CNV breakpoint datasets from diverse CNV classes will tell us more about the factors required for CNV formation.
Supporting Information
Logo plots of top 50-bp motif detected in each control dataset (C1-C20) by NestedMica.
(TIF)
Repeats in 4-kb CNV breakpoint regions (html). The location of tandem repeats (red), G-quadruplexes (green), Alus (light blue) and 50-bp motif (dark blue) sequences are shown for each of the 140 CNV breakpoint regions.
(EPS)
Repeats in 4-kb control regions (html). 140 control regions from control dataset C3 are shown as an example of repeat content in control sequences.
(EPS)
CNV breakpoint resolution. For 140 breakpoint regions, minimum, maximum, mean, and median breakpoint resolution is 0 bp, 7,542 bp, 468 bp, and 101 bp, respectively. The method of breakpoint mapping and the type of rearrangement is described. Insertion length is listed for three sequenced junctions with insertions.
(XLSX)
Distance from breakpoint region to the nearest chromosome end.
(XLSX)
50-bp NestedMica (NM) motifs in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one NM motif and the number of NM motifs per dataset are listed.
(XLSX)
Tandem repeats (TR) in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one TR and the number of TRs per dataset are listed.
(XLSX)
G-quadruplex consensus sequences (G3+N1–7G3+N1–7G3+N1–7G3+) in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one G-quadruplex and the number of G-quadruplexes per dataset are listed.
(XLSX)
GC content of CNV breakpoint region dataset (B) and control datasets (C1-C20). For each dataset, minimum, maximum, mean, and median percent GC were calculated.
(XLSX)
Acknowledgments
We thank Cheryl Strauss for editorial assistance and members of the Rudd laboratory for helpful discussions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant from the National Institutes of Health (grant number MH092902 to MKR) and a grant from the March of Dimes (grant number #12-FY11-203 to MKR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the March of Dimes. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, et al. (2011) A copy number variation morbidity map of developmental delay. Nature genetics 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, et al. (2011) An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine 13: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Itsara A, Cooper GM, Baker C, Girirajan S, Li J, et al. (2009) Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rudd MK, Keene J, Bunke B, Kaminsky EB, Adam MP, et al. (2009) Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum Mol Genet 18: 2957–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo Y, Hermetz KE, Jackson JM, Mulle JG, Dodd A, et al. (2011) Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Human molecular genetics 20: 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hermetz KE, Newman S, Conneely KN, Martin CL, Ballif BC, et al. (2014) Large inverted duplications in the human genome form via a fold-back mechanism. PLoS genetics 10: e1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Rowley JD (2006) Chromatin structural elements and chromosomal translocations in leukemia. DNA repair 5: 1282–1297. [DOI] [PubMed] [Google Scholar]
- 8. Wang G, Vasquez KM (2006) Non-B DNA structure-induced genetic instability. Mutation research 598: 103–119. [DOI] [PubMed] [Google Scholar]
- 9. Sinclair PB, Parker H, An Q, Rand V, Ensor H, et al. (2011) Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy. Human molecular genetics 20: 2591–2602. [DOI] [PubMed] [Google Scholar]
- 10. Sutherland GR (2003) Rare fragile sites. Cytogenet Genome Res 100: 77–84. [DOI] [PubMed] [Google Scholar]
- 11. Glover TW, Arlt MF, Casper AM, Durkin SG (2005) Mechanisms of common fragile site instability. Hum Mol Genet 14 Spec No. 2 R197–205. [DOI] [PubMed] [Google Scholar]
- 12. Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, et al. (1997) Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 88: 367–374. [DOI] [PubMed] [Google Scholar]
- 13. Hewett DR, Handt O, Hobson L, Mangelsdorf M, Eyre HJ, et al. (1998) FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol Cell 1: 773–781. [DOI] [PubMed] [Google Scholar]
- 14. Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, et al. (2003) Molecular basis for expression of common and rare fragile sites. Mol Cell Biol 23: 7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang H, Freudenreich CH (2007) An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell 27: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Human mutation 22: 229–244. [DOI] [PubMed] [Google Scholar]
- 17. Gajecka M, Pavlicek A, Glotzbach CD, Ballif BC, Jarmuz M, et al. (2006) Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet 120: 519–526. [DOI] [PubMed] [Google Scholar]
- 18. Gajecka M, Gentles AJ, Tsai A, Chitayat D, Mackay KL, et al. (2008) Unexpected complexity at breakpoint junctions in phenotypically normal individuals and mechanisms involved in generating balanced translocations t(1;22)(p36;q13). Genome Res 18: 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault AC, et al. (2009) Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet 18: 1924–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, et al. (2009) Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet 18: 3579–3593. [DOI] [PubMed] [Google Scholar]
- 21. Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, et al. (2010) Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson C, Jasin M (2000) Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405: 697–700. [DOI] [PubMed] [Google Scholar]
- 23. Simsek D, Jasin M (2010) Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology 17: 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic acids research 34: W369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Down TA, Hubbard TJ (2005) NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res 33: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Homann OR, Johnson AD (2010) MochiView: versatile software for genome browsing and DNA motif analysis. BMC biology 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smit AF (1996) The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6: 743–748. [DOI] [PubMed] [Google Scholar]
- 28. Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huppert JL, Balasubramanian S (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33: 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 31. Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic acids research 23: 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379. [DOI] [PubMed] [Google Scholar]
- 33. Popescu NC (2003) Genetic alterations in cancer as a result of breakage at fragile sites. Cancer letters 192: 1–17. [DOI] [PubMed] [Google Scholar]
- 34. De S, Michor F (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nature structural & molecular biology 18: 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34: 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, et al. (2008) Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 18: 900–905. [DOI] [PubMed] [Google Scholar]
- 37. Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, et al. (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Costantini M, Clay O, Auletta F, Bernardi G (2006) An isochore map of human chromosomes. Genome research 16: 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Consortium IHGS (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 40.Rudd MK (2007) Subtelomeres: Evolution in the Human Genome. Encyclopedia of Life Sciences: John Wiley & Sons, Ltd.
