Abstract
Background
Acute pyelonephritis (APN) versus acute rejection (AR) is a frequently encountered diagnostic and therapeutic dilemma in kidney transplants. Variable culture results, overlapping histologic features, and persistent graft dysfunction despite antibiotics are frequently encountered. Therefore, we explored the utility of intragraft microRNA profiles to distinguish between allograft APN and AR.
Materials and Methods
Between 2003 and 2011, we identified 49 patients with biopsy features of APN, within the first 2 years posttransplant. MicroRNA profiling was performed on 20 biopsies (normal kidney, n=4; unequivocal AR, n=5; features of APN, n=11).
Results
Only 32% (16/49) of the patients had concomitant positive urine cultures at biopsy, and in 8 of 16 patients, colony count was less than 105 CFU/mL. In 14 of 49 patients, positive urine culture did not coincide with the biopsy, and in 19 of 49 patients, urine cultures were negative. On microRNA profiling, good clustering was seen among the normal kidneys and among AR biopsies. Among the 11 biopsies with features of APN, 4 biopsies showed good clustering with a pattern distinct from AR; (these patients recovered graft function with antibiotics); 7 of 11 biopsies showed heterogeneity in microRNA profiles and variable outcomes with antibiotic treatment. We identified a panel of 25 microRNAs showing statistical difference in expression between AR and APN. MiR-99b, miR-23b let-7b-5p, miR-30a, and miR-145 were validated using qPCR.
Conclusion
Allograft pyelonephritis can be a diagnostic and therapeutic challenge. A gestalt approach is required. In addition to histology and cultures, differential intragraft microRNA expression may prove helpful to distinguish APN from AR in renal allograft biopsies.
Keywords: Acute pyelonephritis, Acute rejection, microRNA, Renal allograft biopsy, NanoString
Kidney transplantation, being an immunocompromised state, predisposes the recipient to a variety of bacterial, viral, and fungal infections. Urinary tract infections (UTIs) are common during the first several months posttransplantation, and these can predispose the patient to allograft pyelonephritis (1–8). The diagnosis of acute pyelonephritis (APN) in the native kidney is usually made based on the classic tetrad—fevers, costovertebral angle tenderness, history of lower urinary tract infection, and microbiological cultures of the urine. The native kidney is therefore rarely biopsied for APN. However, in the context of renal transplantation and immunosuppression, the classic clinical features of fever and pain are frequently subdued, and costovertebral angle tenderness is not an accompanying feature. Blood leukocyte counts can be altered by immunosuppressive medications. APN in the renal allograft is therefore often coincidentally discovered on allograft biopsy, and a definitive diagnosis can only be made if there is positive concomitant urine culture result. Allograft biopsy and urine culture are the best currently available tools to diagnose APN in the renal allograft, but both these methods have pitfalls.
Interstitial inflammation with predominance of polymorphonuclear leukocytes (PMNs) and intratubular PMNs forming microabscesses are considered hallmark histologic features of APN. However, it is common to find PMNs in the inflammatory cell infiltrates of acute rejection (AR) as well. Also, the characteristic finding of neutrophilic tubulitis and tubular microabscesses in APN may not always be demonstrated in a biopsy because of the focal nature of these lesions and sampling issues. Thus, histologic features between APN and acute AR may overlap. Concomitant microbiological urine culture and colony counting by plating measured quantity of urine on culture plates is used to aid in the diagnosis. Colony count of 105 colony forming units per milliliter (CFU/mL) is considered to be diagnostic of true infection (as opposed to contamination by urogenital skin flora) (9). However, we have encountered cases with low colony counts despite biopsy features of APN in kidney transplant recipients. This can make the diagnosis of APN in renal allografts difficult. To further confound the diagnosis, rapid response to antibiotics may not always be achieved despite histologic findings of APN and positive urine culture results.
The purpose of our study was twofold: 1. to retrospectively assess the degree of correlation between histologic features of APN on biopsy, and positive urine culture results in transplant patients; and 2. to explore the potential for intragraft microRNA profiling to distinguish between APN and AR. Intragraft miRNA profiling was performed on a subset of biopsies from our study cohort.
MicroRNAs (miRNAs) are short noncoding RNAs that modulate physiological and pathological processes by inhibiting target gene expression by inducing mRNA degradation and blocking protein translation (10). MiRNAs potentially regulate the expression of thousands of proteins. The miRNA field is being extensively explored to discover new diagnostic biomarkers and potential treatments in cancer (11). MiRNA profiling is also being studied in native kidney disease and renal allografts (12–23, 26, 27, 33, 34).
RESULTS
Review of Biopsies
We identified biopsies from 64 renal allograft recipients, transplanted between 2003 and 2011, in whom the allograft biopsy showed morphologic features of APN—interstitial in flammation with predominance of neutrophils, tubular infiltration with neutrophils, and formation of tubular microabscesses (24, 25) (see Figure S1A, S1B, SDC, http://links.lww.com/TP/A928). Of the 64 recipients, 49 had the biopsy within the first 2 years posttransplant. For our clinicopathologic study, we focused on these 49 patients. (In the remaining 15 patients, the diagnostic biopsy was performed after 2 years posttransplant). Biopsy features, urine culture results, timing of urine culture relative to biopsy, and renal function at 1-year postbiopsy were assessed (Table 1). Many of these 49 allograft recipients had more than one allograft biopsies performed within 1 month for persistent graft dysfunction. We used only the first biopsy (with features of pyelonephritis) for the clinicopathologic study.
TABLE 1.