- 41.Rudd MK (2014) Human and Primate Subtelomeres. In: Louis EJ, Becker MM, editors. Subtelomeres: Springer. pp. 153–164. [Google Scholar]
- 42. Lamb J, Harris PC, Wilkie AO, Wood WG, Dauwerse JG, et al. (1993) De novo truncation of chromosome 16p and healing with (TTAGGG)n in the alpha-thalassemia/mental retardation syndrome (ATR-16). Am J Hum Genet 52: 668–676. [PMC free article] [PubMed] [Google Scholar]
- 43. Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, et al. (1994) Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet 55: 505–512. [PMC free article] [PubMed] [Google Scholar]
- 44. Ballif BC, Yu W, Shaw CA, Kashork CD, Shaffer LG (2003) Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum Mol Genet 12: 2153–2165. [DOI] [PubMed] [Google Scholar]
- 45. Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, et al. (1985) Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science 227: 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pousi B, Hautala T, Heikkinen J, Pajunen L, Kivirikko KI, et al. (1994) Alu-Alu recombination results in a duplication of seven exons in the lysyl hydroxylase gene in a patient with the type VI variant of Ehlers-Danlos syndrome. Am J Hum Genet 55: 899–906. [PMC free article] [PubMed] [Google Scholar]
- 47. Sen SK, Han K, Wang J, Lee J, Wang H, et al. (2006) Human genomic deletions mediated by recombination between Alu elements. American journal of human genetics 79: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beck CR, Garcia-Perez JL, Badge RM, Moran JV (2011) LINE-1 elements in structural variation and disease. Annual review of genomics and human genetics 12: 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS (2006) The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296. [DOI] [PubMed] [Google Scholar]
- 50. Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, et al. (2008) Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. Embo J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, et al. (2009) Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Molecular cell 35: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Logo plots of top 50-bp motif detected in each control dataset (C1-C20) by NestedMica.
(TIF)
Repeats in 4-kb CNV breakpoint regions (html). The location of tandem repeats (red), G-quadruplexes (green), Alus (light blue) and 50-bp motif (dark blue) sequences are shown for each of the 140 CNV breakpoint regions.
(EPS)
Repeats in 4-kb control regions (html). 140 control regions from control dataset C3 are shown as an example of repeat content in control sequences.
(EPS)
CNV breakpoint resolution. For 140 breakpoint regions, minimum, maximum, mean, and median breakpoint resolution is 0 bp, 7,542 bp, 468 bp, and 101 bp, respectively. The method of breakpoint mapping and the type of rearrangement is described. Insertion length is listed for three sequenced junctions with insertions.
(XLSX)
Distance from breakpoint region to the nearest chromosome end.
(XLSX)
50-bp NestedMica (NM) motifs in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one NM motif and the number of NM motifs per dataset are listed.
(XLSX)
Tandem repeats (TR) in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one TR and the number of TRs per dataset are listed.
(XLSX)
G-quadruplex consensus sequences (G3+N1–7G3+N1–7G3+N1–7G3+) in the CNV breakpoint region dataset (B) and control datasets (C1-C20). The number of sequences with at least one G-quadruplex and the number of G-quadruplexes per dataset are listed.
(XLSX)
GC content of CNV breakpoint region dataset (B) and control datasets (C1-C20). For each dataset, minimum, maximum, mean, and median percent GC were calculated.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.