Clinical and laboratory data on the 49 patients transplanted between 2003 and 2011, with biopsy within two years post-transplant, showing features of acute pyelonephritis
| Patient characteristics | Group I—positive urine culture within 10 days before or after biopsy |
Group II—positive urine culture beyond 10 days before or after biopsy |
Group III—urine culture negative |
|---|---|---|---|
| No. patients (n) | 16 | 14 | 19 |
| Sex, males; females | 6; 10 | 5; 9 | 9; 10 |
| Mean age (yr) | 45±13 | 46±19 | 42±15 |
| Patients with biopsy within 1 month posttransplant | 7 (43%) | 4 (28%) | 12 (63%) |
| Patients with colony count below 100,000 CFU/mL | 8 (50%) | 4 (28%) | N/A |
| Graft loss within 1 yr postbiopsy (death censored) | 5 (31%) | 5 (35%) | 0 |
| Baseline serum creatinine before biopsy (mg/dL) | 1.8±1.4 | 1.8±1.0 | 2.3±1.4 |
| Serum creatinine 1 month postbiopsy (mg/dL) | 2.9±1.8 | 3.3±1.8 | 2.13±1.1 |
| Serum creatinine 1 yr postbiopsy (mg/dL) | 2.1±0.8 | 2.1±0.4 | 1.9±1.1 |
| Δserum creatinine (at 1 yr vs. baseline) mg/dl | 0.3 | 0.3 | −0.3 |
| No. of biopsies with overlapping features of APN and AR | 5 | 3 | 4 |
| (6 showed predominant ATN) |
|||
| Patients who received antibiotic treatment in addition to routine prophylaxis for pyelonephritis |
14 | 12 | 11 |
The biopsy was performed during the first 2 years posttransplant.
CFU/mL, colony forming units/milliliter; AR, acute rejection; ATN, acute tubular necrosis.
Allograft Biopsy Findings and Urine Culture Results
The diagnosis of APN in the allograft kidney is difficult, and there are no well-defined criteria as for native kidney pyelonephritis. If there are characteristic histologic features on biopsy and concomitant positive urine cultures, we considered them as “most likely” APN. If the histologic features are suggestive of APN, but there are no concomitant positive urine cultures, we diagnose them as “possible” APN. To consider the urine culture as concomitant, we used an arbitrary cutoff of 10 days between date of urine culture and date of the biopsy. Based on this, we classified these 49 patients into three groups for the purpose of this study.
Group I “most likely APN”—biopsy features of APN and positive urine culture performed 10 days before or after the day of biopsy. Of the 49 patients, 16 (32.6%) fulfilled these criteria.
Group II “possible APN”—biopsy features of APN, no concomitant positive urine culture but had positive urine cultures more than 10 days before or after the biopsy. It is possible that the positive culture so far remote from the biopsy represents a separate episode of urinary tract infection and does not correspond with the findings in the allograft biopsy. Even then, the biopsy features cannot be ignored, and therefore, a diagnosis of “possible APN” was made. Of the 49 patients, 14 patients fulfilled these criteria. These patients had a positive urine culture ranging from 13 days to 7 months from the time of biopsy.
Group III “equivocal APN”—biopsy features of APN but no positive urine cultures at all during the year of the biopsy. Of the 49 patients, 19 patients fulfilled these criteria.
Table 1 shows clinicopathologic and laboratory features of these 49 patients. It is interesting to note that some biopsies from all three groups showed overlapping histologic features of AR. Urine cultures with low colony counts were seen. Ten patients had graft loss within 1 year of biopsy, of which, seven were lost within 1 month of the biopsy (see Table S1, SDC, http://links.lww.com/TP/A928). These were five patients from Group I and five patients from Group II.
All renal allograft recipients at our institution receive trimethoprim/sulfamethoxazole (Bactrim) prophylaxis. Most patients from groups I and II (and 11/19 patients from group III) received additional antimicrobial treatment (usually ciprofloxacin and fluconazole) to treat episode of possible pyelonephritis, in addition to posttransplant prophylaxis for infection (Bactrim, nystatin, and vancyclovir).
Escherichia sp., Klebsiella sp., Enterococcus sp., and Enterobacter sp. were the most commonly found genus in the patients with positive urine cultures. Less common pathogens seen were Staphylococcus epidermidis (n=2), Serratia marcescens (n=1), Providencia rettgeri (n=1), Citrobacter koseri (n=1), Candida albicans (n=3), Candida glabrata (n=1), and Candida lusitaniae (n=1). Candida species were even found alone (without associated bacterial growth) in five patients. The patient with Candida glabrata on urine culture was also found to have Mycoplasma by urine PCR testing data not shown.
MicroRNA NanoString Assay Results
The 20 samples analyzed using NanoString assay include 4 preimplantation transplant biopsies (baseline transplant biopsies B1 to B4), 5 biopsies with unequivocal AR (R1 to R5), and 11 biopsies with histologic features of APN (I1 to I11). The 11 biopsies with histologic features of APN were chosen from all three groups—group I (n=6; I1 to I6), group II (n=2; I7, I8), and group III (n=3; I9 to I11). These are shown in Table 2.
TABLE 2.
Eleven biopsies (from eleven patients) with features of acute pyelonephritis, analyzed using NanoString
| Biopsy sample |
Age | Race | Sex | Transplant type |
Duration from transplant to biopsy |
Biopsy diagnosis | Duration between biopsy and urine culture |
Urine culture result |
CFU/mL | Diagnostic difficulty | Treatment | Baseline S. cr. Before biopsy |
S. cr. 1 month post biopsy |
S. cr. l year post biopsy |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I | I1 | 65 | C | F | Cad | 12 d | Acute pyelonephritis | 10 d before biopsy | Klebsiella pneumoniae |
>100,000 | None | Antibiotics | DGF | 1.2 | 1.49 |
| I2 | 53 | AA | M | Cad | l yr, 4 months |
Acute pyelonephritis | 1 d after biopsy | Enterococcus fecalis |
10,000 to 50,000 |
None | Antibiotics | 2.5 | 2.6 | 3.73 | |
| I3 | 51 | Fil | F | Cad | 1 yr, 3 mo | Acute pyelonephritis | On day of biopsy | Klebsiella oxytoca | >100,000 | None | Antibiotics | 1.4 | 2.4 | 2.4 | |
| I4 | 44 | C | M | Cad | 1 yr, 21 d | Acute Pyelonephritis or rejection or both |
7 d before biopsy | Enterococcus fecalis, Pseudomonas rettgeri |
>100,000 | Poor response to antibiotics. Early graft loss. |
Antibiotics | 1.5 | lost | lost | |
| I5 | 36 | C | F | Cad | 8 d | Acute Pyelonephritis or rejection for both |
10 d before biopsy | Enterococcus fecalis |
>100,000 | Poor response to antibiotics. Early graft loss. |
Antibiotics | DGF | lost | lost | |
| I6 | 57 | C | F | LRD | 6 mo | Acute pyelonephritis | 4 d after biopsy | Candida glabrata; Mycoplasma |
< 50,000 | Earlier cultures negative; only Candida positive. |
Antibiotics, antifungal |
1.4 | 4.2 | 2.4 | |
|
| |||||||||||||||
| Group II | I7 | 30 | AA | F | Cad | 15 d | Smoldering pyelonephritis, possible rejection |
1 mo after biopsy | Klebsiella oxytoca, Citrobacter koseri |
< 50,000 | Poor response to antibiotics; low colony count in culture; Unusual pathogen. |
Antibiotics | DGF | lost | lost |
| I8 | 46 | C | M | Cad | 8 d | Acute pyelonephritis | 2 mo after biopsy | Klebsiella, Enterobacter ESBL |
<1000 | Earlier cultures negative; Low colony count in culture |
Antibiotics, followed by corticosteroids |
DGF | 8.2 | 2.1 | |
|
| |||||||||||||||
| Group Iii | I9 | 61 | AA | F | Cad | 8 d | ATN, patchy inflammation with PMNs, possible pyelonephritis |
2 d before biopsy | Negative | n/a | Negative culture | Antibiotics, followed by corticosteroids |
DGF | 1.6 | 1.1 |
| I10 | 37 | C | M | Cad | 15 d | Acute pyelonephritis or mild rejection |
2 d before biopsy | Negative | n/a | Negative culture | Antibiotics, followed by corticosteroids |
DGF | 1.1 | 2.4 | |
| I11 | 55 | C | F | Cad | 11 d | Acute pyelonephritis | On the day of biopsy |
Negative | n/a | Negative culture | Antibiotics | DGF | 3.7 | expired | |
Culture results, biopsy features, follow-up serum creatinine levels at 1 year are shown.
LRD, living related donor; Cad, cadaveric donor; S. cr., serum creatinine in mg/dL; DGF, delayed graft function (dialysis within first week posttransplant); Fil, Filipino.
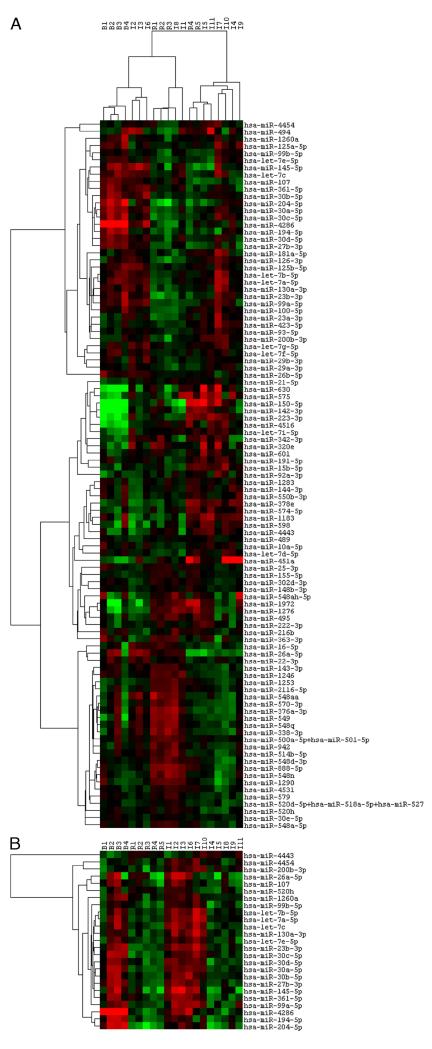
Unsupervised cluster analysis of the miRNA profiles (using top 100 miRNAs) showed good intragroup clustering among the baseline allograft biopsies (B1 to B4) and among the AR biopsies (R1, R2, R3 and R4, R5) (Fig. 1A). Among the 11 biopsies with APN, three biopsies (I2, I3, and I6) clustered together, distinct from AR. All three biopsies were from group I. These patients had concomitant positive urine cultures, and they did recover graft function with antibiotic treatment. So these patients probably had unequivocal APN. Patient with biopsy I1 had similar clinicopathologic characteristics. The miRNA profile of I1 did not show tight clustering with I2, I3, and I6 but did show similarities (shown in Fig. 1B). The remaining biopsies with histologic features of APN-I4 and I5 (group I), I7, I8 (group II) and I9, I10, and I11 (group III) showed heterogeneity in the miRNA profiles showing some similarities to both AR and APN. The miRNA profile of biopsies I4 and I5 appears to cluster closer to the AR profile. These two patients had concomitant positive urine cultures but persistent graft dysfunction despite antibiotic treatment. They underwent repeated biopsies in quick succession, which showed persistent inflammation and ultimately lost their grafts within 1 month. Biopsy I8 also appeared to clustered with AR. This patient received antibiotics without much improvement, requiring a repeat biopsy in 2 weeks, which showed persistent inflammation. He improved with steroid treatment.
FIGURE 1.
A, unsupervised hierarchical cluster analysis showing the normalized top 100 expressed miRNAs in the 20 biopsy samples. Each column represents a biopsy sample. Each row represents a miRNA. The color in each cell reflects the level of expression of the corresponding miRNA in the corresponding sample, relative to the geometric mean of the top 100 most highly expressed miRNAs. Increasing intensities of red mean higher expression, and increasing intensity of green means lower expression. B1 to B4 are pretransplant baseline biopsies representing normal kidney; R1 to R5 represent unequivocal acute rejection (AR); I1 to I11 are biopsies with histologic features of acute pyelonephritis (APN). I1 to I6 (Group I have positive concomitant urine cultures), I7, I8 (Group II have negative concomitant urine cultures but did have positive urine culture one to two months after biopsy), I9 to I11 (Group III have negative urine cultures throughout the year of the biopsy). The degree of relatedness is represented by the dendrogram at the top of the panel. There is good clustering among the four baseline biopsies and among the five biopsies with AR with clearly different profile between normal kidneys and rejection kidneys. Among the biopsies with presumed APN, the biopsies I2, I3, I6 clustered together, and I1 showed some heterogeneity. I8, I4, I5, and I11 clustered closer to AR. Biopsies I7 and I10 clustered together, with some similarities to the APN profile. I9 did not show any specific clustering with either AR or APN. B, hierarchical cluster analysis heat map showing the panel of 25 differentially expressed miRs between AR and APN (two-tailed, two-sample unequal variance Student’s t test, using cutoff P value of 0.05). Overall miRNA expression is lower in AR as compared with baselines and APN. In biopsies I7 and I10, most of these miRNAs showed expression similar to APN. In biopsies I4, I5, I8, I9, and I11, overall miR expression was lower, closer to AR.
In patient with biopsy I7, urine cultures at the time of biopsy were negative but became positive 1 month later. He lost the graft within 1 month despite antibiotic treatment. Biopsies I9, 10, and 11 were from group III. Urine cultures were repeatedly negative. Patients with biopsies I9 and I10, both improved on steroids after repeat biopsy 2 to 4 weeks later showed histologic features of AR. Patient with biopsy I11 expired soon after the biopsy despite antibiotics. The miRNA profile of I11 appeared closer to AR. MiRNA profile of I10 was similar to that of I7 and showed similarity to APN profile. I9 did not cluster well with either AR or APN.
Among the top 100 expressed miRNAs, we tried to identify a panel of miRNAs, which showed statistical difference in expression between AR profile and APN profile. This was difficult to do because the APN samples showed such heterogeneity. However, we used the four biopsies I1, I2, I3, and I6 that clinically behaved as pyelonephritis and compared them with the five AR biopsies using Student’s t test. A panel of 25 miRNAs was identified (Table 3). We also compared the five AR biopsies with all the 11 APN biopsies using t test. Eighteen of the 25 miRs showed statistical difference (shaded). A heat map showing expression pattern of these 25 miRNAs is shown in Figure 1B. Overall, the AR biopsies showed lower expression of these miRNAs as compared with the baselines and the APN biopsies I1, I2, I3, and I6. Biopsies I7 and I10 did show some similarity to the “pyelonephritis profile.” In biopsies I4, I5, I8, I9, and I11, miRNA expression levels are lower, probably intermediate between APN and AR or closer to AR.
TABLE 3.
25 miRs whose expression shows statistically significant difference between biopsies with unequivocal acute rejection and biopsies with unequivocal acute pyelonephritis
| Baseline biopsies (normal kidney) |
Acute rejection |
Acute pyelonephritis |
Infection vs Rejection |
Infection vs baseline |
Rejection vs baseline |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | B1 | B2 | B3 | B4 | R1 | R2 | R3 | R4 | R5 | 11 | 12 | 13 | 16 | t-test | fold change |
t-test | fold change |
t-test | fold change |
| hsa-let-7b-5p | 581.06 | 1081.64 | 2040.35 | 1285.99 | 657.54 | 859.98 | 659.67 | 763.84 | 599.56 | 1328.98 | 2101.49 | 1569.89 | 1398.51 | 0.01163346 | 2.25911148 | 0.72400391 | 1.0887419 | 0.1172651 | 0.48193367 |
| hsa-let-7a-5p | 399.25 | 989.94 | 1590 | 817.81 | 406.48 | 533.13 | 379.73 | 533.02 | 462.2 | 901.23 | 1407.81 | 1159.29 | 1131.94 | 0.00464405 | 2.48441928 | 0.95013566 | 1.01543742 | 0.10094557 | 0.40872224 |
| hsa-miR-4286 | 532.04 | 2307.9 | 2592.49 | 1289.62 | 51.81 | 41.08 | 25.56 | 32.12 | 48.13 | 70.84 | 123.67 | 116.19 | 174.69 | 0.02802506 | 3.05353548 | 0.03871567 | 0.05881129 | 0.03612014 | 0.01926007 |
| hsa-let-7c | 107.83 | 178.71 | 366.66 | 182.68 | 104.41 | 135.74 | 103.45 | 82.1 | 75.2 | 245.71 | 349.63 | 262.08 | 210.03 | 0.0072098 | 2.66383011 | 0.74883758 | 1.09963258 | 0.14463544 | 0.41280132 |
| hsa-miR-30a-5p | 142.59 | 348.01 | 339.05 | 216.55 | 76.51 | 107.16 | 76.68 | 76.15 | 84.22 | 201.77 | 245.48 | 217.53 | 253.45 | 0.00023246 | 2.72815055 | 0.22806171 | 0.76213466 | 0.03393774 | 0.27935946 |
| hsa-miR-99b-5p | 106.05 | 174 | 199.29 | 156.06 | 143.46 | 114.31 | 70.59 | 108.27 | 76.2 | 201.77 | 200.85 | 187.83 | 168.63 | 0.00117114 | 1.85022327 | 0.42416148 | 1.07548881 | 0.00806728 | 0.58127515 |
| hsa-miR-30d-5p | 129.22 | 273.94 | 270.03 | 180.26 | 117.96 | 133.06 | 62.07 | 95.18 | 81.21 | 231.36 | 254.78 | 183.46 | 209.02 | 0.0007053 | 2.24375868 | 0.56987272 | 0.9098836 | 0.02792005 | 0.40551758 |
| hsa-let-7e-5p | 52.58 | 131.68 | 164.78 | 104.04 | 76.51 | 68.76 | 52.34 | 70.2 | 57.15 | 104.02 | 137.62 | 103.96 | 130.26 | 0.00375315 | 1.83045606 | 0.51269815 | 0.8911236 | 0.05213081 | 0.48683146 |
| hsa-miR-23b-3p | 44.56 | 89.35 | 112.15 | 127.03 | 27.9 | 27.68 | 35.3 | 33.31 | 40.1 | 71.74 | 115.3 | 95.22 | 95.93 | 0.00446064 | 2.87745754 | 0.34558916 | 0.86336864 | 0.01635028 | 0.30004566 |
| hsa-miR-30c-5p | 48.12 | 131.68 | 119.92 | 112.51 | 22.32 | 36.61 | 25.56 | 32.12 | 29.08 | 50.22 | 71.6 | 70.76 | 75.73 | 0.00332846 | 2.3020626 | 0.00119164 | 0.55266952 | 0.00087937 | 0.2400758 |
| hsa-miR-200b-3p | 25.84 | 41.15 | 53.49 | 54.44 | 36.66 | 48.22 | 19.47 | 47.59 | 40.1 | 62.77 | 88.34 | 69.89 | 79.77 | 0.00221562 | 1.95773016 | 0.01536073 | 1.51313053 | 0.14668862 | 0.77290046 |
| hsa-miR-194-5p | 56.15 | 148.14 | 154.43 | 116.14 | 32.68 | 46.44 | 25.56 | 28.55 | 34.09 | 63.67 | 76.25 | 60.28 | 68.66 | 0.00027454 | 2.00857638 | 0.01901691 | 0.4815863 | 0.00775707 | 0.23976499 |
| hsa-miR-27b-3p | 49.02 | 88.18 | 101.8 | 65.33 | 29.49 | 34.83 | 25.56 | 22.61 | 21.05 | 58.29 | 84.62 | 75.13 | 60.59 | 0.00316128 | 2.60811367 | 0.29071036 | 0.81850495 | 0.02639645 | 0.31383025 |
| hsa-miR-204-5p | 230.82 | 545.52 | 501.24 | 580.69 | 87.67 | 71.44 | 43.82 | 70.2 | 91.24 | 185.63 | 175.74 | 312.75 | 224.17 | 0.01339315 | 3.08165464 | 0.00045823 | 0.41397124 | 0.00080123 | 0.13433408 |
| hsa-miR-145-5p | 220.12 | 634.88 | 985.23 | 394.39 | 231.13 | 365.25 | 111.97 | 182.04 | 94.25 | 517.42 | 665.43 | 1065.62 | 665.43 | 0.01378771 | 3.69919463 | 0.79851907 | 1.08484736 | 0.09975515 | 0.29326582 |
| hsa-miR-130a-3p | 59.71 | 102.29 | 92.31 | 100.41 | 41.44 | 74.12 | 38.95 | 59.49 | 51.13 | 82.5 | 185.04 | 104.83 | 138.34 | 0.03917099 | 2.40782824 | 0.28043046 | 1.29837124 | 0.00100643 | 0.53922918 |
| hsa-miR-99a-5p | 41.89 | 77.6 | 87.14 | 105.25 | 35.87 | 42.86 | 36.51 | 35.69 | 29.08 | 47.53 | 125.53 | 104.83 | 110.06 | 0.03628405 | 2.69394756 | 0.72928724 | 1.0776788 | 0.0163781 | 0.40003704 |
| hsa-miR-30b-5p | 91.79 | 228.09 | 256.25 | 101.62 | 71.73 | 99.13 | 63.29 | 64.25 | 56.15 | 107.61 | 190.62 | 184.33 | 260.52 | 0.03135471 | 2.61979975 | 0.87568098 | 0.95113834 | 0.11690545 | 0.36305765 |
| hsa-miR-4454 | 24656.7 | 45474.84 | 43213.07 | 37745.05 | 59187.17 | 38481.1 | 29000.97 | 48646.69 | 30316.83 | 57374.89 | 84435.09 | 62653.95 | 58230.54 | 0.02563256 | 1.59686653 | 0.02828314 | 1.55830293 | 0.87508775 | 0.97585046 |
| hsa-miR-1260a | 1114.88 | 2447.8 | 3501.8 | 3850.72 | 1891.32 | 2045.91 | 904.3 | 1754.93 | 1481.85 | 2305.54 | 2465.99 | 2843.62 | 2105.34 | 0.01521725 | 1.50410327 | 0.17557079 | 0.74389076 | 0.03990346 | 0.49457426 |
| hsa-miR-26a-5p | 75.75 | 248.07 | 372.7 | 39.92 | 161.79 | 166.1 | 160.66 | 46.4 | 37.1 | 212.53 | 400.77 | 383.52 | 254.46 | 0.01446137 | 2.73420156 | 0.45466072 | 1.4204241 | 0.39130886 | 0.51950234 |
| hsa-miR-4443 | 75.75 | 47.03 | 63.84 | 79.85 | 115.57 | 118.77 | 114.41 | 89.23 | 101.26 | 50.22 | 69.74 | 87.36 | 76.74 | 0.00943529 | 0.65847304 | 0.5751216 | 1.11705642 | 0.02157864 | 1.69643456 |
| hsa-miR-107 | 70.4 | 132.85 | 161.33 | 73.8 | 65.36 | 90.19 | 54.77 | 45.21 | 26.07 | 92.37 | 155.29 | 124.93 | 80.78 | 0.03326474 | 2.01247337 | 0.77864615 | 0.92403799 | 0.10727801 | 0.45915539 |
| hsa-miR-520h | 46.34 | 77.6 | 125.96 | 50.81 | 94.84 | 75.01 | 86.41 | 82.1 | 56.15 | 104.92 | 104.14 | 103.96 | 88.86 | 0.02817147 | 1.27335175 | 0.55150609 | 1.18492747 | 0.81812055 | 0.93055785 |
| hsa-miR-361-5p | 41.89 | 97.58 | 95.76 | 39.92 | 57.39 | 44.65 | 35.3 | 24.99 | 29.08 | 68.15 | 96.71 | 96.1 | 87.85 | 0.00113492 | 2.27789823 | 0.67562291 | 1.12152748 | 0.16338086 | 0.49235188 |
Comparison using Student’s t test (P<0.05) was performed between AR biopsies (R1 to R5) and unequivocal APN biopsies (I1, 12, 13, and 16), which gave a panel of 25 miRNAs. Comparing AR biopsies with all APN biopsies (I1 to I11) showed differential expression in 18 of the 25 miRNAs and are shown in the gray shaded area.
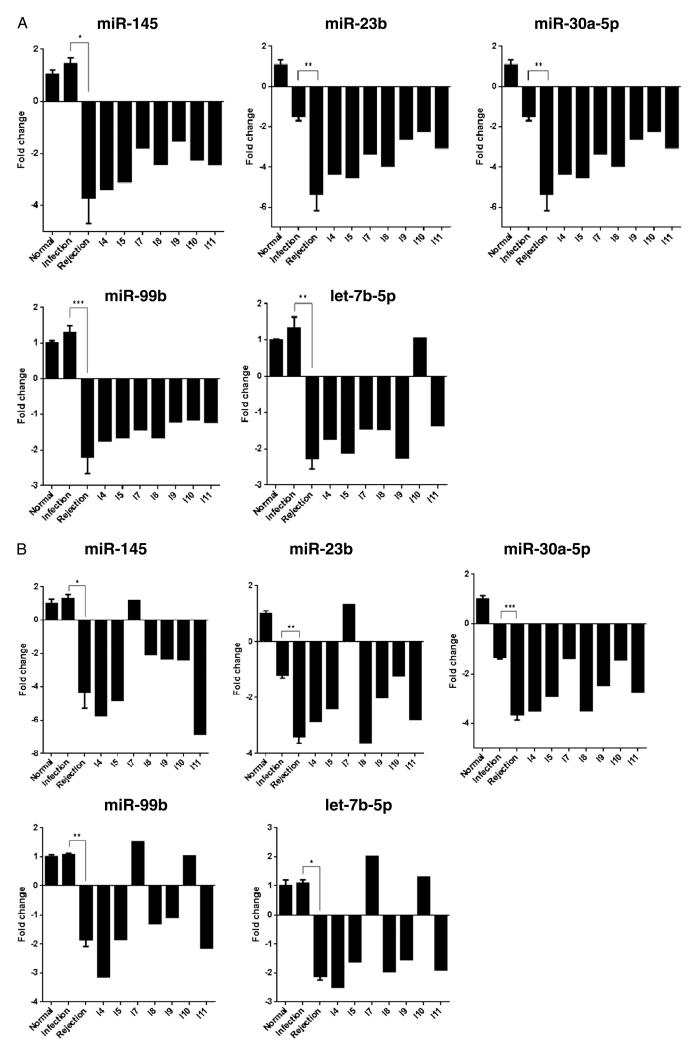
Five of these miRs were further validated using Taqman qPCR (Fig. 2A). These are miR-145, miR-23b, miR-30a-5p, miR-99b, and let-7b-5p. The trend is similar to that seen by NanoString (Fig. 2B), except for biopsy I7. The miRNAs with differential expression between APN and normal kidneys and between AR and normal kidneys, respectively, are shown in the supplemental data (see Figure S2 and Table S3, SDC, http://links.lww.com/TP/A928).
FIGURE 2.
A, taqman real-time PCR validation of miR-145, miR-23b, miR-3a, miR-99b, and let-7b-5p. Fold changes were calculated using the $$Ct method. B, fold change in the miR expression from NanoString data. Baseline bar includes mean of the B2, B3, and B4. Infection bar includes mean of I1, I2, I3, and I6 biopsies. Rejection bar includes mean of R1 to R5 biopsies. The remaining bars represent individual biopsies I4, I5, I7, I8, I9, I10, and I11. The fold induction or downregulation of miRNAs shows similar trends by both the methodologies. Only biopsy I7 shows discrepant results. *P<0.05, ** P<0.01, *** P<0.005.
DISCUSSION
Urinary tract infections represent a common problem in patients with renal transplants. The impact of acute pyelonephritis on prognosis and long-term graft outcome is somewhat controversial (3, 28–30), not surprisingly though, because of numerous factors simultaneously affecting long-term graft survival and function. These include delayed graft function in the posttransplant period, local complications, acute rejection episodes, toxicity of immunosuppressive drugs, and metabolic factors such as obesity, hypertension, diabetes mellitus, and recurrent native disease. Another difficulty, in our opinion, is making a definitive diagnosis of pyelonephritis in renal allografts. Allograft biopsy and urine cultures are the two most objective criteria currently available, neither one being the gold standard. We found that only 32.6% of the biopsies with features of APN had concomitant positive urine cultures. Even in the culture-positive cases, the bacterial colony counts can be lower than the usually accepted cutoff of 105 CFU/mL. Uncommon bacterial species can be found (31) such as Citrobacter koseri and Providencia rettgeri, and sometimes, Candida species alone can be seen. Despite this, one important finding in our study was that patients with concomitant positive urine cultures and (even those with positive cultures temporally separated from the biopsy) had higher rate of early graft loss and rise in mean serum creatinine at 1 year as compared with culture-negative patients.
Biopsy also has its pitfalls. Biopsy findings of APN and AR can be overlapping and difficult to distinguish. Limited accuracy due to sampling error, patchy distribution of inflammation in pyelonephritis, and interobserver variability among pathologists are contributing factors. There are two recent studies highlighting difficulty in the biopsy diagnosis of APN in renal allografts: Gupta et al. (25) and Mohamed et al. (32). We found that in 24% (12/49) of the patients, biopsy showed overlapping biopsy features of APN and AR. In addition, six biopsies in the culture negative group showed predominant acute tubular necrosis (ATN) with a patchy neutrophilic infiltrate. This may represent mainly ATN with mild associated inflammation, but it is also difficult to exclude the possibility of a subtle pyelonephritis. The two conditions can coexist.
Considering all these difficulties, we attempted intragraft microRNA profiling using the NanoString platform to see if we can find more objective biomarkers to distinguish between APN and AR in allograft biopsies. This is a preliminary study and the number of biopsies tested is small. We demonstrated a distinct miRNA profile for normal kidney (baseline pretransplant biopsies), renal allografts with acute rejection, and allografts with unequivocal acute pyelonephritis. However, 7 of the 11 biopsies with presumed APN by histology showed a heterogeneous miRNA profile, probably representing a spectrum between AR and APN. Some are closer to AR (I4, I5, I8, and I11) and some closer to APN (I7, I10), irrespective of urine culture results. Several of these biopsies also had overlapping features of APN and AR. Three of these patients had early graft loss despite intensive antibiotic treatment (I4, I5, I7). Three patients recovered with steroid treatment (I8, I9, I10). Based on this, we would like to draw attention to this subset of transplant patients with biopsy features of APN (and positive cultures at the time of biopsy or subsequently) but poor graft outcome in the early posttransplant period, despite antibiotics. They usually undergo repeated biopsies, which show persistent inflammation and overlapping morphologic features of pyelonephritis and rejection. Whether these patients have concomitant infection and rejection or the initial episode of pyelonephritis predisposes the graft to subsequent rejection or they had rejection all along but were misdiagnosed as pyelonephritis is difficult to determine. Our data suggest that in these patients with overlapping biopsy changes of rejection and pyelonephritis and poor response to antibiotics, careful anti-rejection treatment is warranted. However, antibiotics should also be given, particularly in culture-positive patients. This is a preliminary study. Study on larger sample number is required to understand possible prognostic subsets of APN based on miRNA profiles. Heterogeneity however is expected because these are individual human tissue samples, not tissues from inbred experimental animals.
We identified a panel of 25 miRNAs whose expression significantly differed between rejection and pyelonephritis. We validated five of these miRNAs using qPCR (miR-145, miR-99b, let-7b-5p, miR23b, and miR-30a). These miRNAs were found to be downregulated in AR as compared with normal kidney and APN. Another recent study described downregulation of miR-99b, miR-30c, and miR-23b in acute cellular rejection (23). Interestingly, miR-145 and miR-99b have been implicated in neutrophil differentiation and are involved in the temporal expression of genes in the different stages of myeloid maturation. Both these miRNAs are up-regulated in mature stage of peripheral neutrophils (35–37). In our study, expression of miR-145 and miR-99b was mildly increased in the APN biopsies compared with baselines, which may be a manifestation of neutrophil-predominant inflammation in APN. MiR-23b has recently been a focus of study in IL-17 associated autoimmune disease in humans and mouse models. Overexpression of miR-23b was found to suppress several proinflammatory signaling pathways including IL-17, tumor necrosis factor >, IL-1-induced NF-κB, TGF-β-activated kinases and several others in human lupus and rheumatoid arthritis as well as mouse models (38, 39, 40). Both miR-23b and miR-30a have been found to be downregulated in inflammatory cells in the lesions of lupus and rheumatoid arthritis in humans and in mouse models of lupus, rheumatoid arthritis, and multiple sclerosis (38), thereby causing upregulation of inflammatory cytokines. We found MiR-23b to be significantly down-regulated in our AR biopsies (but not in APN), probably triggering alloimmune inflammatory pathways in AR but not in APN. MiR-23b may be a new therapeutic target for inflammatory autoimmune diseases.
Earlier studies have published a predictive miRNA signature for AR (14, 15). Anglicheau et al. described significant alterations in let-7c, miR-10a-5p, miR-10b-5p, miR-125a, miR-30a-5p, miR-30b, miR-30c, miR-142, miR-155, and miR-223 as compared with normal kidney. We agree with their findings. However, we found that miR-10a-5p, miR-10b-5p, miR-125a, miR-142, and miR-223 (and 12 additional miRNAs) were similarly altered in APN as well and did not significantly differ between AR and APN and therefore may not be exclusive for AR. MiR-21 was also upregulated in both AR and APN. This suggests that many of the miRNA alterations are not specific to either APN or AR but may be altered in multiple pathologic conditions of the kidney. MiR-21 is widely studied in models of interstitial fibrosis and tubular atrophy in the kidney and reported to be overexpressed in diseased kidneys (34).
In summary, diagnosis of APN versus AR in the early posttransplant period can be difficult. There is no single gold standard method. As in all aspects of renal pathology, a gestalt approach including clinical history, biopsy findings, culture results, immunosuppressive drug levels, C4d staining, and donor-specific antibody results should be used to render the best possible diagnosis. Patients whose biopsies show over-lapping histologic features and persistent dysfunction despite antibiotics may portend a bad outcome. Their miRNA profiles showed a spectrum between that of AR and APN. MiRNA profiling can become a useful ancillary test but needs extensive validation and optimization. Testing on larger sample numbers is required to expand on this experience.
MATERIALS AND METHODS
Biopsies for miRNA Analysis by NanoString Assay
A total of 20 renal allograft biopsy samples were analyzed for miRNA expression. All the biopsies for miRNA testing were carefully reviewed. We did not include biopsies with moderate to extensive chronic allograft injury, since that may alter the miRNA profile. The APN biopsies were from our study cohort. The preimplantation (baseline) biopsies and biopsies with unequivocal AR were selected from our biopsy archives (see Table S2, SDC, http://links.lww.com/TP/A928). These served as controls.
MicroRNA NanoString Assay
RNA Isolation
Formalin-fixed paraffin-embedded (FFPE) tissue sections from the allograft biopsies were deparaffinized using xylene, and total RNA was prepared using the RNeasy FFPE kit (Qiagen Valencia, CA). Total RNA quality and quantity were assessed by Nanodrop spectrophotometry (ThermoFisher Scientific, Waltham, MA).
MicroRNA Expression Profiling
The digital multiplexed NanoString nCounter human microRNA expression assay (NanoString Technologies) was performed with 100 ng total RNA as input material (24, 25).
Data Analysis
All data analysis was performed using the nSolver software analysis (available for download from NanoString Technologies). For normalization purposes, we adopted two methods—geometric mean of the top 100 most highly expressed miRNAs, and global sum method of stably expressed miRNA. The data were expressed in terms of absolute number of miRNAs in each sample (24, 25).
The NanoString code set consists of a total of 800 mature human miRNAs. Using the top 100 expressed miRNAs, heat maps were created to show median-centered expression of each miRNA and clustering pattern between biopsies, using Cluster 3.0 and JavaTreeView software algorithms. Among the top 100 expressed miRNAs, we identified 25 miRNAs, which showed statistically significant difference in expression between AR and APN using Student’s t test (two-tailed, two-sample unequal variance T test, using cutoff P value of 0.05) (Table 3).
Quantitative PCR Validation
Total RNA from the various FFPE samples were reverse transcribed into cDNA and PCR amplified in triplicates using predesigned primers and probes (Applied Biosystems, Foster City, CA), with RNU44 and U6 as the normalizing gene. Fold changes were calculated using the ΔΔCt method.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Hansjuerg Alder in the Comprehensive Cancer Core Facility for helpful advice in the analysis for the NanoString miRNA data.
Footnotes
The authors report no funding or conflict of interest.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Takai K, Tollemar J, Wilczek HE, et al. Urinary tract infections following renal transplantation. Clin Transplant. 1998;12:19. [PubMed] [Google Scholar]
- 2.Schmaldienst S, Dittrich E, Hörl WL. Urinary tract infections after renal transplantation. Curr Opin Urol. 2002;12:125. doi: 10.1097/00042307-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Pelle G, Vimont S, Levy PP, et al. Acute pyelonephritis represents a risk factor impairing long term kidney graft function. Am J Transplant. 2007;7:889. doi: 10.1111/j.1600-6143.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- 4.Ekberg H, Tedesco-Silva H, Demirbas A, et al. Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med. 2007;357:2562. doi: 10.1056/NEJMoa067411. [DOI] [PubMed] [Google Scholar]
- 5.Fiorante S, Lopez-Medrano F, Lizasoain M, et al. Systematic screening and treatment of asymptomatic bacteriuria in renal transplant recipients. Kidney Int. 2010;78:774. doi: 10.1038/ki.2010.286. [DOI] [PubMed] [Google Scholar]
- 6.Sharifian M, Rees L, Trompeter RS. High incidence of bacteriuria following renal transplantation in children. Nephrol Dial Transplant. 1998;13:432. doi: 10.1093/oxfordjournals.ndt.a027842. [DOI] [PubMed] [Google Scholar]
- 7.Golebiewska J, Debska-Ślizien A, Komarnicka J, et al. Urinary tract infections in renal transplant recipients. Transplant Proc. 2011;43:2985. doi: 10.1016/j.transproceed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Lee JR, Bang H, Dadhania D, et al. Independent risk factors for urinary tract infection and for subsequent bacteremia or acute cellular rejection: a single-center report of 1166 kidney allograft recipients. Transplantation. 2013;96:732. doi: 10.1097/TP.0b013e3182a04997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical Microbiology Procedures Handbook. 3rd ed. ASM Press; Lynn Garcia: 2010. [Google Scholar]
- 10.Griffiths-Jones S, Grocock RJ, von Dongen S, et al. miRBAse: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research. 2006:34. doi: 10.1093/nar/gkj112. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sand M, Skrygan M, Sand D, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 12.Dai Y, Sui W, Lan H, et al. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 13.Jordan Yz Li, Yong TY, Michael MZ, et al. Review: the role of microRNAs in kidney disease. Nephrology. 2010;15:599. doi: 10.1111/j.1440-1797.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- 14.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA. 2009;106:5330. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui W, Dal Y, Huang Y, et al. Microarray analysis of microRNA expression in acute rejection after renal transplantation. Transpl. Immunol. 2008;19:81. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Mas VR, Dumur CI, Scian MJ, et al. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13:11. doi: 10.1111/j.1600-6143.2012.04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Øzbay A, Tørring C, Plsen R, et al. Transcriptional profiles in urine during acute rejection, bacteriuria, CMV infection ans stable graft function after transplantation. Scand J Immunol. 2009;69:357. doi: 10.1111/j.1365-3083.2009.02226.x. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Dov IZ, Muthukumar T, Morozov P, et al. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation. 2012;94:1086. doi: 10.1097/TP.0b013e3182751efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplantation. 2011;11:2110. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Xiao Y, Ge W, et al. miR-200b inhibits TGF-beta1-induced epithelial-mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis. 2013;4:e541. doi: 10.1038/cddis.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiguo S, Hua L, Wujian P, et al. Genomics, available online after Molecular dysfunction in acute rejection after renal transplantation revealed by integrated analysis of transcription factor, microRNA and long noncoding RNA. 2013 May 15; doi: 10.1016/j.ygeno.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Wilflingseder J, Regele H, Perco P, et al. MiRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation. 2013;95:835. doi: 10.1097/TP.0b013e318280b385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meehan SM, Nadasdy T. Tubulointerstitial diseases. In: Zhou XJ, Laszik Z, Nadasdy T, D’Agati V, Silva FG, editors. Silva’s Diagnostic Renal Pathology. Cambridge University Press; New York, NY: 2009. p. 407. [Google Scholar]
- 25.Gupta G, Shapiro R, Girnita A, et al. Neutrophilic tubulitis as a marker for urinary tract infection in renal allograft biopsies with C4d deposition. Transplantation. 2009;87:1013. doi: 10.1097/TP.0b013e31819ca304. [DOI] [PubMed] [Google Scholar]
- 26.Alder H, Taccioli C, Chen H, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736. doi: 10.1093/carcin/bgs204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waggott D, Chu K, Yin S, et al. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28:1546. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giral M, Pascuariello G, Karam G, et al. Acute graft pyelonephritis and long-term kidney allograft outcome. Kidney Int. 2002;61:1880. doi: 10.1046/j.1523-1755.2002.00323.x. [DOI] [PubMed] [Google Scholar]
- 29.Kamath NS, John GT, Neelakantan N, et al. Acute graft pyelonephritis following renal transplantation. Transpl Infect Dis. 2006;8:140. doi: 10.1111/j.1399-3062.2006.00148.x. [DOI] [PubMed] [Google Scholar]
- 30.Fiorante S, Fernández-Ruiz M, Lopez-Medrano F, et al. Acute graft pyelonephritis in renal transplant recipients: incidence, risk factors and long term outcome. Nephrol Dial Transplant. 2011;26:1065. doi: 10.1093/ndt/gfq531. [DOI] [PubMed] [Google Scholar]
- 31.Gupta A, Gupta P, Khaira A. Actinobaculum Schaalii pyelonephritis in a kidney allograft recipient. Iran J Kidney Dis. 2012;6:386. [PubMed] [Google Scholar]
- 32.Mohamed N, Aggarwal V, Cole E, et al. Histopathologic detection of rejection in acute allograft pyelonephritis. Transplantation. 2012;94:e46. doi: 10.1097/TP.0b013e318265c4b8. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Liu J, Liu C, et al. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic A-cells. Diabetes. 2013;62:887. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez IG, Grafals M, Portilla D, et al. MicroRNAs as potential therapeutic targets in kidney disease. J Formosan Med Assoc. 2013;112:237. doi: 10.1016/j.jfma.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batliner J, Buehrer E, Fey MF, et al. Inhibition of the miR-143/145 cluster attenuated neutrophil differentiation of APL cells. Leukemia Res. 2012;36:236. doi: 10.1016/j.leukres.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu C, Kim J, Stepanowsky P, et al. Differential expression of miR-145 in children with Kawasaki disease. Plos One. 2013;8:e58159. doi: 10.1371/journal.pone.0058159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen MT, Hother C, Häger M, et al. MicroRNA profiling in human neutrophils during bone marrow granulopoiesis and in vivo exudation. Plos One. 2013;8:e58454. doi: 10.1371/journal.pone.0058454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu S, Pan W, Song X, et al. The microRNA miR-23b suppresses IL-17 associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 2013;18:1077. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 39.Hu R, O’Connell RM. MiR-23b is a safeguard against autoimmunity. News and Views. Nat Med. 2012;18:1009. doi: 10.1038/nm.2849. [DOI] [PubMed] [Google Scholar]
- 40.Bordon Y. MicroRNA 23-b keeps TABs on tissue inflammation. Research Highlights. Nat Rev Immunol. 2012;8:438. doi: 10.1038/nrrheum.2012.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.